

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145109.3 - phase: 0
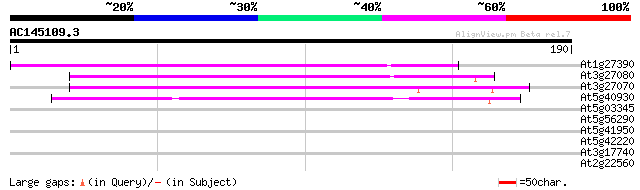
(190 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g27390 putative protein import receptor 107 5e-24
At3g27080 TOM20, putative 106 6e-24
At3g27070 TOM20, putative 100 4e-22
At5g40930 protein import receptor TOM20, mitochondrial-like 80 5e-16
At5g03345 unknown protein 29 1.7
At5g56290 peroxisomal targeting signal type 1 receptor 28 2.2
At5g41950 unknown protein 28 2.2
At5g42220 unknown protein 28 3.8
At3g17740 unknown protein 28 3.8
At2g22560 hypothetical protein 27 8.5
>At1g27390 putative protein import receptor
Length = 208
Score = 107 bits (266), Expect = 5e-24
Identities = 58/152 (38%), Positives = 84/152 (55%), Gaps = 1/152 (0%)
Query: 1 MDYPLRKFKFHLPMKHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQV 60
M++ F+ + +H +E +Y +P D+ENL +WG ALL LSQ P++ +
Sbjct: 1 MEFSTADFERLIMFEHARKNSEAQYKNDPLDSENLLKWGGALLELSQFQPIPEAKLMLND 60
Query: 61 SISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
+ISKLEEA ++NP W + A T A D +AK HFD A YF+RA +DP +
Sbjct: 61 AISKLEEALTINPGKHQALWCIANAYTAHAFYVHDPEEAKEHFDKATEYFQRAENEDPGN 120
Query: 121 PTYQISLELPDTKDHEQHPKIVNHGLGQQSKG 152
TY+ SL+ K E H + +N G+GQQ G
Sbjct: 121 DTYRKSLD-SSLKAPELHMQFMNQGMGQQILG 151
>At3g27080 TOM20, putative
Length = 202
Score = 106 bits (265), Expect = 6e-24
Identities = 61/149 (40%), Positives = 82/149 (54%), Gaps = 6/149 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE Y NP DA+NLTRWG LL LSQ HS D+ IQ +I+K EEA ++P + W
Sbjct: 20 AENTYKSNPLDADNLTRWGGVLLELSQFHSISDAKQMIQEAITKFEEALLIDPKKDEAVW 79
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQHPK 140
+G A T A LTPD +AK +FD A +F++A + P + Y SLE+ K + H +
Sbjct: 80 CIGNAYTSFAFLTPDETEAKHNFDLATQFFQQAVDEQPDNTHYLKSLEM-TAKAPQLHAE 138
Query: 141 IVNHGLGQQSKGSSSA-----TKVIKHYK 164
GLG Q G A +K +K+ K
Sbjct: 139 AYKQGLGSQPMGRVEAPAPPSSKAVKNKK 167
>At3g27070 TOM20, putative
Length = 188
Score = 100 bits (250), Expect = 4e-22
Identities = 65/164 (39%), Positives = 87/164 (52%), Gaps = 8/164 (4%)
Query: 21 AEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHW 80
AE+ Y NP DA+NL RWG ALL LSQ + DSL IQ +ISKLE+A ++P D W
Sbjct: 14 AEETYKLNPEDADNLMRWGEALLELSQFQNVIDSLKMIQDAISKLEDAILIDPMKHDAVW 73
Query: 81 LLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKDHEQ--- 137
LG A T A LTPD A+L+F A ++F A Q P + Y SLE+ D
Sbjct: 74 CLGNAYTSYARLTPDDTQARLNFGLAYLFFGIAVAQQPDNQVYHKSLEMADKAPQLHTGF 133
Query: 138 HPKIVNHGLGQQSKGSSSATKVIKH-----YKFLVFLFTLLVIG 176
H + LG + + KV+K+ K++V + +L IG
Sbjct: 134 HKNRLLSLLGGVETLAIPSPKVVKNKKSSDEKYIVMGWVILAIG 177
>At5g40930 protein import receptor TOM20, mitochondrial-like
Length = 187
Score = 80.5 bits (197), Expect = 5e-16
Identities = 56/161 (34%), Positives = 80/161 (48%), Gaps = 9/161 (5%)
Query: 15 KHECILAEQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPN 74
+H +AE Y KNP DAENLTRW ALL LSQ + P + I +I KL EA ++P
Sbjct: 14 EHARKVAEATYVKNPLDAENLTRWAGALLELSQFQTEPKQM--ILEAILKLGEALVIDPK 71
Query: 75 NPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSDPTYQISLELPDTKD 134
D WL+G A L+ D +A +F+ A +F+ A + P Y+ SL L
Sbjct: 72 KHDALWLIGNAHLSFGFLSSDQTEASDNFEKASQFFQLAVEEQPESELYRKSLTLA---- 127
Query: 135 HEQHPKIVNHGLGQQSKGSSSATKVIK--HYKFLVFLFTLL 173
+ P++ G S S+ K K +K+ VF + +L
Sbjct: 128 -SKAPELHTGGTAGPSSNSAKTMKQKKTSEFKYDVFGWVIL 167
>At5g03345 unknown protein
Length = 104
Score = 28.9 bits (63), Expect = 1.7
Identities = 20/56 (35%), Positives = 28/56 (49%), Gaps = 2/56 (3%)
Query: 65 LEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKRAFRQDPSD 120
+EE FS P N + ++G+AL M A LT +H DS + + F D SD
Sbjct: 32 MEEEFSRPPINVILELIIGLALCMWAALTFPGKFLSIHPDSDE--NRAVFLPDNSD 85
>At5g56290 peroxisomal targeting signal type 1 receptor
Length = 728
Score = 28.5 bits (62), Expect = 2.2
Identities = 20/68 (29%), Positives = 31/68 (45%), Gaps = 12/68 (17%)
Query: 53 DSLHTIQVSISKLEEAFSLNPNNPDVHWLLGMALTMQALLTPDSHDAKLHFDSADVYFKR 112
DSL+ ++ EA LNP + DVH +LG+ ++ FD A F+
Sbjct: 568 DSLYHADIA-RLFNEASQLNPEDADVHIVLGVL-----------YNLSREFDRAITSFQT 615
Query: 113 AFRQDPSD 120
A + P+D
Sbjct: 616 ALQLKPND 623
>At5g41950 unknown protein
Length = 565
Score = 28.5 bits (62), Expect = 2.2
Identities = 31/117 (26%), Positives = 46/117 (38%), Gaps = 10/117 (8%)
Query: 22 EQRYNKNPHDAENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEEAFSLNPNNPDVHWL 81
E+ N + ++ L WG+ L LSQ + ++ +ISK A L + +
Sbjct: 306 EKAVQLNWNSSQALNNWGLVLQELSQIVPAREKEKVVRTAISKFRAAIRLQFDFHRAIYN 365
Query: 82 LG---MALTMQALLTPDSHDAK------LHFDSADVYFKRAFRQDPSDPTYQISLEL 129
LG L L T S + K L+ SA +Y A PS Y +L L
Sbjct: 366 LGTVLYGLAEDTLRTGGSGNGKDMPPGELYSQSA-IYIAAAHSLKPSYSVYSSALRL 421
>At5g42220 unknown protein
Length = 879
Score = 27.7 bits (60), Expect = 3.8
Identities = 12/26 (46%), Positives = 17/26 (65%)
Query: 52 PDSLHTIQVSISKLEEAFSLNPNNPD 77
PDSL T+ I+++E+A S N PD
Sbjct: 251 PDSLDTLMEFINRMEQALSQNGYQPD 276
>At3g17740 unknown protein
Length = 1149
Score = 27.7 bits (60), Expect = 3.8
Identities = 17/53 (32%), Positives = 29/53 (54%), Gaps = 1/53 (1%)
Query: 27 KNPHDAENLTRWGVALLRLSQAHSFPD-SLHTIQVSISKLEEAFSLNPNNPDV 78
+ PHDA+ + ++S S L T++ IS LE+AF LNP++ ++
Sbjct: 287 ERPHDAKAWLAFADFQDKVSSMQSQKGVRLQTLEKKISILEKAFELNPDSEEL 339
>At2g22560 hypothetical protein
Length = 891
Score = 26.6 bits (57), Expect = 8.5
Identities = 16/50 (32%), Positives = 24/50 (48%), Gaps = 2/50 (4%)
Query: 20 LAEQRYNKNPHDA--ENLTRWGVALLRLSQAHSFPDSLHTIQVSISKLEE 67
L E+++ N + ENL W Q S+ S+ +Q ISKLE+
Sbjct: 660 LVEEQFRLNIDELLEENLDFWLRFSTAFGQIQSYDTSIEDLQAEISKLEQ 709
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.417
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,418,234
Number of Sequences: 26719
Number of extensions: 169422
Number of successful extensions: 385
Number of sequences better than 10.0: 10
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 376
Number of HSP's gapped (non-prelim): 11
length of query: 190
length of database: 11,318,596
effective HSP length: 94
effective length of query: 96
effective length of database: 8,807,010
effective search space: 845472960
effective search space used: 845472960
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 57 (26.6 bits)
Medicago: description of AC145109.3


