

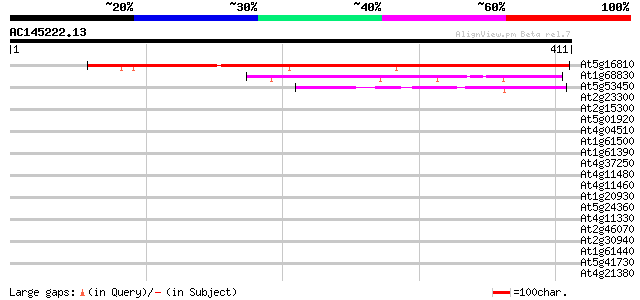
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145222.13 + phase: 0
(411 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g16810 unknown protein 485 e-137
At1g68830 putative protein kinase 48 9e-06
At5g53450 unknown protein 42 8e-04
At2g23300 putative receptor-like protein kinase 34 0.17
At2g15300 putative receptor-like protein kinase 33 0.22
At5g01920 unknown protein 33 0.29
At4g04510 putative receptor-like protein kinase 33 0.37
At1g61500 32 0.64
At1g61390 S-like receptor protein kinase; member of gene cluster... 32 0.64
At4g37250 receptor kinase like protein 32 0.83
At4g11480 serine/threonine kinase-like protein 32 0.83
At4g11460 serine/threonine kinase-like protein 32 0.83
At1g20930 putative cell division control protein, cdc2 kinase 32 0.83
At5g24360 unknown protein 31 1.1
At4g11330 MAP kinase (ATMPK5) 31 1.1
At2g46070 mitogen-activated protein kinase like protein 31 1.1
At2g30940 putative protein kinase 31 1.1
At1g61440 S-like receptor protein kinase; member of gene cluster... 31 1.1
At5g41730 putative protein 31 1.4
At4g21380 receptor-like serine/threonine protein kinase ARK3 31 1.4
>At5g16810 unknown protein
Length = 418
Score = 485 bits (1249), Expect = e-137
Identities = 252/367 (68%), Positives = 299/367 (80%), Gaps = 16/367 (4%)
Query: 58 ITNSDWFQVGRPIGNYGFMNVTT------SSTDQYSLG--GGLKTQDVQEGSVKIRLYEG 109
IT++D F+VGR IG+YGFMNVT+ S +Y+ G LK+QD+ EG+VKIRLY+G
Sbjct: 54 ITSADSFEVGRLIGSYGFMNVTSYTGLQSGSDFEYTSDDIGRLKSQDIGEGAVKIRLYQG 113
Query: 110 RVSQGPLKQTPVLFKVYPGTRAGGVVADMMAANELNSHMFLQSSSKGISQHLMLLLGGFE 169
R+SQGP + TPV+FKVYPG RAGGV ADMMAANELN+H FLQS K + +L+LL+GGFE
Sbjct: 114 RISQGPFRGTPVVFKVYPGQRAGGVEADMMAANELNAHSFLQS--KSLPANLLLLVGGFE 171
Query: 170 TTTGEQWLAFRDYGKSTAADYAKVASEKVSKLSS---WNSFERGQSMKRRRRFIIKLLQG 226
T GEQWLAFRD GK +AADYA+ ASEK ++ S WN +E+ Q +KRRR F+IK+LQG
Sbjct: 172 TQLGEQWLAFRDGGKDSAADYAQTASEKTTRARSQGVWNPYEKEQMIKRRRNFVIKILQG 231
Query: 227 ALRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELED---S 283
A++GLA+MHD+DRLHQSLGP S+ LNT +E EA YLIPRLRDLAFSV +R S LE+ S
Sbjct: 232 AMKGLAFMHDNDRLHQSLGPSSIVLNTPAEREAIYLIPRLRDLAFSVDIRPSCLEEGATS 291
Query: 284 GPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLLFAYLAFVPFCEAGVMDGLSLQRL 343
G L+E LW RA+AA A+T EKRAFGIADDIYEAGLLFAYLAFVPFCEAGV D LSLQRL
Sbjct: 292 GSLSEQLWRRANAAGAYTVFEKRAFGIADDIYEAGLLFAYLAFVPFCEAGVTDSLSLQRL 351
Query: 344 LENTFRLDLEATREYCIADDKLVNAIEFLDLGDGAGWELLQAMLNADFRKRPTAEAVLSH 403
LENTFRLD+EA REYC+AD++L A++FLDLGD AGWELLQAMLNAD+RKRP AEAVLSH
Sbjct: 352 LENTFRLDIEAVREYCLADERLEEAVKFLDLGDRAGWELLQAMLNADYRKRPMAEAVLSH 411
Query: 404 RFMTGEV 410
RF+ G V
Sbjct: 412 RFLNGVV 418
>At1g68830 putative protein kinase
Length = 562
Score = 48.1 bits (113), Expect = 9e-06
Identities = 60/247 (24%), Positives = 102/247 (41%), Gaps = 18/247 (7%)
Query: 174 EQWLAFRDYGKSTAADY--AKVASEKVSKLSSWNSFERGQSMKRRRRFIIKLLQGALRGL 231
E WL ++ G+ST A +K V + + + ++R + I +++ L L
Sbjct: 208 EYWLLWKYEGESTLAGLMQSKEFPYNVETIILGKVQDLPKGLERENKIIQTIMRQLLFAL 267
Query: 232 AYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDLA---------FSVSVRYSELED 282
+H +H+ + P ++ + S S + DL F + RY+ E
Sbjct: 268 DGLHSTGIIHRDVKPQNIIFSEGSRSFKIIDLGAAADLRVGINYIPKEFLLDPRYAAPEQ 327
Query: 283 SGPLTEGLWARASAASAFTYLEKRAFGIAD--DIYEAGLLFAYLAFVPFCEAGVMDGLSL 340
T+ A ++ +A + D DIY GL+F +AF + +
Sbjct: 328 YIMSTQTPSAPSAPVAAALSPVLWQMNLPDRFDIYSIGLIFLQMAFPSLRSDSNL--IQF 385
Query: 341 QRLLENTFRLDLEATREYCI--ADDKLVNAIEFLDLGDGAGWELLQAMLNADFRKRPTAE 398
R L+ DL A R+ A L E +DL G GWELL +M+ R+R +A+
Sbjct: 386 NRQLKRC-DYDLTAWRKLVEPRASADLRRGFELVDLDGGIGWELLTSMVRYKARQRISAK 444
Query: 399 AVLSHRF 405
A L+H +
Sbjct: 445 AALAHPY 451
>At5g53450 unknown protein
Length = 670
Score = 41.6 bits (96), Expect = 8e-04
Identities = 44/205 (21%), Positives = 84/205 (40%), Gaps = 31/205 (15%)
Query: 210 GQSMKRRRRFIIKLLQGALRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDL 269
G ++ R+ R I L++ L G+ Y+H H H L +V ++ + D
Sbjct: 222 GPAVSRQLRLIRTLMRDILIGVNYLHSHGLAHTELRLENVHISPV-------------DR 268
Query: 270 AFSVSVRYSELEDSGPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLLFAYLAFVPF 329
V + + + +G + S ++A++ +++R IA D+ G + A +
Sbjct: 269 HIKVGILGNAADFNGDVP-------STSNAYSTMDRRQMMIAFDMRCVGFMMAKMVLQE- 320
Query: 330 CEAGVMDGLSLQRLLENTFR-LDLEATREYCIA-----DDKLVNAIEFLDLGDGAGWELL 383
+MD L +L + D + RE+ + + ++ LD GAGW LL
Sbjct: 321 ----LMDPLIFAKLKSFLAKGNDPSSLREFFVTTLNTNSESGNTGVQILDRNWGAGWHLL 376
Query: 384 QAMLNADFRKRPTAEAVLSHRFMTG 408
++ +R + L H F+ G
Sbjct: 377 SLLIATRPSERISCLDALKHPFLCG 401
>At2g23300 putative receptor-like protein kinase
Length = 773
Score = 33.9 bits (76), Expect = 0.17
Identities = 13/31 (41%), Positives = 22/31 (70%)
Query: 221 IKLLQGALRGLAYMHDHDRLHQSLGPFSVSL 251
+K+++G RGLAY+HD +H +L P ++ L
Sbjct: 560 LKIVKGLARGLAYLHDKKHVHGNLKPSNILL 590
>At2g15300 putative receptor-like protein kinase
Length = 744
Score = 33.5 bits (75), Expect = 0.22
Identities = 25/104 (24%), Positives = 49/104 (47%), Gaps = 9/104 (8%)
Query: 184 KSTAADYAKVASEKVSKLSSWNSFERGQSMKRRRRFIIKLLQGALRGLAYMHDHDRLHQS 243
K +DY + +S +S+ +S + + R +KL +G RG+AY+HD +H +
Sbjct: 543 KLLISDYVPNGNLPLSSISAKSSSFSHKPLSFEAR--LKLARGIARGIAYIHDKKHVHGN 600
Query: 244 LGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELEDSGPLT 287
+ ++ L++ E P + D+ + + L GPL+
Sbjct: 601 IKANNILLDSEFE-------PVITDMGLDRIMTSAHLLTDGPLS 637
>At5g01920 unknown protein
Length = 495
Score = 33.1 bits (74), Expect = 0.29
Identities = 75/319 (23%), Positives = 116/319 (35%), Gaps = 46/319 (14%)
Query: 114 GPLKQTPVLFKVYPGTRAGGVVADMMAANELNSHMFLQSSSKGISQHLMLLLGGFET--- 170
G KQ +L KV G R + F S+ LG F
Sbjct: 177 GEFKQRVILKKVKVGVRGAEEFGEY-------EEWFNYRLSRAAPDTCAEFLGSFVADKT 229
Query: 171 ----TTGEQWLAFRDYGKSTAADYAKVAS--EKVSKLSSWNSFERGQSMKRRRRFIIKLL 224
T G +WL +R G ADY K S + + + +S+KRR I +++
Sbjct: 230 NTMFTKGGKWLVWRFEGDRDLADYMKDRSFPSNLESIMFGRVLQGVESVKRRALIIKQIM 289
Query: 225 QGALRGLAYMHDHDRLHQSLGPFSV------SLNTISESEAPYL------IPR--LRDLA 270
+ + L +H +H+ + P ++ + I A L IP L D
Sbjct: 290 RQIITSLRKIHGTGIVHRDVKPANLVVTKKGQIKLIDFGAAADLRIGKNYIPERTLLDPD 349
Query: 271 FSVSVRYSELEDS-GPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLLFAYLAFVPF 329
+ Y E++ P E + AA L + D+Y AG++ +A VP
Sbjct: 350 YCPPELYVLPEETPSPPPEPI-----AALLSPILWQLNSPDLFDMYSAGIVLLQMA-VPT 403
Query: 330 CEAGVMDGLSLQRLLENTFRLDLEATREYCIADDKLVNAIEFLDLGDGAGWELLQAMLN- 388
+ GL L + DL RE L LDL G GW+L+ +++
Sbjct: 404 LRSTA--GLKNFNLEIKSVEYDLNRWRERTRTRPDL----SILDLDSGRGWDLVTKLISE 457
Query: 389 --ADFRKRPTAEAVLSHRF 405
+ R R +A A L H +
Sbjct: 458 RGSLRRGRLSAAAALRHPY 476
>At4g04510 putative receptor-like protein kinase
Length = 648
Score = 32.7 bits (73), Expect = 0.37
Identities = 22/102 (21%), Positives = 47/102 (45%), Gaps = 14/102 (13%)
Query: 222 KLLQGALRGLAYMHDHDRL---HQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYS 278
++++G RGL Y+H+ +L H+ L ++ L+ Y+ P++ D + R
Sbjct: 440 RIIEGVARGLVYLHEDSQLRIIHRDLKASNILLDA-------YMNPKVADFGMA---RLF 489
Query: 279 ELEDSGPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLL 320
++ + +T + A Y+ R F + D+Y G++
Sbjct: 490 NMDQTRAVTRKV-VGTFGYMAPEYVRNRTFSVKTDVYSFGVV 530
>At1g61500
Length = 804
Score = 32.0 bits (71), Expect = 0.64
Identities = 26/98 (26%), Positives = 41/98 (41%), Gaps = 8/98 (8%)
Query: 223 LLQGALRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELED 282
++QG RGL Y+H RL VS + E + P++ D + + +E +D
Sbjct: 593 IIQGIARGLLYLHHDSRLRVIHRDLKVSNILLDEK----MNPKISDFGLARMYQGTEYQD 648
Query: 283 SGPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLL 320
+ G S A+T + F DIY G+L
Sbjct: 649 NTRRVVGTLGYMSPEYAWTGM----FSEKSDIYSFGVL 682
>At1g61390 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 831
Score = 32.0 bits (71), Expect = 0.64
Identities = 27/105 (25%), Positives = 45/105 (42%), Gaps = 10/105 (9%)
Query: 216 RRRFIIKLLQGALRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSV 275
++RF ++QG RGL Y+H RL VS + E +IP++ D +
Sbjct: 617 QKRF--NIIQGVARGLLYLHRDSRLRVIHRDLKVSNILLDEK----MIPKISDFGLARMS 670
Query: 276 RYSELEDSGPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLL 320
+ ++ +D+ G + A+T F DIY G+L
Sbjct: 671 QGTQYQDNTRRVVGTLGYMAPEYAWT----GVFSEKSDIYSFGVL 711
>At4g37250 receptor kinase like protein
Length = 768
Score = 31.6 bits (70), Expect = 0.83
Identities = 12/31 (38%), Positives = 21/31 (67%)
Query: 221 IKLLQGALRGLAYMHDHDRLHQSLGPFSVSL 251
+K+ +G RGLAY+H+ +H +L P ++ L
Sbjct: 555 LKIAKGIARGLAYLHEKKHVHGNLKPSNILL 585
>At4g11480 serine/threonine kinase-like protein
Length = 656
Score = 31.6 bits (70), Expect = 0.83
Identities = 38/166 (22%), Positives = 67/166 (39%), Gaps = 26/166 (15%)
Query: 216 RRRFIIKLLQGALRGLAYMHDHDRL---HQSLGPFSVSLNTISESEAPYLIPRLRDLAFS 272
+RR+ ++ G RGL Y+H RL H+ + ++ L+ + P++ D +
Sbjct: 426 KRRY--NIIGGITRGLLYLHQDSRLTIIHRDIKASNILLDA-------DMNPKIADFGMA 476
Query: 273 VSVRYSELEDSGPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLLFAYL-------A 325
+ R + ED+ G + Y+ F D+Y G+L + +
Sbjct: 477 RNFRVDQTEDNTRRVVGTFGYMPP----EYVTHGQFSTKSDVYSFGVLILEIVCGKKNSS 532
Query: 326 FVPFCEAGVMDGLSLQRLLENTFRLDL--EATREYCIADDKLVNAI 369
F ++G + RL N LDL A E C +DK++ I
Sbjct: 533 FYKIDDSGGNLVTHVWRLWNNDSPLDLIDPAIEESC-DNDKVIRCI 577
>At4g11460 serine/threonine kinase-like protein
Length = 700
Score = 31.6 bits (70), Expect = 0.83
Identities = 48/205 (23%), Positives = 81/205 (39%), Gaps = 31/205 (15%)
Query: 124 KVYPGTRAGGVVADMMAANELNSHMFLQSSSKGIS----QHLMLL-LGGFETTTGEQWLA 178
+VY GT + G + + + L+ ++ + QH L+ L GF E+ L
Sbjct: 359 EVYKGTLSNGTEVAVKRLSRTSDQGELEFKNEVLLVAKLQHRNLVRLLGFALQGEEKILV 418
Query: 179 FRDYGKSTAADYAKVASEKVSKLSSWNSFERGQSMKRRRRFIIKLLQGALRGLAYMHDHD 238
F ++ + + DY S +K +GQ RR II G RGL Y+H
Sbjct: 419 F-EFVPNKSLDYFLFGSTNPTK--------KGQLDWTRRYNIIG---GITRGLLYLHQDS 466
Query: 239 RL---HQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELEDSGPLTEGLWARAS 295
RL H+ + ++ L+ + P++ D + + R + EDS G +
Sbjct: 467 RLTIIHRDIKASNILLDA-------DMNPKIADFGMARNFRDHQTEDSTGRVVGTFGYMP 519
Query: 296 AASAFTYLEKRAFGIADDIYEAGLL 320
Y+ F D+Y G+L
Sbjct: 520 P----EYVAHGQFSTKSDVYSFGVL 540
>At1g20930 putative cell division control protein, cdc2 kinase
Length = 315
Score = 31.6 bits (70), Expect = 0.83
Identities = 53/196 (27%), Positives = 83/196 (42%), Gaps = 30/196 (15%)
Query: 223 LLQGALRGLAYMHDHDRLHQSLGPFSVSLN--TISESEAPYLIPRLRDLAFSVSVRYSEL 280
L+ +G+A+ H H LH+ L P ++ ++ T++ A + R AF++ ++
Sbjct: 127 LMYQLCKGMAFCHGHGVLHRDLKPHNLLMDRKTMTLKIADLGLAR----AFTLPMKKYTH 182
Query: 281 EDSGPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLLFAYLAFVPFCEAGVMDGLSL 340
E LT LW RA L + D++ G +FA L AG + L
Sbjct: 183 E---ILT--LWYRAPE----VLLGATHYSTGVDMWSVGCIFAELVTKQAIFAGDSE---L 230
Query: 341 QRLLENTFRLDLEATREYCIADDKLVNAIEF-----LDLG------DGAGWELLQAMLNA 389
Q+LL FRL E KL + E+ L L D AG +LL ML
Sbjct: 231 QQLL-RIFRLLGTPNEEVWPGVSKLKDWHEYPQWKPLSLSTAVPNLDEAGLDLLSKMLEY 289
Query: 390 DFRKRPTAEAVLSHRF 405
+ KR +A+ + H +
Sbjct: 290 EPAKRISAKKAMEHPY 305
>At5g24360 unknown protein
Length = 881
Score = 31.2 bits (69), Expect = 1.1
Identities = 54/229 (23%), Positives = 92/229 (39%), Gaps = 41/229 (17%)
Query: 185 STAADYAKVASEKVSKLSSWNSFERGQSMKRRRR------FIIKLLQGALRGLAYMHDHD 238
S+A + +AS + + FE G+ ++ + ++KL++ + GL ++HD
Sbjct: 544 SSALLESPMASSSIHSIQINPIFENGKGVELWKENGHPSPVLLKLMRDIVAGLVHLHDIG 603
Query: 239 RLHQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELEDSGPLTE---GLWARAS 295
+H+ L P +V + S L +L D+ S + D+ LT GL + +S
Sbjct: 604 IVHRDLKPQNVLIVKNSS-----LCAKLSDMGISKRLP----ADTSALTRNSTGLGSGSS 654
Query: 296 AASAFTYLEKRAFGIADDIYEAG-LLFAYLAFVPFCEAGVMDGLSLQRLLENTFRLDLEA 354
A L A D++ G +LF FC G + + + D+
Sbjct: 655 GWQAPEQLRNERQTRAVDLFSLGCVLF-------FCMTGG------KHPYGDNYERDVNV 701
Query: 355 TREYCIADDKLVNAIEFLDLGDGAGWELLQAMLNADFRKRPTAEAVLSH 403
+ D K + IE L LL +LN D RP A+ V+ H
Sbjct: 702 -----LNDQKDLFLIESLP----EAVHLLTGLLNPDPNLRPRAQDVMHH 741
>At4g11330 MAP kinase (ATMPK5)
Length = 373
Score = 31.2 bits (69), Expect = 1.1
Identities = 26/95 (27%), Positives = 40/95 (41%), Gaps = 13/95 (13%)
Query: 228 LRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELEDSGPLT 287
LRGL Y+H + LH+ L P ++ LN+ + L+ F ++ SE E
Sbjct: 151 LRGLKYIHSANVLHRDLKPSNLLLNSNCD---------LKITDFGLARTTSETEYMTEYV 201
Query: 288 EGLWARASAASAFTYLEKRAFGIADDIYEAGLLFA 322
W RA L + A D++ G +FA
Sbjct: 202 VTRWYRAPE----LLLNSSEYTSAIDVWSVGCIFA 232
>At2g46070 mitogen-activated protein kinase like protein
Length = 372
Score = 31.2 bits (69), Expect = 1.1
Identities = 17/39 (43%), Positives = 25/39 (63%), Gaps = 3/39 (7%)
Query: 218 RFIIKLLQGALRGLAYMHDHDRLHQSLGPFSVSLNTISE 256
RF++ L LRGL Y+H + LH+ L P +V LN+ +E
Sbjct: 145 RFLVYQL---LRGLKYVHSANILHRDLRPSNVLLNSKNE 180
>At2g30940 putative protein kinase
Length = 445
Score = 31.2 bits (69), Expect = 1.1
Identities = 22/78 (28%), Positives = 34/78 (43%), Gaps = 14/78 (17%)
Query: 201 LSSWNSFERGQSMKRRRRFIIKLLQGALRGLAYMHDHDR---LHQSLGPFSVSLN----- 252
L W G++ R +K++QG +GLAY+H+ HQ + P + L+
Sbjct: 248 LHEWLHGSAGRNRPLTWRKRMKIIQGVAKGLAYIHEDIEPKITHQDIRPSKILLDYQWNP 307
Query: 253 ------TISESEAPYLIP 264
I S+ P LIP
Sbjct: 308 KILDVGFIGHSDIPTLIP 325
>At1g61440 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 792
Score = 31.2 bits (69), Expect = 1.1
Identities = 26/98 (26%), Positives = 40/98 (40%), Gaps = 8/98 (8%)
Query: 223 LLQGALRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDLAFSVSVRYSELED 282
++QG +RGL Y+H RL VS + E + P++ D + + S+ +D
Sbjct: 580 IIQGIVRGLLYLHRDSRLRVIHRDLKVSNILLDEK----MNPKISDFGLARLFQGSQYQD 635
Query: 283 SGPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLL 320
G S A+T F DIY G+L
Sbjct: 636 KTRRVVGTLGYMSPEYAWT----GVFSEKSDIYSFGVL 669
>At5g41730 putative protein
Length = 711
Score = 30.8 bits (68), Expect = 1.4
Identities = 20/82 (24%), Positives = 34/82 (41%), Gaps = 7/82 (8%)
Query: 216 RRRF------IIKLLQGALRGLAYMHDHDRLHQSLGPFSVSLNTISESEAPYLIPRLRDL 269
RRR+ +I ++ RG+ Y+H +D H L P ++ L S +E Y ++
Sbjct: 326 RRRYLFSIPVVIDIMLQIARGMEYLHGNDIFHGDLNPMNIHLKERSHTEG-YFHAKICGF 384
Query: 270 AFSVSVRYSELEDSGPLTEGLW 291
S V+ G +W
Sbjct: 385 GLSSVVKAQSSSKPGTPDPVIW 406
>At4g21380 receptor-like serine/threonine protein kinase ARK3
Length = 850
Score = 30.8 bits (68), Expect = 1.4
Identities = 24/117 (20%), Positives = 47/117 (39%), Gaps = 14/117 (11%)
Query: 207 FERGQSMKRRRRFIIKLLQGALRGLAYMHDHDR---LHQSLGPFSVSLNTISESEAPYLI 263
F++ ++ K + ++ G RGL Y+H R +H+ L ++ L+ Y+
Sbjct: 612 FDKSRNSKLNWQMRFDIINGIARGLLYLHQDSRFRIIHRDLKASNILLD-------KYMT 664
Query: 264 PRLRDLAFSVSVRYSELEDSGPLTEGLWARASAASAFTYLEKRAFGIADDIYEAGLL 320
P++ D + E E + G + S Y F + D++ G+L
Sbjct: 665 PKISDFGMARIFGRDETEANTRKVVGTYGYMSP----EYAMDGIFSMKSDVFSFGVL 717
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.135 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,639,647
Number of Sequences: 26719
Number of extensions: 345991
Number of successful extensions: 1197
Number of sequences better than 10.0: 54
Number of HSP's better than 10.0 without gapping: 16
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 1167
Number of HSP's gapped (non-prelim): 61
length of query: 411
length of database: 11,318,596
effective HSP length: 102
effective length of query: 309
effective length of database: 8,593,258
effective search space: 2655316722
effective search space used: 2655316722
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC145222.13


