

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.4 - phase: 0
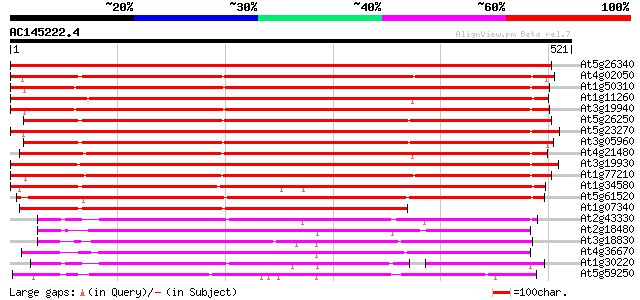
(521 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g26340 hexose transporter - like protein 842 0.0
At4g02050 putative hexose transporter 604 e-173
At1g50310 hexose transporter, putative 590 e-169
At1g11260 glucose transporter 588 e-168
At3g19940 putative monosaccharide transport protein, STP4 582 e-166
At5g26250 hexose transporter - like protein 582 e-166
At5g23270 monosaccharide transporter 576 e-164
At3g05960 putative hexose transporter 574 e-164
At4g21480 glucose transporter 556 e-158
At3g19930 monosaccharide transport protein, STP4 553 e-158
At1g77210 unknown protein 553 e-158
At1g34580 monosaccharide transporter like protein 488 e-138
At5g61520 monosaccharide transporter STP3 474 e-134
At1g07340 hexose transporter like protein 363 e-100
At2g43330 membrane transporter like protein 211 6e-55
At2g18480 putative sugar transporter 210 2e-54
At3g18830 sugar transporter like protein 207 1e-53
At4g36670 sugar transporter like protein 204 1e-52
At1g30220 unknown protein 197 9e-51
At5g59250 D-xylose-H+ symporter - like protein 187 9e-48
>At5g26340 hexose transporter - like protein
Length = 526
Score = 842 bits (2175), Expect = 0.0
Identities = 415/504 (82%), Positives = 462/504 (91%), Gaps = 1/504 (0%)
Query: 1 MAGGGFATSGGG-EFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPA 59
M GGGFATS G EFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSM FL+KFFP
Sbjct: 1 MTGGGFATSANGVEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMPDFLEKFFPV 60
Query: 60 VYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFF 119
VYRK V A DSNYCKYDNQGLQLFTSSLYLA LT+TFFASYTTRT+GRRLTMLIAG F
Sbjct: 61 VYRKVVAGADKDSNYCKYDNQGLQLFTSSLYLAGLTATFFASYTTRTLGRRLTMLIAGVF 120
Query: 120 FIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVT 179
FI GVA NA AQ+LAMLI GRILLGCGVGFANQAVP+FLSEIAP+RIRG LNILFQLNVT
Sbjct: 121 FIIGVALNAGAQDLAMLIAGRILLGCGVGFANQAVPLFLSEIAPTRIRGGLNILFQLNVT 180
Query: 180 IGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGK 239
IGILFANLVNYGT KI GGWGWRLSLGLAGIPALLLTVGA++V +TPNSL+ERGRL+EGK
Sbjct: 181 IGILFANLVNYGTAKIKGGWGWRLSLGLAGIPALLLTVGALLVTETPNSLVERGRLDEGK 240
Query: 240 AVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGI 299
AVL++IRGTDN+EPEF +L EASR+AKEVK FRNLL+ Q+++A+ L+IFQQ TGI
Sbjct: 241 AVLRRIRGTDNVEPEFADLLEASRLAKEVKHPFRNLLQRRNRPQLVIAVALQIFQQCTGI 300
Query: 300 NAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMF 359
NAIMFYAPVLF+T+GF +DASLYSAV+TGAVNVLST+VSIY VDK+GRR+LLLEAGVQMF
Sbjct: 301 NAIMFYAPVLFSTLGFGSDASLYSAVVTGAVNVLSTLVSIYSVDKVGRRVLLLEAGVQMF 360
Query: 360 LSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLET 419
SQ+VIAIILG+KVTD S +LSKG+AI VV+++CT+V+AFAWSWGPLGWLIPSETFPLET
Sbjct: 361 FSQVVIAIILGVKVTDTSTNLSKGFAILVVVMICTYVAAFAWSWGPLGWLIPSETFPLET 420
Query: 420 RSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIP 479
RSAGQSVTVCVN+LFTF+IAQAFLSMLCHFKFGIF+FFS WVLIMS+FV+FL+PETKNIP
Sbjct: 421 RSAGQSVTVCVNLLFTFIIAQAFLSMLCHFKFGIFIFFSAWVLIMSVFVMFLLPETKNIP 480
Query: 480 IEEMTERVWKQHWFWKRFMEDDNE 503
IEEMTERVWK+HWFW RFM+D N+
Sbjct: 481 IEEMTERVWKKHWFWARFMDDHND 504
>At4g02050 putative hexose transporter
Length = 513
Score = 604 bits (1558), Expect = e-173
Identities = 297/517 (57%), Positives = 386/517 (74%), Gaps = 16/517 (3%)
Query: 1 MAGGGFATSG-----GGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKK 55
MAGG F +G +++ K+T VII+C++AA GG +FGYD+G+SGGVTSM FL++
Sbjct: 1 MAGGSFGPTGVAKERAEQYQGKVTSYVIIACLVAAIGGSIFGYDIGISGGVTSMDEFLEE 60
Query: 56 FFPAVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLI 115
FF VY K + +SNYCKYDNQGL FTSSLYLA L ST AS TR GRR +++
Sbjct: 61 FFHTVYEKK--KQAHESNYCKYDNQGLAAFTSSLYLAGLVSTLVASPITRNYGRRASIVC 118
Query: 116 AGFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQ 175
G F+ G NA A NLAML+ GRI+LG G+GF NQAVP++LSE+AP+ +RG LN++FQ
Sbjct: 119 GGISFLIGSGLNAGAVNLAMLLAGRIMLGVGIGFGNQAVPLYLSEVAPTHLRGGLNMMFQ 178
Query: 176 LNVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRL 235
L TIGI AN+VNYGT ++ WGWRLSLGLA PALL+T+G + +TPNSL+ERG
Sbjct: 179 LATTIGIFTANMVNYGTQQLKP-WGWRLSLGLAAFPALLMTLGGYFLPETPNSLVERGLT 237
Query: 236 EEGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQ 295
E G+ VL K+RGT+N+ E ++ +AS +A +K FRN+L+ Q++MAI + +FQ
Sbjct: 238 ERGRRVLVKLRGTENVNAELQDMVDASELANSIKHPFRNILQKRHRPQLVMAICMPMFQI 297
Query: 296 FTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAG 355
TGIN+I+FYAPVLF T+GF +ASLYS+ +TGAV VLST +SI VD+LGRR LL+ G
Sbjct: 298 LTGINSILFYAPVLFQTMGFGGNASLYSSALTGAVLVLSTFISIGLVDRLGRRALLITGG 357
Query: 356 VQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETF 415
+QM + Q+++A+ILG+K D + +LSKGY++ VVI +C FV AF WSWGPLGW IPSE F
Sbjct: 358 IQMIICQVIVAVILGVKFGD-NQELSKGYSVIVVIFICLFVVAFGWSWGPLGWTIPSEIF 416
Query: 416 PLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPET 475
PLETRSAGQS+TV VN+LFTF+IAQAFL +LC FKFGIFLFF+GWV +M+IFV FL+PET
Sbjct: 417 PLETRSAGQSITVAVNLLFTFIIAQAFLGLLCAFKFGIFLFFAGWVTVMTIFVYFLLPET 476
Query: 476 KNIPIEEMTERVWKQHWFWKRF------MEDDNEKVS 506
K +PIEEMT +W +HWFWK+ +ED+++ VS
Sbjct: 477 KGVPIEEMT-LLWSKHWFWKKVLPDATNLEDESKNVS 512
>At1g50310 hexose transporter, putative
Length = 517
Score = 590 bits (1520), Expect = e-169
Identities = 292/506 (57%), Positives = 384/506 (75%), Gaps = 8/506 (1%)
Query: 1 MAGGGFATSGGG---EFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFF 57
MAGG F + GGG +E +T VI++CI+AA GGL+FGYD+G+SGGVTSM FL KFF
Sbjct: 1 MAGGAFVSEGGGGGNSYEGGVTVFVIMTCIVAAMGGLLFGYDLGISGGVTSMEEFLSKFF 60
Query: 58 PAVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAG 117
P V K + EA ++ YCK+DNQ LQLFTSSLYLAAL S+F AS TR GR+++M + G
Sbjct: 61 PEV-DKQMHEARRETAYCKFDNQLLQLFTSSLYLAALASSFVASAVTRKYGRKISMFVGG 119
Query: 118 FFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLN 177
F+ G FNA A N+AMLIVGR+LLG GVGFANQ+ PV+LSE+AP++IRGALNI FQ+
Sbjct: 120 VAFLIGSLFNAFATNVAMLIVGRLLLGVGVGFANQSTPVYLSEMAPAKIRGALNIGFQMA 179
Query: 178 VTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEE 237
+TIGIL ANL+NYGT++++ GWR+SLGLA +PA+++ +G+ V+ DTPNS++ERG+ E+
Sbjct: 180 ITIGILIANLINYGTSQMAKN-GWRVSLGLAAVPAVIMVIGSFVLPDTPNSMLERGKYEQ 238
Query: 238 GKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEK-GGQVIMAIGLEIFQQF 296
+ +L+KIRG DN++ EF +LC+A AK+V ++N+ + K ++ + FQQ
Sbjct: 239 AREMLQKIRGADNVDEEFQDLCDACEAAKKVDNPWKNIFQQAKYRPALVFCSAIPFFQQI 298
Query: 297 TGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGV 356
TGIN IMFYAPVLF T+GF +DASL SAVITGAVNV+ST+VSIY VD+ GRR+L LE G+
Sbjct: 299 TGINVIMFYAPVLFKTLGFADDASLISAVITGAVNVVSTLVSIYAVDRYGRRILFLEGGI 358
Query: 357 QMFLSQIVIAIILGIKV-TDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETF 415
QM +SQIV+ ++G+K T S L+ A +++ +C +V+ FAWSWGPLGWL+PSE
Sbjct: 359 QMIVSQIVVGTLIGMKFGTTGSGTLTPATADWILAFICLYVAGFAWSWGPLGWLVPSEIC 418
Query: 416 PLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPET 475
PLE R AGQ++ V VNM FTF+I Q FL+MLCH KFG+F FF G V +M++F+ FL+PET
Sbjct: 419 PLEIRPAGQAINVSVNMFFTFLIGQFFLTMLCHMKFGLFYFFGGMVAVMTVFIYFLLPET 478
Query: 476 KNIPIEEMTERVWKQHWFWKRFMEDD 501
K +PIEEM RVWKQH FWKR+M DD
Sbjct: 479 KGVPIEEM-GRVWKQHPFWKRYMPDD 503
>At1g11260 glucose transporter
Length = 522
Score = 588 bits (1515), Expect = e-168
Identities = 292/503 (58%), Positives = 376/503 (74%), Gaps = 5/503 (0%)
Query: 1 MAGGGFATSGGGE-FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPA 59
M GGF G + + K+TP V+ +C++AA GGL+FGYD+G+SGGVTSM FLK+FFP+
Sbjct: 1 MPAGGFVVGDGQKAYPGKLTPFVLFTCVVAAMGGLIFGYDIGISGGVTSMPSFLKRFFPS 60
Query: 60 VYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFF 119
VYRK +A + YC+YD+ L +FTSSLYLAAL S+ AS TR GRRL+ML G
Sbjct: 61 VYRKQQEDASTNQ-YCQYDSPTLTMFTSSLYLAALISSLVASTVTRKFGRRLSMLFGGIL 119
Query: 120 FIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVT 179
F AG N A+++ MLIVGRILLG G+GFANQAVP++LSE+AP + RGALNI FQL++T
Sbjct: 120 FCAGALINGFAKHVWMLIVGRILLGFGIGFANQAVPLYLSEMAPYKYRGALNIGFQLSIT 179
Query: 180 IGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGK 239
IGIL A ++NY KI GGWGWRLSLG A +PAL++T+G++V+ DTPNS+IERG+ EE K
Sbjct: 180 IGILVAEVLNYFFAKIKGGWGWRLSLGGAVVPALIITIGSLVLPDTPNSMIERGQHEEAK 239
Query: 240 AVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGI 299
L++IRG D++ EF +L AS+ ++ ++ +RNLL+ + + MA+ + FQQ TGI
Sbjct: 240 TKLRRIRGVDDVSQEFDDLVAASKESQSIEHPWRNLLRRKYRPHLTMAVMIPFFQQLTGI 299
Query: 300 NAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMF 359
N IMFYAPVLFNT+GF DASL SAV+TG+VNV +T+VSIY VD+ GRR L LE G QM
Sbjct: 300 NVIMFYAPVLFNTIGFTTDASLMSAVVTGSVNVAATLVSIYGVDRWGRRFLFLEGGTQML 359
Query: 360 LSQIVIAIILGIK--VTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPL 417
+ Q V+A +G K V +L K YAI VV +C +V+ FAWSWGPLGWL+PSE FPL
Sbjct: 360 ICQAVVAACIGAKFGVDGTPGELPKWYAIVVVTFICIYVAGFAWSWGPLGWLVPSEIFPL 419
Query: 418 ETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKN 477
E RSA QS+TV VNM+FTF+IAQ FL+MLCH KFG+FL F+ +V++MSIFV +PETK
Sbjct: 420 EIRSAAQSITVSVNMIFTFIIAQIFLTMLCHLKFGLFLVFAFFVVVMSIFVYIFLPETKG 479
Query: 478 IPIEEMTERVWKQHWFWKRFMED 500
IPIEEM + VW+ HW+W RF+ED
Sbjct: 480 IPIEEMGQ-VWRSHWYWSRFVED 501
>At3g19940 putative monosaccharide transport protein, STP4
Length = 514
Score = 582 bits (1501), Expect = e-166
Identities = 283/505 (56%), Positives = 381/505 (75%), Gaps = 7/505 (1%)
Query: 1 MAGGGFATSGGG---EFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFF 57
MAGG F + GGG +E +T VI++CI+AA GGL+FGYD+G+SGGVTSM FL KFF
Sbjct: 1 MAGGAFVSEGGGGGRSYEGGVTAFVIMTCIVAAMGGLLFGYDLGISGGVTSMEEFLTKFF 60
Query: 58 PAVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAG 117
P V + +A D+ YCK+DNQ LQLFTSSLYLAAL ++F AS TR GR+++M I G
Sbjct: 61 PQV-ESQMKKAKHDTAYCKFDNQMLQLFTSSLYLAALVASFMASVITRKHGRKVSMFIGG 119
Query: 118 FFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLN 177
F+ G FNA A N++MLI+GR+LLG GVGFANQ+ PV+LSE+AP++IRGALNI FQ+
Sbjct: 120 LAFLIGALFNAFAVNVSMLIIGRLLLGVGVGFANQSTPVYLSEMAPAKIRGALNIGFQMA 179
Query: 178 VTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEE 237
+TIGIL ANL+NYGT+K++ GWR+SLGLA +PA+++ +G+ ++ DTPNS++ERG+ EE
Sbjct: 180 ITIGILVANLINYGTSKMAQH-GWRVSLGLAAVPAVVMVIGSFILPDTPNSMLERGKNEE 238
Query: 238 GKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFT 297
K +LKKIRG DN++ EF +L +A AK+V+ ++N+++ + +I + FQQ T
Sbjct: 239 AKQMLKKIRGADNVDHEFQDLIDAVEAAKKVENPWKNIMESKYRPALIFCSAIPFFQQIT 298
Query: 298 GINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQ 357
GIN IMFYAPVLF T+GF +DA+L SAVITG VN+LST VSIY VD+ GRR+L LE G+Q
Sbjct: 299 GINVIMFYAPVLFKTLGFGDDAALMSAVITGVVNMLSTFVSIYAVDRYGRRLLFLEGGIQ 358
Query: 358 MFLSQIVIAIILGIKV-TDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFP 416
MF+ Q+++ +G + T + L+ A +++ +C +V+ FAWSWGPLGWL+PSE P
Sbjct: 359 MFICQLLVGSFIGARFGTSGTGTLTPATADWILAFICVYVAGFAWSWGPLGWLVPSEICP 418
Query: 417 LETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETK 476
LE R AGQ++ V VNM FTF+I Q FL+MLCH KFG+F FF+ V IM++F+ FL+PETK
Sbjct: 419 LEIRPAGQAINVSVNMFFTFLIGQFFLTMLCHMKFGLFYFFASMVAIMTVFIYFLLPETK 478
Query: 477 NIPIEEMTERVWKQHWFWKRFMEDD 501
+PIEEM RVWKQHWFWK+++ +D
Sbjct: 479 GVPIEEM-GRVWKQHWFWKKYIPED 502
>At5g26250 hexose transporter - like protein
Length = 507
Score = 582 bits (1500), Expect = e-166
Identities = 290/490 (59%), Positives = 370/490 (75%), Gaps = 4/490 (0%)
Query: 14 FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSN 73
F+AK+T V I I+AA GGL+FGYD+G+SGGVT+M FLK+FFP+VY + + ++N
Sbjct: 14 FDAKMTVYVFICVIIAAVGGLIFGYDIGISGGVTAMDDFLKEFFPSVYERK--KHAHENN 71
Query: 74 YCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNL 133
YCKYDNQ LQLFTSSLYLAAL ++FFAS T +GRR TM +A FF+ GV A A N+
Sbjct: 72 YCKYDNQFLQLFTSSLYLAALVASFFASATCSKLGRRPTMQLASIFFLIGVGLAAGAVNI 131
Query: 134 AMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTN 193
MLI+GRILLG GVGF NQAVP+FLSEIAP+R+RG LNI+FQL VTIGIL AN+VNY T+
Sbjct: 132 YMLIIGRILLGFGVGFGNQAVPLFLSEIAPARLRGGLNIVFQLMVTIGILIANIVNYFTS 191
Query: 194 KISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEP 253
I +GWR++LG AGIPAL+L G++++ +TP SLIER + +EGK LKKIRG ++++
Sbjct: 192 SIHP-YGWRIALGGAGIPALILLFGSLLICETPTSLIERNKTKEGKETLKKIRGVEDVDE 250
Query: 254 EFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTV 313
E+ + A +A++VK + L+K ++ + L+ FQQFTGINAIMFYAPVLF TV
Sbjct: 251 EYESIVHACDIARQVKDPYTKLMKPASRPPFVIGMLLQFFQQFTGINAIMFYAPVLFQTV 310
Query: 314 GFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKV 373
GF NDA+L SAV+TG +NVLST V I+ VDK GRR LLL++ V M + Q+VI IIL K
Sbjct: 311 GFGNDAALLSAVVTGTINVLSTFVGIFLVDKTGRRFLLLQSSVHMLICQLVIGIILA-KD 369
Query: 374 TDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNML 433
D + L++ A+ VVI VC +V FAWSWGPLGWLIPSETFPLETR+ G ++ V NM
Sbjct: 370 LDVTGTLARPQALVVVIFVCVYVMGFAWSWGPLGWLIPSETFPLETRTEGFALAVSCNMF 429
Query: 434 FTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQHWF 493
FTFVIAQAFLSMLC K GIF FFSGW+++M +F LF VPETK + I++M + VWK HW+
Sbjct: 430 FTFVIAQAFLSMLCAMKSGIFFFFSGWIVVMGLFALFFVPETKGVSIDDMRDSVWKLHWY 489
Query: 494 WKRFMEDDNE 503
WKRFM +++E
Sbjct: 490 WKRFMLEEDE 499
>At5g23270 monosaccharide transporter
Length = 514
Score = 576 bits (1484), Expect = e-164
Identities = 291/514 (56%), Positives = 374/514 (72%), Gaps = 5/514 (0%)
Query: 1 MAGGGFATSGG--GEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFP 58
MAGG F G G++E ++T V+I+CI+AA GGL+FGYD+G+SGGV SM FL KFFP
Sbjct: 1 MAGGAFIDESGHGGDYEGRVTAFVMITCIVAAMGGLLFGYDIGISGGVISMEDFLTKFFP 60
Query: 59 AVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGF 118
V R+ + G ++ YCKYDN+ L LFTSSLYLAAL ++F AS TR GR+++M+I
Sbjct: 61 DVLRQMQNKRGRETEYCKYDNELLTLFTSSLYLAALFASFLASTITRLFGRKVSMVIGSL 120
Query: 119 FFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNV 178
F++G N A NL MLI+GR+ LG GVGFANQ+VP++LSE+AP++IRGALNI FQL +
Sbjct: 121 AFLSGALLNGLAINLEMLIIGRLFLGVGVGFANQSVPLYLSEMAPAKIRGALNIGFQLAI 180
Query: 179 TIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEG 238
TIGIL AN+VNY T K+ G GWRLSLGLAG+PA+++ VG + DTPNS++ERG E+
Sbjct: 181 TIGILAANIVNYVTPKLQNGIGWRLSLGLAGVPAVMMLVGCFFLPDTPNSILERGNKEKA 240
Query: 239 KAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTG 298
K +L+KIRGT +E EF ELC A AK+VK + N+++ Q+ + FQQ TG
Sbjct: 241 KEMLQKIRGTMEVEHEFNELCNACEAAKKVKHPWTNIMQARYRPQLTFCTFIPFFQQLTG 300
Query: 299 INAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQM 358
IN IMFYAPVLF T+GF NDASL SAVITG VNVLSTIVSIY VDK GRR L L+ G QM
Sbjct: 301 INVIMFYAPVLFKTIGFGNDASLISAVITGLVNVLSTIVSIYSVDKFGRRALFLQGGFQM 360
Query: 359 FLSQIVIAIILGIKVTDHSD-DLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPL 417
++QI + ++G K + + +LS A ++ L+C +V+ FAWSWGPLGWL+PSE PL
Sbjct: 361 IVTQIAVGSMIGWKFGFNGEGNLSGVDADIILALICLYVAGFAWSWGPLGWLVPSEICPL 420
Query: 418 ETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKN 477
E RSAGQS+ V VNM FTF I Q FL+MLCH KFG+F FF+G VLIM+IF+ FL+PETK
Sbjct: 421 EIRSAGQSLNVSVNMFFTFFIGQFFLTMLCHMKFGLFYFFAGMVLIMTIFIYFLLPETKG 480
Query: 478 IPIEEMTERVWKQHWFWKRFM-EDDNEKVSNADY 510
+PIEEM +VWK+H +W ++ DD + V + Y
Sbjct: 481 VPIEEM-GKVWKEHRYWGKYSNNDDGDDVDDDAY 513
>At3g05960 putative hexose transporter
Length = 507
Score = 574 bits (1480), Expect = e-164
Identities = 290/494 (58%), Positives = 367/494 (73%), Gaps = 6/494 (1%)
Query: 14 FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSN 73
FEAK+T V I ++AA GGL+FGYD+G+SGGV++M FLK+FFPAV+ + + ++N
Sbjct: 13 FEAKMTVYVFICVMIAAVGGLIFGYDIGISGGVSAMDDFLKEFFPAVWERK--KHVHENN 70
Query: 74 YCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNL 133
YCKYDNQ LQLFTSSLYLAAL ++F AS T +GRR TM A FF+ GV A A NL
Sbjct: 71 YCKYDNQFLQLFTSSLYLAALVASFVASATCSKLGRRPTMQFASIFFLIGVGLTAGAVNL 130
Query: 134 AMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTN 193
MLI+GR+ LG GVGF NQAVP+FLSEIAP+++RG LNI+FQL VTIGIL AN+VNY T
Sbjct: 131 VMLIIGRLFLGFGVGFGNQAVPLFLSEIAPAQLRGGLNIVFQLMVTIGILIANIVNYFTA 190
Query: 194 KISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEP 253
+ +GWR++LG AGIPA++L G++++++TP SLIER + EEGK L+KIRG D+I
Sbjct: 191 TVHP-YGWRIALGGAGIPAVILLFGSLLIIETPTSLIERNKNEEGKEALRKIRGVDDIND 249
Query: 254 EFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPVLFNTV 313
E+ + A +A +VK +R LLK I+ + L++FQQFTGINAIMFYAPVLF TV
Sbjct: 250 EYESIVHACDIASQVKDPYRKLLKPASRPPFIIGMLLQLFQQFTGINAIMFYAPVLFQTV 309
Query: 314 GFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKV 373
GF +DA+L SAVITG++NVL+T V IY VD+ GRR LLL++ V M + Q++I IIL K
Sbjct: 310 GFGSDAALLSAVITGSINVLATFVGIYLVDRTGRRFLLLQSSVHMLICQLIIGIILA-KD 368
Query: 374 TDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNML 433
+ L + A+ VVI VC +V FAWSWGPLGWLIPSETFPLETRSAG +V V NM
Sbjct: 369 LGVTGTLGRPQALVVVIFVCVYVMGFAWSWGPLGWLIPSETFPLETRSAGFAVAVSCNMF 428
Query: 434 FTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQHWF 493
FTFVIAQAFLSMLC + GIF FFSGW+++M +F F +PETK I I++M E VWK HWF
Sbjct: 429 FTFVIAQAFLSMLCGMRSGIFFFFSGWIIVMGLFAFFFIPETKGIAIDDMRESVWKPHWF 488
Query: 494 WKRFM--EDDNEKV 505
WKR+M EDD+ +
Sbjct: 489 WKRYMLPEDDHHDI 502
>At4g21480 glucose transporter
Length = 508
Score = 556 bits (1433), Expect = e-158
Identities = 281/493 (56%), Positives = 368/493 (73%), Gaps = 8/493 (1%)
Query: 10 GGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAG 69
G E+ K+T V ++CI+AA GGL+FGYD+G+SGGVT+M F +KFFP+VY K +
Sbjct: 11 GKKEYPGKLTLYVTVTCIVAAMGGLIFGYDIGISGGVTTMDSFQQKFFPSVYEKQKKDH- 69
Query: 70 LDSN-YCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNA 128
DSN YC++D+ L LFTSSLYLAAL S+ ASY TR GR+++ML+ G F AG N
Sbjct: 70 -DSNQYCRFDSVSLTLFTSSLYLAALCSSLVASYVTRQFGRKISMLLGGVLFCAGALLNG 128
Query: 129 AAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLV 188
A + MLIVGR+LLG G+GF NQ+VP++LSE+AP + RGALNI FQL++TIGIL AN++
Sbjct: 129 FATAVWMLIVGRLLLGFGIGFTNQSVPLYLSEMAPYKYRGALNIGFQLSITIGILVANVL 188
Query: 189 NYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGT 248
N+ +KIS WGWRLSLG A +PAL++TVG++++ DTPNS+IERG+ +A L+KIRG
Sbjct: 189 NFFFSKIS--WGWRLSLGGAVVPALIITVGSLILPDTPNSMIERGQFRLAEAKLRKIRGV 246
Query: 249 DNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINAIMFYAPV 308
D+I+ E +L AS +K V+ +RNLL+ + + MAI + FQQ TGIN IMFYAPV
Sbjct: 247 DDIDDEINDLIIASEASKLVEHPWRNLLQRKYRPHLTMAILIPAFQQLTGINVIMFYAPV 306
Query: 309 LFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAII 368
LF T+GF +DA+L SAV+TG VNV +T+VSIY VDK GRR L LE G QM +SQ+ +A
Sbjct: 307 LFQTIGFGSDAALISAVVTGLVNVGATVVSIYGVDKWGRRFLFLEGGFQMLISQVAVAAA 366
Query: 369 LGIK--VTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSV 426
+G K V L K YAI VV+ +C +V+AFAWSWGPLGWL+PSE FPLE RSA QS+
Sbjct: 367 IGAKFGVDGTPGVLPKWYAIVVVLFICIYVAAFAWSWGPLGWLVPSEIFPLEIRSAAQSI 426
Query: 427 TVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMTER 486
TV VNM+FTF+IAQ FL MLCH KFG+F+FF+ +V++MSIFV +PET+ +PIEEM R
Sbjct: 427 TVSVNMIFTFLIAQVFLMMLCHLKFGLFIFFAFFVVVMSIFVYLFLPETRGVPIEEM-NR 485
Query: 487 VWKQHWFWKRFME 499
VW+ HW+W +F++
Sbjct: 486 VWRSHWYWSKFVD 498
>At3g19930 monosaccharide transport protein, STP4
Length = 514
Score = 553 bits (1426), Expect = e-158
Identities = 268/511 (52%), Positives = 374/511 (72%), Gaps = 5/511 (0%)
Query: 1 MAGGGFA-TSGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPA 59
MAGG + T G + K+TP V ++C + A GGL+FGYD+G+SGGVTSM PFL++FFP
Sbjct: 1 MAGGFVSQTPGVRNYNYKLTPKVFVTCFIGAFGGLIFGYDLGISGGVTSMEPFLEEFFPY 60
Query: 60 VYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFF 119
VY+K +++ ++ YC++D+Q L LFTSSLY+AAL S+ FAS TR GR+ +M + GF
Sbjct: 61 VYKK--MKSAHENEYCRFDSQLLTLFTSSLYVAALVSSLFASTITRVFGRKWSMFLGGFT 118
Query: 120 FIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVT 179
F G AFN AQN+AML++GRILLG GVGFANQ+VPV+LSE+AP +RGA N FQ+ +
Sbjct: 119 FFIGSAFNGFAQNIAMLLIGRILLGFGVGFANQSVPVYLSEMAPPNLRGAFNNGFQVAII 178
Query: 180 IGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGK 239
GI+ A ++NY T ++ G GWR+SLGLA +PA+++ +GA+++ DTPNSLIERG EE K
Sbjct: 179 FGIVVATIINYFTAQMKGNIGWRISLGLACVPAVMIMIGALILPDTPNSLIERGYTEEAK 238
Query: 240 AVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGI 299
+L+ IRGT+ ++ EF +L +AS +K+VK ++N++ Q+IM + FQQ TGI
Sbjct: 239 EMLQSIRGTNEVDEEFQDLIDASEESKQVKHPWKNIMLPRYRPQLIMTCFIPFFQQLTGI 298
Query: 300 NAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMF 359
N I FYAPVLF T+GF + ASL SA++TG + +L T VS++ VD+ GRR+L L+ G+QM
Sbjct: 299 NVITFYAPVLFQTLGFGSKASLLSAMVTGIIELLCTFVSVFTVDRFGRRILFLQGGIQML 358
Query: 360 LSQIVIAIILGIKV-TDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLE 418
+SQI I ++G+K + ++ K A +V L+C +V+ FAWSWGPLGWL+PSE PLE
Sbjct: 359 VSQIAIGAMIGVKFGVAGTGNIGKSDANLIVALICIYVAGFAWSWGPLGWLVPSEISPLE 418
Query: 419 TRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNI 478
RSA Q++ V VNM FTF++AQ FL+MLCH KFG+F FF+ +V+IM+IF+ ++PETKN+
Sbjct: 419 IRSAAQAINVSVNMFFTFLVAQLFLTMLCHMKFGLFFFFAFFVVIMTIFIYLMLPETKNV 478
Query: 479 PIEEMTERVWKQHWFWKRFMEDDNEKVSNAD 509
PIEEM RVWK HWFW +F+ D+ + A+
Sbjct: 479 PIEEM-NRVWKAHWFWGKFIPDEAVNMGAAE 508
>At1g77210 unknown protein
Length = 504
Score = 553 bits (1426), Expect = e-158
Identities = 282/508 (55%), Positives = 367/508 (71%), Gaps = 9/508 (1%)
Query: 1 MAGGGFATSGGGE----FEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKF 56
MAGG GG + +E +IT I +CI+ + GG +FGYD+GVSGGVTSM FLK+F
Sbjct: 1 MAGGALTDEGGLKRAHLYEHRITSYFIFACIVGSMGGSLFGYDLGVSGGVTSMDDFLKEF 60
Query: 57 FPAVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIA 116
FP +Y++ + +++YCKYDNQ L LFTSSLY A L STF ASY TR GRR ++L+
Sbjct: 61 FPGIYKRKQMHLN-ETDYCKYDNQILTLFTSSLYFAGLISTFGASYVTRIYGRRGSILVG 119
Query: 117 GFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQL 176
F G NAAA+N+ MLI+GRI LG G+GF NQAVP++LSE+AP++IRG +N LFQL
Sbjct: 120 SVSFFLGGVINAAAKNILMLILGRIFLGIGIGFGNQAVPLYLSEMAPAKIRGTVNQLFQL 179
Query: 177 NVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLE 236
IGIL ANL+NY T +I WGWRLSLGLA +PA+L+ +G +V+ +TPNSL+E+G+LE
Sbjct: 180 TTCIGILVANLINYKTEQIH-PWGWRLSLGLATVPAILMFLGGLVLPETPNSLVEQGKLE 238
Query: 237 EGKAVLKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQ-VIMAIGLEIFQQ 295
+ KAVL K+RGT+NIE EF +L EAS A+ VK FRNLL Q VI AIGL FQQ
Sbjct: 239 KAKAVLIKVRGTNNIEAEFQDLVEASDAARAVKNPFRNLLARRNRPQLVIGAIGLPAFQQ 298
Query: 296 FTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAG 355
TG+N+I+FYAPV+F ++GF ASL S+ IT A V++ I+S+Y DK GRR LLLEA
Sbjct: 299 LTGMNSILFYAPVMFQSLGFGGSASLISSTITNAALVVAAIMSMYSADKFGRRFLLLEAS 358
Query: 356 VQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETF 415
V+MF +V+ + L +K + +L K + +V+L+C FV A+ SWGP+GWL+PSE F
Sbjct: 359 VEMFCYMVVVGVTLALKFGE-GKELPKSLGLILVVLICLFVLAYGRSWGPMGWLVPSELF 417
Query: 416 PLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPET 475
PLETRSAGQSV VCVN+ FT +IAQ FL LCH K+GIFL F+G +L M FV FL+PET
Sbjct: 418 PLETRSAGQSVVVCVNLFFTALIAQCFLVSLCHLKYGIFLLFAGLILGMGSFVYFLLPET 477
Query: 476 KNIPIEEMTERVWKQHWFWKRFMEDDNE 503
K +PIEE+ +W+QHW WK+++ED +E
Sbjct: 478 KQVPIEEV-YLLWRQHWLWKKYVEDVDE 504
>At1g34580 monosaccharide transporter like protein
Length = 506
Score = 488 bits (1257), Expect = e-138
Identities = 246/505 (48%), Positives = 342/505 (67%), Gaps = 13/505 (2%)
Query: 1 MAGGGFA--TSGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFP 58
MAGGG A S G +AKIT V++SCI+AA+ GL+FGYD+G+SGGVT+M PFL+KFFP
Sbjct: 1 MAGGGLALDVSSAGNIDAKITAAVVMSCIVAASCGLIFGYDIGISGGVTTMKPFLEKFFP 60
Query: 59 AVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGF 118
+V +K + YC YD+Q L FTSSLY+A L ++ AS T GRR TM++ GF
Sbjct: 61 SVLKKA--SEAKTNVYCVYDSQLLTAFTSSLYVAGLVASLVASRLTAAYGRRTTMILGGF 118
Query: 119 FFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNV 178
F+ G N A N+AMLI GRILLG GVGF NQA PV+LSE+AP R RGA NI F +
Sbjct: 119 TFLFGALINGLAANIAMLISGRILLGFGVGFTNQAAPVYLSEVAPPRWRGAFNIGFSCFI 178
Query: 179 TIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEG 238
++G++ ANL+NYGT+ S GWR+SLGLA +PA ++TVG + + DTP+SL+ RG+ +E
Sbjct: 179 SMGVVAANLINYGTD--SHRNGWRISLGLAAVPAAIMTVGCLFISDTPSSLLARGKHDEA 236
Query: 239 KAVLKKIRGTDNI---EPEFLELCEASRVAKEVKQA--FRNLLKGEKGGQVIMAIGLEIF 293
L K+RG +NI E E EL +S++A E + + +L+ +++A+ + F
Sbjct: 237 HTSLLKLRGVENIADVETELAELVRSSQLAIEARAELFMKTILQRRYRPHLVVAVVIPCF 296
Query: 294 QQFTGINAIMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLE 353
QQ TGI FYAPVLF +VGF + +L + I G VN+ S ++S +D+ GRR L +
Sbjct: 297 QQLTGITVNAFYAPVLFRSVGFGSGPALIATFILGFVNLGSLLLSTMVIDRFGRRFLFIA 356
Query: 354 AGVQMFLSQIVIAIILGIKVTDHSD-DLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPS 412
G+ M L QI +A++L + V D ++ KGYA+ VV+L+C + + F WSWGPL WL+PS
Sbjct: 357 GGILMLLCQIAVAVLLAVTVGATGDGEMKKGYAVTVVVLLCIYAAGFGWSWGPLSWLVPS 416
Query: 413 ETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLV 472
E FPL+ R AGQS++V VN TF ++Q FL+ LC FK+G FLF+ GW+ M+IFV+ +
Sbjct: 417 EIFPLKIRPAGQSLSVAVNFAATFALSQTFLATLCDFKYGAFLFYGGWIFTMTIFVIMFL 476
Query: 473 PETKNIPIEEMTERVWKQHWFWKRF 497
PETK IP++ M + VW++HW+W+RF
Sbjct: 477 PETKGIPVDSMYQ-VWEKHWYWQRF 500
>At5g61520 monosaccharide transporter STP3
Length = 514
Score = 474 bits (1219), Expect = e-134
Identities = 237/496 (47%), Positives = 345/496 (68%), Gaps = 16/496 (3%)
Query: 7 ATSGGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVL 66
+ SGG KIT V+ SC+MAA GG++FGYD+GVSGGV SM PFLK+FFP VY+
Sbjct: 14 SVSGG-----KITYFVVASCVMAAMGGVIFGYDIGVSGGVMSMGPFLKRFFPKVYKLQEE 68
Query: 67 E----AGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIA 122
+ +++YC +++Q L FTSSLY++ L +T AS TR+ GR+ ++ + G F+A
Sbjct: 69 DRRRRGNSNNHYCLFNSQLLTSFTSSLYVSGLIATLLASSVTRSWGRKPSIFLGGVSFLA 128
Query: 123 GVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGI 182
G A +AQN+AMLI+ R+LLG GVGFANQ+VP++LSE+AP++ RGA++ FQL + IG
Sbjct: 129 GAALGGSAQNVAMLIIARLLLGVGVGFANQSVPLYLSEMAPAKYRGAISNGFQLCIGIGF 188
Query: 183 LFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIER-GRLEEGKAV 241
L AN++NY T I GW R+SL A IPA +LT+G++ + +TPNS+I+ G + + + +
Sbjct: 189 LSANVINYETQNIKHGW--RISLATAAIPASILTLGSLFLPETPNSIIQTTGDVHKTELM 246
Query: 242 LKKIRGTDNIEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQVIMAIGLEIFQQFTGINA 301
L+++RGT++++ E +L EAS + AF LL+ + +++MA+ + FQQ TGIN
Sbjct: 247 LRRVRGTNDVQDELTDLVEASSGSDTDSNAFLKLLQRKYRPELVMALVIPFFQQVTGINV 306
Query: 302 IMFYAPVLFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLS 361
+ FYAPVL+ TVGF SL S ++TG V ST++S+ VD++GR+ L L G+QM +S
Sbjct: 307 VAFYAPVLYRTVGFGESGSLMSTLVTGIVGTSSTLLSMLVVDRIGRKTLFLIGGLQMLVS 366
Query: 362 QIVIAIILGIKVTD-HSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETR 420
Q+ I +I + V D H + +GY VV+LVC +V+ F WSWGPLGWL+PSE FPLE R
Sbjct: 367 QVTIGVI--VMVADVHDGVIKEGYGYAVVVLVCVYVAGFGWSWGPLGWLVPSEIFPLEIR 424
Query: 421 SAGQSVTVCVNMLFTFVIAQAFLSMLCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPI 480
S QSVTV V+ +FTF +AQ+ MLC F+ GIF F+ GW+++M++ V +PETKN+PI
Sbjct: 425 SVAQSVTVAVSFVFTFAVAQSAPPMLCKFRAGIFFFYGGWLVVMTVAVQLFLPETKNVPI 484
Query: 481 EEMTERVWKQHWFWKR 496
E++ +W++HWFW+R
Sbjct: 485 EKVV-GLWEKHWFWRR 499
>At1g07340 hexose transporter like protein
Length = 383
Score = 363 bits (931), Expect = e-100
Identities = 190/361 (52%), Positives = 253/361 (69%), Gaps = 5/361 (1%)
Query: 10 GGGEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAG 69
G F AK+T V + C++AA GGLMFGYD+G+SGGVTSM FL FFP VY K
Sbjct: 11 GTKAFPAKLTGQVFLCCVIAAVGGLMFGYDIGISGGVTSMDTFLLDFFPHVYEKK--HRV 68
Query: 70 LDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAA 129
++NYCK+D+Q LQLFTSSLYLA + ++F +SY +R GR+ T+++A FF+ G N +
Sbjct: 69 HENNYCKFDDQLLQLFTSSLYLAGIFASFISSYVSRAFGRKPTIMLASIFFLVGAILNLS 128
Query: 130 AQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVN 189
AQ L MLI GRILLG G+GF NQ VP+F+SEIAP+R RG LN++FQ +TIGIL A+ VN
Sbjct: 129 AQELGMLIGGRILLGFGIGFGNQTVPLFISEIAPARYRGGLNVMFQFLITIGILAASYVN 188
Query: 190 YGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTD 249
Y T+ + GWR SLG A +PAL+L +G+ + +TP SLIERG+ E+GK VL+KIRG +
Sbjct: 189 YLTSTLKN--GWRYSLGGAAVPALILLIGSFFIHETPASLIERGKDEKGKQVLRKIRGIE 246
Query: 250 NIEPEFLELCEASRVAKEVKQAFRNLL-KGEKGGQVIMAIGLEIFQQFTGINAIMFYAPV 308
+IE EF E+ A+ VA +VK F+ L K E ++ L+ FQQFTGIN +MFYAPV
Sbjct: 247 DIELEFNEIKYATEVATKVKSPFKELFTKSENRPPLVCGTLLQFFQQFTGINVVMFYAPV 306
Query: 309 LFNTVGFKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAII 368
LF T+G ++ASL S V+T VN ++T++S+ VD GRR LL+E +QM +Q+ I I
Sbjct: 307 LFQTMGSGDNASLISTVVTNGVNAIATVISLLVVDFAGRRCLLMEGALQMTATQMTIGGI 366
Query: 369 L 369
L
Sbjct: 367 L 367
>At2g43330 membrane transporter like protein
Length = 509
Score = 211 bits (538), Expect = 6e-55
Identities = 140/471 (29%), Positives = 232/471 (48%), Gaps = 32/471 (6%)
Query: 27 IMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDNQGLQLFT 86
+ A GGL+FGYD GV G ++K F V + + L+ +
Sbjct: 36 VTAGIGGLLFGYDTGVISGALL---YIKDDFEVVKQSSFLQ---------------ETIV 77
Query: 87 SSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCG 146
S + A+ + GR+ L A F AG AAA + +LI GR+L+G G
Sbjct: 78 SMALVGAMIGAAAGGWINDYYGRKKATLFADVVFAAGAIVMAAAPDPYVLISGRLLVGLG 137
Query: 147 VGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLG 206
VG A+ PV+++E +PS +RG L L +T G + LVN ++ G W W LG
Sbjct: 138 VGVASVTAPVYIAEASPSEVRGGLVSTNVLMITGGQFLSYLVNSAFTQVPGTWRW--MLG 195
Query: 207 LAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVAK 266
++G+PA++ + + + ++P L + R E VL + +E E L A K
Sbjct: 196 VSGVPAVIQFILMLFMPESPRWLFMKNRKAEAIQVLARTYDISRLEDEIDHLSAAEEEEK 255
Query: 267 EVKQ--AFRNLLKGEKGGQVIMA-IGLEIFQQFTGINAIMFYAPVLFNTVGF-KNDASLY 322
+ K+ + ++ + ++ +A GL+ FQQFTGIN +M+Y+P + GF N +L+
Sbjct: 256 QRKRTVGYLDVFRSKELRLAFLAGAGLQAFQQFTGINTVMYYSPTIVQMAGFHSNQLALF 315
Query: 323 SAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDDLSK 382
++I A+N T+V IYF+D GR+ L L + I+ +IL + S+ S
Sbjct: 316 LSLIVAAMNAAGTVVGIYFIDHCGRKKLALSS----LFGVIISLLILSVSFFKQSETSSD 371
Query: 383 G--YAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQ 440
G Y V+ + ++ FA GP+ W + SE +P + R ++ VN + ++AQ
Sbjct: 372 GGLYGWLAVLGLALYIVFFAPGMGPVPWTVNSEIYPQQYRGICGGMSATVNWISNLIVAQ 431
Query: 441 AFLSMLCHFKFGI-FLFFSGWVLIMSIFVLFLVPETKNIPIEEMTERVWKQ 490
FL++ G+ FL +G ++ IFV+ VPET+ + E+ E++WK+
Sbjct: 432 TFLTIAEAAGTGMTFLILAGIAVLAVIFVIVFVPETQGLTFSEV-EQIWKE 481
>At2g18480 putative sugar transporter
Length = 508
Score = 210 bits (534), Expect = 2e-54
Identities = 142/479 (29%), Positives = 235/479 (48%), Gaps = 48/479 (10%)
Query: 27 IMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDNQGLQLFT 86
I+A+ ++FGYD GV G F++ + K ++ +++
Sbjct: 27 IVASIISIIFGYDTGVMSGAQI---FIR------------------DDLKINDTQIEVLA 65
Query: 87 SSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCG 146
L L AL + A T+ +GRR T+ ++ F+ G N +L+VGR + G G
Sbjct: 66 GILNLCALVGSLTAGKTSDVIGRRYTIALSAVIFLVGSVLMGYGPNYPVLMVGRCIAGVG 125
Query: 147 VGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLG 206
VGFA PV+ +EI+ + RG L L +L +++GIL + NY K++ GWRL LG
Sbjct: 126 VGFALMIAPVYSAEISSASHRGFLTSLPELCISLGILLGYVSNYCFGKLTLKLGWRLMLG 185
Query: 207 LAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGT-DNIEPEFLELCEASRV- 264
+A P+L+L G + ++P L+ +GRLEE K ++ + T + E F ++ A+ V
Sbjct: 186 IAAFPSLILAFGITRMPESPRWLVMQGRLEEAKKIMVLVSNTEEEAEERFRDILTAAEVD 245
Query: 265 AKEVKQAFRNLLKGEKGGQV----------------IMAIGLEIFQQFTGINAIMFYAPV 308
E+K+ + K G V I A+G+ F+ TGI A++ Y+P
Sbjct: 246 VTEIKEVGGGVKKKNHGKSVWRELVIKPRPAVRLILIAAVGIHFFEHATGIEAVVLYSPR 305
Query: 309 LFNTVG-FKNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEA--GVQMFLSQIVI 365
+F G D L + V G I++ + +DK+GRR LLL + G+ L+ + +
Sbjct: 306 IFKKAGVVSKDKLLLATVGVGLTKAFFIIIATFLLDKVGRRKLLLTSTGGMVFALTSLAV 365
Query: 366 AIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQS 425
++ + + + LS ++ FV+ F+ GP+ W+ SE FPL R+ G S
Sbjct: 366 SLTMVQRFGRLAWALS-----LSIVSTYAFVAFFSIGLGPITWVYSSEIFPLRLRAQGAS 420
Query: 426 VTVCVNMLFTFVIAQAFLSMLCHFKF-GIFLFFSGWVLIMSIFVLFLVPETKNIPIEEM 483
+ V VN + ++ +FLSM G+F F+G + F F++PETK +P+EEM
Sbjct: 421 IGVAVNRIMNATVSMSFLSMTKAITTGGVFFVFAGIAVAAWWFFFFMLPETKGLPLEEM 479
>At3g18830 sugar transporter like protein
Length = 539
Score = 207 bits (526), Expect = 1e-53
Identities = 143/481 (29%), Positives = 235/481 (48%), Gaps = 47/481 (9%)
Query: 27 IMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDNQGLQLFT 86
I+A+ ++ GYD+GV G +Y K L K ++ + +
Sbjct: 41 ILASMTSILLGYDIGVMSGAM------------IYIKRDL---------KINDLQIGILA 79
Query: 87 SSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVGRILLGCG 146
SL + +L + A T+ +GRR T+++AG F AG + N A L+ GR + G G
Sbjct: 80 GSLNIYSLIGSCAAGRTSDWIGRRYTIVLAGAIFFAGAILMGLSPNYAFLMFGRFIAGIG 139
Query: 147 VGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGWGWRLSLG 206
VG+A PV+ +E++P+ RG LN ++ + GI+ + N + + GWRL LG
Sbjct: 140 VGYALMIAPVYTAEVSPASSRGFLNSFPEVFINAGIMLGYVSNLAFSNLPLKVGWRLMLG 199
Query: 207 LAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELCEASRVA- 265
+ +P+++L +G + + ++P L+ +GRL + K VL K +D+ L L + A
Sbjct: 200 IGAVPSVILAIGVLAMPESPRWLVMQGRLGDAKRVLDKT--SDSPTEATLRLEDIKHAAG 257
Query: 266 ------KEVKQAFRNLLKGEKGGQ-------------VIMAIGLEIFQQFTGINAIMFYA 306
+V Q R GE + +I AIG+ FQQ +GI+A++ ++
Sbjct: 258 IPADCHDDVVQVSRRNSHGEGVWRELLIRPTPAVRRVMIAAIGIHFFQQASGIDAVVLFS 317
Query: 307 PVLFNTVGFKND-ASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVI 365
P +F T G K D L + V G V +V+ + +D++GRR LLL + M LS +
Sbjct: 318 PRIFKTAGLKTDHQQLLATVAVGVVKTSFILVATFLLDRIGRRPLLLTSVGGMVLS--LA 375
Query: 366 AIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQS 425
A+ + + D S+ + + V T+V+ F+ GP+ W+ SE FPL RS G S
Sbjct: 376 ALGTSLTIIDQSEKKVMWAVVVAIATVMTYVATFSIGAGPITWVYSSEIFPLRLRSQGSS 435
Query: 426 VTVCVNMLFTFVIAQAFLSM-LCHFKFGIFLFFSGWVLIMSIFVLFLVPETKNIPIEEMT 484
+ V VN + + VI+ +FL M G F F G + +F +PET+ +E+M
Sbjct: 436 MGVVVNRVTSGVISISFLPMSKAMTTGGAFYLFGGIATVAWVFFYTFLPETQGRMLEDMD 495
Query: 485 E 485
E
Sbjct: 496 E 496
>At4g36670 sugar transporter like protein
Length = 493
Score = 204 bits (518), Expect = 1e-52
Identities = 139/492 (28%), Positives = 234/492 (47%), Gaps = 43/492 (8%)
Query: 12 GEFEAKITPIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLD 71
GE A + + I+A+ ++FGYD GV G +E L
Sbjct: 7 GEKPAGVNRFALQCAIVASIVSIIFGYDTGVMSGAM----------------VFIEEDLK 50
Query: 72 SNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQ 131
+N + +++ T L L AL + A T+ +GRR T+++A F+ G
Sbjct: 51 TNDVQ-----IEVLTGILNLCALVGSLLAGRTSDIIGRRYTIVLASILFMLGSILMGWGP 105
Query: 132 NLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYG 191
N +L+ GR G GVGFA PV+ +EIA + RG L L L ++IGIL +VNY
Sbjct: 106 NYPVLLSGRCTAGLGVGFALMVAPVYSAEIATASHRGLLASLPHLCISIGILLGYIVNYF 165
Query: 192 TNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGT-DN 250
+K+ GWRL LG+A +P+L+L G + + ++P LI +GRL+EGK +L+ + + +
Sbjct: 166 FSKLPMHIGWRLMLGIAAVPSLVLAFGILKMPESPRWLIMQGRLKEGKEILELVSNSPEE 225
Query: 251 IEPEFLELCEASRVAKEVKQAFRNLLKGEKGGQ-----------------VIMAIGLEIF 293
E F ++ A+ + + + + G+ ++ A+G+ F
Sbjct: 226 AELRFQDIKAAAGIDPKCVDDVVKMEGKKTHGEGVWKELILRPTPAVRRVLLTALGIHFF 285
Query: 294 QQFTGINAIMFYAPVLFNTVGFKNDASLYSAVI-TGAVNVLSTIVSIYFVDKLGRRMLLL 352
Q +GI A++ Y P +F G L+ I G + + +DK+GRR LLL
Sbjct: 286 QHASGIEAVLLYGPRIFKKAGITTKDKLFLVTIGVGIMKTTFIFTATLLLDKVGRRKLLL 345
Query: 353 EAGVQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSAFAWSWGPLGWLIPS 412
+ M ++ ++ G+ + ++ + ++ +FV+ F+ GP+ W+ S
Sbjct: 346 TSVGGMVIALTMLG--FGLTMAQNAGGKLAWALVLSIVAAYSFVAFFSIGLGPITWVYSS 403
Query: 413 ETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHFKF-GIFLFFSGWVLIMSIFVLFL 471
E FPL+ R+ G S+ V VN + ++ +FLS+ G F F+G + F FL
Sbjct: 404 EVFPLKLRAQGASLGVAVNRVMNATVSMSFLSLTSAITTGGAFFMFAGVAAVAWNFFFFL 463
Query: 472 VPETKNIPIEEM 483
+PETK +EE+
Sbjct: 464 LPETKGKSLEEI 475
>At1g30220 unknown protein
Length = 580
Score = 197 bits (502), Expect = 9e-51
Identities = 118/359 (32%), Positives = 192/359 (52%), Gaps = 30/359 (8%)
Query: 20 PIVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKKFFPAVYRKTVLEAGLDSNYCKYDN 79
P V+ A GGL+FGYD GV G +++ F +V R T L+
Sbjct: 26 PYVLRLAFSAGIGGLLFGYDTGVISGALL---YIRDDFKSVDRNTWLQ------------ 70
Query: 80 QGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLIAGFFFIAGVAFNAAAQNLAMLIVG 139
++ S A+ + +GRR +L+A F F+ G AAA N ++L+VG
Sbjct: 71 ---EMIVSMAVAGAIVGAAIGGWANDKLGRRSAILMADFLFLLGAIIMAAAPNPSLLVVG 127
Query: 140 RILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQLNVTIGILFANLVNYGTNKISGGW 199
R+ +G GVG A+ P+++SE +P++IRGAL +T G + L+N ++G W
Sbjct: 128 RVFVGLGVGMASMTAPLYISEASPAKIRGALVSTNGFLITGGQFLSYLINLAFTDVTGTW 187
Query: 200 GWRLSLGLAGIPALLLTVGAIVVVDTPNSLIERGRLEEGKAVLKKIRGTDNIEPEFLELC 259
W LG+AGIPALL V + ++P L +GR EE KA+L++I +++E E L
Sbjct: 188 RW--MLGIAGIPALLQFVLMFTLPESPRWLYRKGREEEAKAILRRIYSAEDVEQEIRALK 245
Query: 260 EA--SRVAKEVKQAFRNLLKGEKGGQV----IMAIGLEIFQQFTGINAIMFYAPVLFNTV 313
++ + + +E N++K K V I +GL++FQQF GIN +M+Y+P +
Sbjct: 246 DSVETEILEEGSSEKINMIKLCKAKTVRRGLIAGVGLQVFQQFVGINTVMYYSPTIVQLA 305
Query: 314 GF-KNDASLYSAVITGAVNVLSTIVSIYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGI 371
GF N +L +++T +N +I+SIYF+D++GR+ LL+ + +F I + I+ G+
Sbjct: 306 GFASNRTALLLSLVTAGLNAFGSIISIYFIDRIGRKKLLI---ISLFGVIISLGILTGV 361
Score = 58.5 bits (140), Expect = 9e-09
Identities = 35/114 (30%), Positives = 58/114 (50%), Gaps = 4/114 (3%)
Query: 387 FVVILVCTFVSAFAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSML 446
F ++ + ++ F+ G + W++ SE +PL R + N + ++AQ+FLS+
Sbjct: 455 FALLGLGLYIIFFSPGMGTVPWIVNSEIYPLRFRGICGGIAATANWISNLIVAQSFLSLT 514
Query: 447 CHFKFG-IFLFFSGWVLIMSIFVLFLVPETKNIPIEE---MTERVWKQHWFWKR 496
FL F +I +FV+ VPETK +P+EE M ER + FWK+
Sbjct: 515 EAIGTSWTFLIFGVISVIALLFVMVCVPETKGMPMEEIEKMLERRSMEFKFWKK 568
>At5g59250 D-xylose-H+ symporter - like protein
Length = 558
Score = 187 bits (476), Expect = 9e-48
Identities = 159/513 (30%), Positives = 243/513 (46%), Gaps = 52/513 (10%)
Query: 3 GGGFATSGG-GEFEAKITP------IVIISCIMAATGGLMFGYDVGVSGGVTSMHPFLKK 55
GG FA SG + A P VI+ I A GGL+FGYD+G + G T L
Sbjct: 72 GGEFADSGEVADSLASDAPESFSWSSVILPFIFPALGGLLFGYDIGATSGAT-----LSL 126
Query: 56 FFPAVYRKTVLEAGLDSNYCKYDNQGLQLFTSSLYLAALTSTFFASYTTRTMGRRLTMLI 115
PA+ T + + L L S AL + +GRR ++I
Sbjct: 127 QSPALSGTT---------WFNFSPVQLGLVVSGSLYGALLGSISVYGVADFLGRRRELII 177
Query: 116 AGFFFIAGVAFNAAAQNLAMLIVGRILLGCGVGFANQAVPVFLSEIAPSRIRGALNILFQ 175
A ++ G A +L +L+VGR+L G G+G A P++++E PS+IRG L L +
Sbjct: 178 AAVLYLLGSLITGCAPDLNILLVGRLLYGFGIGLAMHGAPLYIAETCPSQIRGTLISLKE 237
Query: 176 LNVTIGILFANLVNYGTNKISGGWGWRLSLGLAGIPALLLTVGAIVVVDTPNSLIER--- 232
L + +GIL + G+ +I GWR G ALL+ +G + +P L+ R
Sbjct: 238 LFIVLGILLG--FSVGSFQIDVVGGWRYMYGFGTPVALLMGLGMWSLPASPRWLLLRAVQ 295
Query: 233 --GRLEEGK----AVLKKIRGT---DNIEPEFLELCEAS-RVAKEVKQAFRNLLKGEKGG 282
G+L+E K L K+RG D I + ++ S + A E +++ N L+ +G
Sbjct: 296 GKGQLQEYKEKAMLALSKLRGRPPGDKISEKLVDDAYLSVKTAYEDEKSGGNFLEVFQGP 355
Query: 283 Q---VIMAIGLEIFQQFTGINAIMFYAPVLFNTVGFKNDA-SLYSAVITGAVNVLSTIVS 338
+ + GL +FQQ TG ++++YA + T GF A + +VI G +L T V+
Sbjct: 356 NLKALTIGGGLVLFQQITGQPSVLYYAGSILQTAGFSAAADATRVSVIIGVFKLLMTWVA 415
Query: 339 IYFVDKLGRRMLLLEAGVQMFLSQIVIAIILGIKVTDHSDDLSKGYAIFVVILVCTFVSA 398
+ VD LGRR LL+ V I L + + G+ + V + +V
Sbjct: 416 VAKVDDLGRRPLLIGG---------VSGIALSLFLLSAYYKFLGGFPLVAVGALLLYVGC 466
Query: 399 FAWSWGPLGWLIPSETFPLETRSAGQSVTVCVNMLFTFVIAQAFLSMLCHF--KFGIFLF 456
+ S+GP+ WL+ SE FPL TR G S+ V N ++ AF S L F +FL
Sbjct: 467 YQISFGPISWLMVSEIFPLRTRGRGISLAVLTNFGSNAIVTFAF-SPLKEFLGAENLFLL 525
Query: 457 FSGWVLIMSIFVLFLVPETKNIPIEEMTERVWK 489
F G L+ +FV+ +VPETK + +EE+ ++ K
Sbjct: 526 FGGIALVSLLFVILVVPETKGLSLEEIESKILK 558
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.142 0.425
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,829,312
Number of Sequences: 26719
Number of extensions: 446786
Number of successful extensions: 1769
Number of sequences better than 10.0: 85
Number of HSP's better than 10.0 without gapping: 57
Number of HSP's successfully gapped in prelim test: 28
Number of HSP's that attempted gapping in prelim test: 1430
Number of HSP's gapped (non-prelim): 115
length of query: 521
length of database: 11,318,596
effective HSP length: 104
effective length of query: 417
effective length of database: 8,539,820
effective search space: 3561104940
effective search space used: 3561104940
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC145222.4


