

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.8 + phase: 0
(209 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
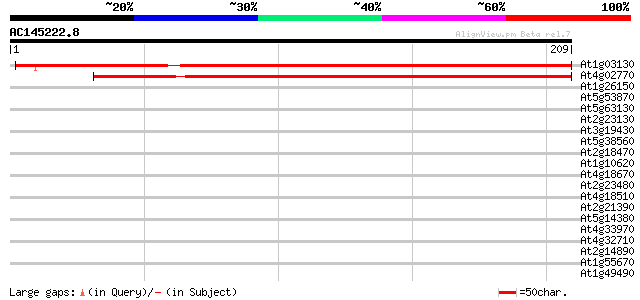
At1g03130 putative photosystem I reaction center subunit II prec... 293 4e-80
At4g02770 putative photosystem I reaction center subunit II prec... 288 1e-78
At1g26150 Pto kinase interactor, putative 37 0.010
At5g53870 predicted GPI-anchored protein 31 0.41
At5g63130 unknown protein 30 0.70
At2g23130 arabinogalactan-protein AGP17 30 0.91
At3g19430 putative late embryogenesis abundant protein 29 1.6
At5g38560 putative protein 29 2.0
At2g18470 putative protein kinase 29 2.0
At1g10620 putative serine/threonine protein kinase emb|CAA18823.1 29 2.0
At4g18670 extensin-like protein 28 2.7
At2g23480 hypothetical protein 28 2.7
At4g18510 unknown protein 28 4.5
At2g21390 coatomer alpha subunit 28 4.5
At5g14380 agp6 27 5.9
At4g33970 extensin-like protein 27 5.9
At4g32710 putative protein kinase 27 5.9
At2g14890 arabinogalactan-protein AGP9 27 5.9
At1g55670 photosystem I subunit V precursor, putative 27 5.9
At1g49490 hypothetical protein 27 5.9
>At1g03130 putative photosystem I reaction center subunit II
precursor
Length = 204
Score = 293 bits (750), Expect = 4e-80
Identities = 150/208 (72%), Positives = 167/208 (80%), Gaps = 5/208 (2%)
Query: 3 MATQAS-LFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEA 61
MATQA+ +F+P +++ + L S +P + T I A ++ +A+ KE
Sbjct: 1 MATQAAGIFSPAITTTTSAVKKLHLFSSSHRPKSLSFTKTAIRAEKTESSSAAPAVKE-- 58
Query: 62 PAAPVGFTPPELDPNTPSPIFGGSTGGLLRKAQVEEFYVITWESPKEQIFEMPTGGAAIM 121
APVGFTPP+LDPNTPSPIF GSTGGLLRKAQVEEFYVITW SPKEQIFEMPTGGAAIM
Sbjct: 59 --APVGFTPPQLDPNTPSPIFAGSTGGLLRKAQVEEFYVITWNSPKEQIFEMPTGGAAIM 116
Query: 122 REGPNLLKLARKEQCLALGNRLRSKYKIKYQFYRVFPNGEVQYLHPKDGVYPEKVNPGRQ 181
REGPNLLKLARKEQCLALG RLRSKYKI YQFYRVFPNGEVQYLHPKDGVYPEK NPGR+
Sbjct: 117 REGPNLLKLARKEQCLALGTRLRSKYKITYQFYRVFPNGEVQYLHPKDGVYPEKANPGRE 176
Query: 182 GVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
GVG N RSIGKNVSPIEVKFTGKQ +D+
Sbjct: 177 GVGLNMRSIGKNVSPIEVKFTGKQSYDL 204
>At4g02770 putative photosystem I reaction center subunit II
precursor
Length = 208
Score = 288 bits (738), Expect = 1e-78
Identities = 141/178 (79%), Positives = 149/178 (83%), Gaps = 3/178 (1%)
Query: 32 KPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLR 91
K + FT + D AA+ +EAP VGFTPP+LDPNTPSPIF GSTGGLLR
Sbjct: 34 KSLSFTKTAIRAEKTDSSAAAAAAPATKEAP---VGFTPPQLDPNTPSPIFAGSTGGLLR 90
Query: 92 KAQVEEFYVITWESPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGNRLRSKYKIKY 151
KAQVEEFYVITW SPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALG RLRSKYKI Y
Sbjct: 91 KAQVEEFYVITWNSPKEQIFEMPTGGAAIMREGPNLLKLARKEQCLALGTRLRSKYKITY 150
Query: 152 QFYRVFPNGEVQYLHPKDGVYPEKVNPGRQGVGQNFRSIGKNVSPIEVKFTGKQPFDV 209
QFYRVFPNGEVQYLHPKDGVYPEK NPGR+GVG N RSIGKNVSPIEVKFTGKQ +D+
Sbjct: 151 QFYRVFPNGEVQYLHPKDGVYPEKANPGREGVGLNMRSIGKNVSPIEVKFTGKQSYDL 208
>At1g26150 Pto kinase interactor, putative
Length = 760
Score = 36.6 bits (83), Expect = 0.010
Identities = 23/68 (33%), Positives = 28/68 (40%), Gaps = 5/68 (7%)
Query: 13 PLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPE 72
PLSSP P P + S P T +S E + + T+ PA PV PPE
Sbjct: 67 PLSSPPPEPSPPSPSLTGPPPT-----TIPVSPPPEPSPPPPLPTEAPPPANPVSSPPPE 121
Query: 73 LDPNTPSP 80
P P P
Sbjct: 122 SSPPPPPP 129
>At5g53870 predicted GPI-anchored protein
Length = 370
Score = 31.2 bits (69), Expect = 0.41
Identities = 23/79 (29%), Positives = 32/79 (40%)
Query: 1 MAMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEE 60
+A A+ S PP SS P + P + S IS P + T S A S +
Sbjct: 160 VAPASAPSKSQPPRSSVSPAQPPKSSSPISHTPALSPSHATSHSPATPSPSPKSPSPVSH 219
Query: 61 APAAPVGFTPPELDPNTPS 79
+P+ TP +TPS
Sbjct: 220 SPSHSPAHTPSHSPAHTPS 238
>At5g63130 unknown protein
Length = 192
Score = 30.4 bits (67), Expect = 0.70
Identities = 13/38 (34%), Positives = 23/38 (60%), Gaps = 2/38 (5%)
Query: 46 ADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPIFG 83
+ KTE++ ++ + +P +P TP PN+PSP +G
Sbjct: 114 SSHKTESSPSSSGDRSPKSPFSVTPS--PPNSPSPAYG 149
>At2g23130 arabinogalactan-protein AGP17
Length = 185
Score = 30.0 bits (66), Expect = 0.91
Identities = 21/65 (32%), Positives = 29/65 (44%), Gaps = 6/65 (9%)
Query: 19 PWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKE---EAPAAPVGFTPPELDP 75
P PST KP + + + E TEA + T E EAP +P + P++ P
Sbjct: 28 PIHSPSTSPH---KPKPTSPAISPAAPTPESTEAPAKTPVEAPVEAPPSPTPASTPQISP 84
Query: 76 NTPSP 80
PSP
Sbjct: 85 PAPSP 89
>At3g19430 putative late embryogenesis abundant protein
Length = 550
Score = 29.3 bits (64), Expect = 1.6
Identities = 22/74 (29%), Positives = 28/74 (37%), Gaps = 6/74 (8%)
Query: 11 TPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAA----SVTTKEEAPAAPV 66
TPP+S P P PS S P+ T S + + T +P PV
Sbjct: 94 TPPVSPPPPTPTPSVPS--PTPPVSPPPPTPTPSVPSPTPPVSPPPPTPTPSVPSPTPPV 151
Query: 67 GFTPPELDPNTPSP 80
PP P+ PSP
Sbjct: 152 SPPPPTPTPSVPSP 165
Score = 28.9 bits (63), Expect = 2.0
Identities = 22/70 (31%), Positives = 26/70 (36%), Gaps = 4/70 (5%)
Query: 11 TPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTP 70
TPP+S P P PS S P+ T S T +P PV P
Sbjct: 130 TPPVSPPPPTPTPSVPS--PTPPVSPPPPTPTPSVPSPTPPVP--TDPMPSPPPPVSPPP 185
Query: 71 PELDPNTPSP 80
P P+ PSP
Sbjct: 186 PTPTPSVPSP 195
Score = 28.5 bits (62), Expect = 2.7
Identities = 20/69 (28%), Positives = 30/69 (42%), Gaps = 12/69 (17%)
Query: 12 PPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPP 71
PP+S P P PS S + P T T + + + VT P+ P +PP
Sbjct: 179 PPVSPPPPTPTPSVPSPPDVTP---TPPTPSVPSPPD------VTPTPPTPSVP---SPP 226
Query: 72 ELDPNTPSP 80
++ P P+P
Sbjct: 227 DVTPTPPTP 235
Score = 27.7 bits (60), Expect = 4.5
Identities = 20/73 (27%), Positives = 28/73 (37%), Gaps = 3/73 (4%)
Query: 11 TPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEA---PAAPVG 67
TPP+S P P PS S P + T SV + + P P
Sbjct: 148 TPPVSPPPPTPTPSVPSPTPPVPTDPMPSPPPPVSPPPPTPTPSVPSPPDVTPTPPTPSV 207
Query: 68 FTPPELDPNTPSP 80
+PP++ P P+P
Sbjct: 208 PSPPDVTPTPPTP 220
>At5g38560 putative protein
Length = 681
Score = 28.9 bits (63), Expect = 2.0
Identities = 13/42 (30%), Positives = 19/42 (44%)
Query: 44 SAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGS 85
S+ T + T+ P+AP TPP P +P P+ S
Sbjct: 14 SSNSSTTAPPPLQTQPTTPSAPPPVTPPPSPPQSPPPVVSSS 55
>At2g18470 putative protein kinase
Length = 633
Score = 28.9 bits (63), Expect = 2.0
Identities = 24/87 (27%), Positives = 35/87 (39%), Gaps = 6/87 (6%)
Query: 1 MAMATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEE 60
MA + +++ T SSP P PS + + P + + D + +T
Sbjct: 1 MASSPESAPPTNSTSSPSP---PSNTNSTTSSPPAPSPPSPTPPQGDSSSSPPPDSTSPP 57
Query: 61 APAAPVGFTPPELDPNTPSPIFGGSTG 87
AP AP PP N+PSP G G
Sbjct: 58 APQAP---NPPNSSNNSPSPPSQGGGG 81
>At1g10620 putative serine/threonine protein kinase emb|CAA18823.1
Length = 718
Score = 28.9 bits (63), Expect = 2.0
Identities = 23/83 (27%), Positives = 35/83 (41%), Gaps = 9/83 (10%)
Query: 6 QASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAA--------DEKTEAASVTT 57
QA LF +S P QP+ + + + I ++TT+ A D +T T
Sbjct: 7 QADLFGKTIS-PFVASQPTNVGGFTDQKIIGGSETTQPPATSPPSPPSPDTQTSPPPATA 65
Query: 58 KEEAPAAPVGFTPPELDPNTPSP 80
+ P P TPP P++P P
Sbjct: 66 AQPPPNQPPNTTPPPTPPSSPPP 88
>At4g18670 extensin-like protein
Length = 839
Score = 28.5 bits (62), Expect = 2.7
Identities = 22/78 (28%), Positives = 30/78 (38%), Gaps = 18/78 (23%)
Query: 11 TPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTP 70
+PP ++P P P T+ TT + + T P++P TP
Sbjct: 457 SPPSTTPSPGSPP-------------TSPTTPTPGGSPPSSPTTPTPGGSPPSSPTTPTP 503
Query: 71 ---PELDPNTPSPIFGGS 85
P P TPSP GGS
Sbjct: 504 GGSPPSSPTTPSP--GGS 519
>At2g23480 hypothetical protein
Length = 705
Score = 28.5 bits (62), Expect = 2.7
Identities = 18/68 (26%), Positives = 26/68 (37%)
Query: 19 PWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTP 78
P K+ + + ++ K K+ A + T A T A A T P P T
Sbjct: 48 PAKKEKVQAPAKKEKVQAPAKKAKVQATAKTTATAQTTATAMATTAAPTTTAPTTAPTTE 107
Query: 79 SPIFGGST 86
SP+ ST
Sbjct: 108 SPMLDDST 115
>At4g18510 unknown protein
Length = 75
Score = 27.7 bits (60), Expect = 4.5
Identities = 13/49 (26%), Positives = 21/49 (42%)
Query: 32 KPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSP 80
+P++ K+ EA S T +++ A G +P L P P P
Sbjct: 24 RPLEGARVGVKVRGLSPSIEATSPTVEDDQAAGSHGKSPERLSPGGPDP 72
>At2g21390 coatomer alpha subunit
Length = 1218
Score = 27.7 bits (60), Expect = 4.5
Identities = 17/52 (32%), Positives = 23/52 (43%), Gaps = 2/52 (3%)
Query: 40 TTKISAADEKTEAASVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTGGLLR 91
T + + E ++ + P+ P G TP L P PSPI G LLR
Sbjct: 763 TASVHGLTDIAERLAIELGDNVPSLPEGKTPSLLMP--PSPIMCGGDWPLLR 812
>At5g14380 agp6
Length = 150
Score = 27.3 bits (59), Expect = 5.9
Identities = 28/109 (25%), Positives = 38/109 (34%), Gaps = 30/109 (27%)
Query: 5 TQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAA----------- 53
T A+ F S P K PS + P K T TTK +A K AA
Sbjct: 14 TIATAFAADAPSASPKKSPSPTA----APTKAPTATTKAPSAPTKAPAAAPKSSSASSPK 69
Query: 54 ---------------SVTTKEEAPAAPVGFTPPELDPNTPSPIFGGSTG 87
S ++ ++ AP +PP P++ S G S G
Sbjct: 70 ASSPAAEGPVPEDDYSASSPSDSAEAPTVSSPPAPTPDSTSAADGPSDG 118
>At4g33970 extensin-like protein
Length = 699
Score = 27.3 bits (59), Expect = 5.9
Identities = 19/72 (26%), Positives = 31/72 (42%), Gaps = 9/72 (12%)
Query: 11 TPPLSSP--KPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGF 68
+P L++P KP PS +P++ + D+ + + VT + P APV
Sbjct: 473 SPVLATPVDKPSPVPS-------RPVQKPQPPKESPQPDDPYDQSPVTKRRSPPPAPVNS 525
Query: 69 TPPELDPNTPSP 80
PP + P P
Sbjct: 526 PPPPVYSPPPPP 537
>At4g32710 putative protein kinase
Length = 731
Score = 27.3 bits (59), Expect = 5.9
Identities = 23/72 (31%), Positives = 28/72 (37%), Gaps = 6/72 (8%)
Query: 10 FTPPLSSPKPWKQPSTLSFISL----KPIKFTTKTTKISAADEKTEAASVTTKEEAPAA- 64
F PP++S P P+T S KP TT S K A+VT PA
Sbjct: 172 FPPPINSSPPNPSPNTPSLPETSPPPKPPLSTTPFPSSSTPPPKKSPAAVTLPFFGPAGQ 231
Query: 65 -PVGFTPPELDP 75
P G P + P
Sbjct: 232 LPDGTVAPPIGP 243
Score = 26.9 bits (58), Expect = 7.7
Identities = 24/84 (28%), Positives = 29/84 (33%), Gaps = 10/84 (11%)
Query: 4 ATQASLFTPPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPA 63
A S PPLSSP P P LS + P + S E+ E P
Sbjct: 34 APPLSPLPPPLSSPPPLPSPPPLSAPTASPPPLPVE----SPPSPPIESPPPPLLESPP- 88
Query: 64 APVGFTPPELDPNTPSPIFGGSTG 87
PP P+ PSP +G
Sbjct: 89 -----PPPLESPSPPSPHVSAPSG 107
>At2g14890 arabinogalactan-protein AGP9
Length = 191
Score = 27.3 bits (59), Expect = 5.9
Identities = 17/69 (24%), Positives = 30/69 (42%), Gaps = 3/69 (4%)
Query: 12 PPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPP 71
PP++SP P P + P T +++ + A + TTK ++P+ +PP
Sbjct: 88 PPVASPPP---PVASPPPATPPPVATPPPAPLASPPAQVPAPAPTTKPDSPSPSPSSSPP 144
Query: 72 ELDPNTPSP 80
+ P P
Sbjct: 145 LPSSDAPGP 153
>At1g55670 photosystem I subunit V precursor, putative
Length = 160
Score = 27.3 bits (59), Expect = 5.9
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 5/58 (8%)
Query: 3 MATQASLFTPP--LSSPKPWKQPSTLSFISLKPIKF---TTKTTKISAADEKTEAASV 55
MAT AS P S+ K P+++SF L+P++ ++ K+S K+ +A V
Sbjct: 1 MATSASALLSPTTFSTAISHKNPNSISFHGLRPLRLGGSSSALPKLSTTGRKSSSAVV 58
>At1g49490 hypothetical protein
Length = 847
Score = 27.3 bits (59), Expect = 5.9
Identities = 14/69 (20%), Positives = 29/69 (41%), Gaps = 11/69 (15%)
Query: 12 PPLSSPKPWKQPSTLSFISLKPIKFTTKTTKISAADEKTEAASVTTKEEAPAAPVGFTPP 71
PP+ +P P + P+ S +TT++ +++ + + + +AP TP
Sbjct: 648 PPMGAPTPTQAPTPSS-----------ETTQVPTPSSESDQSQILSPVQAPTPVQSSTPS 696
Query: 72 ELDPNTPSP 80
P+P
Sbjct: 697 SEPTQVPTP 705
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.314 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,231,701
Number of Sequences: 26719
Number of extensions: 241718
Number of successful extensions: 633
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 7
Number of HSP's successfully gapped in prelim test: 14
Number of HSP's that attempted gapping in prelim test: 608
Number of HSP's gapped (non-prelim): 35
length of query: 209
length of database: 11,318,596
effective HSP length: 95
effective length of query: 114
effective length of database: 8,780,291
effective search space: 1000953174
effective search space used: 1000953174
T: 11
A: 40
X1: 16 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC145222.8


