

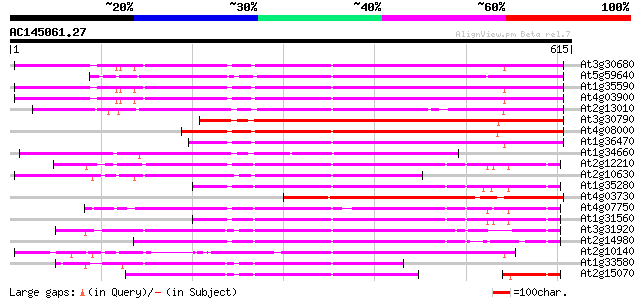
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145061.27 - phase: 0 /pseudo
(615 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g30680 hypothetical protein 449 e-126
At5g59640 serine/threonine-specific protein kinase - like 441 e-124
At1g35590 CACTA-element, putative 439 e-123
At4g03900 putative transposon protein 436 e-122
At2g13010 pseudogene 411 e-115
At3g30790 hypothetical protein 379 e-105
At4g08000 putative transposon protein 369 e-102
At1g36470 hypothetical protein 362 e-100
At1g34660 hypothetical protein 360 e-99
At2g12210 putative TNP2-like transposon protein 301 7e-82
At2g10630 En/Spm transposon hypothetical protein 1 301 9e-82
At1g35280 hypothetical protein 281 8e-76
At4g03730 putative transposon protein 280 2e-75
At4g07750 putative transposon protein 265 4e-71
At1g31560 putative protein 265 4e-71
At3g31920 hypothetical protein 254 1e-67
At2g14980 pseudogene 250 1e-66
At2g10140 putative protein 248 9e-66
At1g33580 En/Spm-like transposon protein, putative 176 3e-44
At2g15070 En/Spm-like transposon protein 171 1e-42
>At3g30680 hypothetical protein
Length = 1116
Score = 449 bits (1154), Expect = e-126
Identities = 248/634 (39%), Positives = 359/634 (56%), Gaps = 50/634 (7%)
Query: 6 FHRSWMYDRKYPGKRKLKAAFVDGVRDFVAYAMAQDAFQ-LEGGIRCSCVKCTC-RLIRS 63
F+R WMY R L A +V GV +F+ +A +Q Q G C C C + I S
Sbjct: 13 FYRDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHIIS 72
Query: 64 PKDVLNHLKDLGFMENYYVWIYHGEQEPTNNTEFDVNMHASSSEARMECENF----GVME 119
+ V +HL GFM +YYVW HGE E ++++ S ++ EN V+E
Sbjct: 73 GRRVSSHLFSQGFMPDYYVWYKHGE-------ELNMDIGTSYTDRTYFSENHEEVGNVVE 125
Query: 120 D----MVGDAVGVNLSYNEG--------------GEEETIPNEKALKFYKMMQEVNKPLF 161
D MV DA N+ Y++ EE + N KFY +++ N PL+
Sbjct: 126 DPYVDMVNDAFNFNVGYDDNVGHDDCYHHDGSYQNVEEPVRNHSN-KFYDLLEGANNPLY 184
Query: 162 EGSSD--SKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEK 219
+G + S+LS++ RL+ ++++++E+ + ++ D P + S Y+ +KL+
Sbjct: 185 DGCREGQSQLSLASRLMHNKAEYNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLMRN 244
Query: 220 LGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSM 279
LGL T D C N CMLF+ K D C+FC A R++ DD +++ +V M
Sbjct: 245 LGLPYHTIDVCKNNCMLFW-----KEDEKEDQCRFCGAQRWKPKDD---RRRTKVPYSRM 296
Query: 280 FYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHN 339
+YLPI +L+R++ S TA M WH ++ G M HP D W++F ++H FAE+P N
Sbjct: 297 WYLPIADQLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRN 356
Query: 340 VRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGI 399
V LGLC+DGF P+ + + +S WP++LTPYN+P MCM+ YLFL+ + GP +
Sbjct: 357 VYLGLCTDGFNPF--SMSRNHSLWPVILTPYNLPPRMCMNTEYLFLTILNSGPNHPRASL 414
Query: 400 DVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTINDFPAYGMLS*WSTHGRLAC 458
DV++QPLI++L LW GV YD++ + F ++A L+WTI+DFPAY MLS W+THG+L+C
Sbjct: 415 DVFLQPLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTTHGKLSC 474
Query: 459 PHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLFTKGK-TEKDGPPPRTTSEE 517
P CME TK+ + +GRK FDCHR FLP H R+ K F KG+ + PP T E+
Sbjct: 475 PVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEYPPESLTGEQ 534
Query: 518 V-YRRVNELGLLKFTDVGKRVRHPK---YGNEHHWTKRSIFWDLPYWKDNLLRHNFDVMH 573
V Y R+ + K DVG K YG EH+W K SI W+L YWKD LRHN DVMH
Sbjct: 535 VYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMH 594
Query: 574 IEKNFCDNIIHIVMNVSAKTKDNEKARMDLPLHC 607
EKNF DNI++ ++ V K+KDN +R+D+ +C
Sbjct: 595 TEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYC 628
>At5g59640 serine/threonine-specific protein kinase - like
Length = 986
Score = 441 bits (1135), Expect = e-124
Identities = 235/524 (44%), Positives = 318/524 (59%), Gaps = 21/524 (4%)
Query: 88 EQEPTNNTEFDVNMHASSSEARMECENFGVMEDMVGDAVGVNLSYNEGGEEETIPNEKAL 147
E++ NT + H S+++ E + V +MV DA G S + EE PN +A
Sbjct: 2 EEDNKKNTSWQ---HICSTKSHTEPVDPYV--EMVSDAFGSTESEFDQMREED-PNFEAK 55
Query: 148 KFYKMMQEVNKPLFEGSSD--SKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTPVKDN 205
KFY ++ +P+++G + SKLS++ RL++ +D ++++ + IM++ P +N
Sbjct: 56 KFYDILDAAKQPIYDGCKEGLSKLSLAARLMSLKTDNNLSQNCMDSIAQIMQEYLPEGNN 115
Query: 206 LPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDD 265
P S+YE +KL+ LGL + D C + CM+F+ E K + C FC RY
Sbjct: 116 SPKSYYEIKKLMRSLGLPYQKIDVCQDNCMIFW-KETEKEE----YCLFCKKDRYRPTQK 170
Query: 266 AGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKH 325
G +K + + MFYLPI RL+RL+ S +TA M WH + + G M HP DG AWKH
Sbjct: 171 IG---QKSIPYRQMFYLPIADRLKRLYQSHNTAKHMRWHAEHLASDGEMGHPSDGEAWKH 227
Query: 326 FDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFL 385
F +VH DFA P NV LGLC+DGF P+ S YS WP++LTPYN+P EMCM + ++FL
Sbjct: 228 FHKVHPDFASKPRNVYLGLCTDGFNPF-GMSGHNYSLWPVILTPYNLPPEMCMKQEFMFL 286
Query: 386 SCIVPGPTSSLDGIDVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTINDFPAY 444
+ +VPGP +D+++QPLI++L LW NGV YDI+ K+ F ++ LMWTI+DFPAY
Sbjct: 287 TILVPGPNHPKRSLDIFLQPLIEELKDLWVNGVEAYDISTKQNFLLKVVLMWTISDFPAY 346
Query: 445 GMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLFTKGKT 504
GMLS W+THGRLACP+C +QT A W +GRK FDCHR FLP H +R K F KGK
Sbjct: 347 GMLSGWTTHGRLACPYCSDQTGAFWLKNGRKTCWFDCHRCFLPVNHSYRGNKKDFKKGKV 406
Query: 505 EKDGPPPRTTSEEVYRRVNELGLLKFTDV-GKRVRHPKYGNEHHWTKRSIFWDLPYWKDN 563
+D P T EE+Y V L K D G R YG H+W K+SI W+L YWKD
Sbjct: 407 VEDSKPEILTGEELYNEV--CCLPKTVDCGGNHGRLEGYGKTHNWHKQSILWELSYWKDL 464
Query: 564 LLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNEKARMDLPLHC 607
LRHN DVMHIEKN DN I ++NV KTKDN K+R+DL +C
Sbjct: 465 KLRHNLDVMHIEKNVLDNFIKTLLNVQGKTKDNIKSRLDLQENC 508
>At1g35590 CACTA-element, putative
Length = 1180
Score = 439 bits (1130), Expect = e-123
Identities = 246/634 (38%), Positives = 355/634 (55%), Gaps = 50/634 (7%)
Query: 6 FHRSWMYDRKYPGKRKLKAAFVDGVRDFVAYAMAQDAFQ-LEGGIRCSCVKCTC-RLIRS 63
F+R WMY R L A +V GV +F+ +A +Q Q G C C C + I S
Sbjct: 13 FYRDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFHCPCSVCKNEKHIIS 72
Query: 64 PKDVLNHLKDLGFMENYYVWIYHGEQEPTNNTEFDVNMHASSSEARMECENF----GVME 119
+ V +HL GFM +YYVW HGE E ++++ S ++ EN V+E
Sbjct: 73 GRRVSSHLFSQGFMPDYYVWYKHGE-------ELNMDIGTSYTDRTYFSENHEEVGNVVE 125
Query: 120 D----MVGDAVGVNLSYNEG--------------GEEETIPNEKALKFYKMMQEVNKPLF 161
D MV DA N+ Y++ EE + N KFY +++ N L+
Sbjct: 126 DPYVDMVNDAFNFNVGYDDNVGHDDCYHHDGSYQNVEEPVRNHSN-KFYDLLEGANNLLY 184
Query: 162 EGSSD--SKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEK 219
+G + S+LS++ RL+ ++++++E+ + ++ D P + S Y+ +KL+
Sbjct: 185 DGCREGQSQLSLASRLMHNKAEYNMSEKLVDSVCEMFTDFLPEGNQATTSHYQTEKLMHN 244
Query: 220 LGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSM 279
LGL T D C N CMLF+ K D C+FC A R++ DD +++ +V M
Sbjct: 245 LGLPYHTIDVCKNNCMLFW-----KEDEKEDQCRFCGAQRWKPKDD---RRRTKVPYSRM 296
Query: 280 FYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHN 339
+YLPI RL+R++ TA M WH ++ G M HP D W++F ++H FAE+P N
Sbjct: 297 WYLPIADRLKRMYQCHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPRN 356
Query: 340 VRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGI 399
V LGLC+DGF P+ + +S WP++LTPYN+P MCM+ YLFL+ + GP +
Sbjct: 357 VYLGLCTDGFNPF--GMSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNYGPNHPRASL 414
Query: 400 DVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTINDFPAYGMLS*WSTHGRLAC 458
DV++QPLI++L LW GV Y+++ + F ++A L+WTI+DFPAY MLS W+THG+L+C
Sbjct: 415 DVFLQPLIEELKELWCTGVDAYEVSLSQNFNLKAVLLWTISDFPAYNMLSGWTTHGKLSC 474
Query: 459 PHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLFTKGK-TEKDGPPPRTTSEE 517
P CME TK+ + GRK FDCHR FLP H R+ K F KG+ + PP T E+
Sbjct: 475 PVCMESTKSFYLPIGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEYPPESLTGEQ 534
Query: 518 V-YRRVNELGLLKFTDVGKRVRHPK---YGNEHHWTKRSIFWDLPYWKDNLLRHNFDVMH 573
V Y R+ + K DVG K YG EH+W K SI W+L YWKD LRHN DVMH
Sbjct: 535 VYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMH 594
Query: 574 IEKNFCDNIIHIVMNVSAKTKDNEKARMDLPLHC 607
EKNF DNI++ ++ V K+KDN +R+D+ +C
Sbjct: 595 TEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYC 628
>At4g03900 putative transposon protein
Length = 817
Score = 436 bits (1122), Expect = e-122
Identities = 246/634 (38%), Positives = 355/634 (55%), Gaps = 50/634 (7%)
Query: 6 FHRSWMYDRKYPGKRKLKAAFVDGVRDFVAYAMAQDAFQ-LEGGIRCSCVKCTC-RLIRS 63
F+R WMY R L A +V GV +F+ +A +Q Q G C C C + I S
Sbjct: 13 FYRDWMYKRFDEVTGNLSAEYVAGVEEFMTFANSQPIVQSCRGKFYCPCSVCKNEKHIIS 72
Query: 64 PKDVLNHLKDLGFMENYYVWIYHGEQEPTNNTEFDVNMHASSSEARMECENF----GVME 119
+ V +HL FM +YYVW HGE E ++++ S ++ EN V+E
Sbjct: 73 GRRVSSHLFSHEFMPDYYVWYKHGE-------ELNMDIGTSYTDRTYFSENHEEVGNVVE 125
Query: 120 D----MVGDAVGVNLSYNEG--------------GEEETIPNEKALKFYKMMQEVNKPLF 161
D MV DA N+ Y++ EE + N KFY +++ N PL+
Sbjct: 126 DPYVDMVNDAFNFNVGYDDNVGHDDNYHHDGSYQNVEEPVRNHSN-KFYDLLEGANNPLY 184
Query: 162 EGSSD--SKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEK 219
+G + S+LS++ RL+ ++++++E+ + ++ P + S Y+ +KL+
Sbjct: 185 DGCREGQSQLSLASRLMHNKAEYNMSEKLVDSVCEMFTAFLPEGNQATTSHYQTEKLMRN 244
Query: 220 LGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSM 279
LGL T D N CMLF+ K D C+FC A R++ DD +++ +V M
Sbjct: 245 LGLPYHTIDVYKNNCMLFW-----KEDEKEDQCRFCGAQRWKPKDD---RRRTKVPYSRM 296
Query: 280 FYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHN 339
+YLPI RL+R++ S TA M WH ++ G M HP D W++F ++H FAE+PHN
Sbjct: 297 WYLPIADRLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHPRFAEEPHN 356
Query: 340 VRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGI 399
V LGL +DGF P+ + +S WP++LTPYN+P MCM+ YLFL+ + GP +
Sbjct: 357 VYLGLYTDGFNPF--GMSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPNHPRASL 414
Query: 400 DVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTINDFPAYGMLS*WSTHGRLAC 458
DV++QPLI++L LW GV YD++ + F ++A L+WTI+DFPAY MLS W+THG+L+C
Sbjct: 415 DVFLQPLIEELKELWCTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTTHGKLSC 474
Query: 459 PHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLFTKGK-TEKDGPPPRTTSEE 517
P CME TK+ + +GRK FDCHR FLP H R+ K F KG+ + PP T E+
Sbjct: 475 PVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRNKKDFLKGRDASSEYPPESLTGEQ 534
Query: 518 V-YRRVNELGLLKFTDVGKRVRHPK---YGNEHHWTKRSIFWDLPYWKDNLLRHNFDVMH 573
V Y R+ + K DVG K YG EH+W K SI W+L YWKD LRHN DVMH
Sbjct: 535 VYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHKESILWELSYWKDLNLRHNIDVMH 594
Query: 574 IEKNFCDNIIHIVMNVSAKTKDNEKARMDLPLHC 607
EKNF DNI++ ++ V K+KDN +R+D+ +C
Sbjct: 595 TEKNFLDNIMNTLLGVKGKSKDNIMSRLDIEKYC 628
>At2g13010 pseudogene
Length = 1040
Score = 411 bits (1057), Expect = e-115
Identities = 236/606 (38%), Positives = 334/606 (54%), Gaps = 50/606 (8%)
Query: 26 FVDGVRDFVAYAMAQDAFQLEGG-IRCSCVKCTCRLIRSPKDVLNHLKDLGFMENYYVWI 84
+ D +R F+ Y + F E G I C C KC V HL + GFM NYYVW+
Sbjct: 188 YYDELRGFM-YQTTNEPFARENGTILCPCRKCLNEKYLEFNVVKKHLYNRGFMPNYYVWL 246
Query: 85 YHGEQEPTNNTEFDVNMHASSS-------EARMECENFGV-------MEDMVGDAVGVNL 130
HG+ + E + + SS+ + FG DMV +A +
Sbjct: 247 RHGDVVEIAH-ELVIQIMVSSNLVILIMMRIMQVPDQFGYGYNQENRFHDMVTNAFHETI 305
Query: 131 -SYNEGGEEETIPNEKALKFYKMMQEVNKPLFEG--SSDSKLSMSVRLLAAASDWSVAEE 187
S+ E EE PN A FY M+ N+P++EG +SKLS++ R++ +D +++E+
Sbjct: 306 ASFPENISEE--PNVDAQHFYDMLDAANQPIYEGCREGNSKLSLASRMMTIKADNNLSEK 363
Query: 188 GSECYTDIMRDTTPVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDG 247
+ + +++++ P + S+YE QKLV GL + D C++ CM+F+ ++
Sbjct: 364 CMDSWAELIKEYLPPDNISAESYYEIQKLVSSHGLPSEMIDVCIDYCMIFWGDDVN---- 419
Query: 248 ALVACKFCNAPRYEVCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYN 307
L C+FC PRY+ + RV K M+YLPI RL+RL+ S TA KM WH +
Sbjct: 420 -LQECRFCGKPRYQTTGG-----RTRVPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQH 473
Query: 308 KPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILL 367
G + HP D AWKHF V+ DFA + NV LGLC+DGF P+ YS WP++L
Sbjct: 474 TTADGEITHPSDAKAWKHFQTVYPDFANECRNVYLGLCTDGFSPF-GLCGRQYSLWPVIL 532
Query: 368 TPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLW-NGVSTYDIARKK 426
TPY +P +MCM ++FLS +VPGP +DV++QPLI +L +LW GV T+D + K+
Sbjct: 533 TPYKLPPDMCMQSEFMFLSILVPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQ 592
Query: 427 IFRMRAALMWTINDFPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFL 486
F MRA LMWTI+DFPAY MLS W+ HGRL P + K WF+ CHR FL
Sbjct: 593 NFNMRAMLMWTISDFPAYRMLSGWTMHGRLYVP--TKDRKPCWFN---------CHRRFL 641
Query: 487 PATHEFRKKKNLFTKGKTEKDGPPPRTTSEEVYRRVNELGLLKFTDVGKRVRHP-----K 541
P H +R+ K LF K K + PP + +++ +++ G + G +
Sbjct: 642 PLHHPYRRNKKLFRKNKIVRLLPPSYVSGTDLFEQIDYYGAQETCKRGGNWHTTANMPDE 701
Query: 542 YGNEHHWTKRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNEKARM 601
YG+ H+W K+SIFW LPYWKD LLRHN VMHIEKNF DNI++ ++N+ KTKD K+R+
Sbjct: 702 YGSTHNWYKQSIFWQLPYWKDLLLRHNLYVMHIEKNFFDNIMNTLLNLQGKTKDTLKSRL 761
Query: 602 DLPLHC 607
DL C
Sbjct: 762 DLAEIC 767
>At3g30790 hypothetical protein
Length = 953
Score = 379 bits (972), Expect = e-105
Identities = 191/405 (47%), Positives = 256/405 (63%), Gaps = 17/405 (4%)
Query: 209 SFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGS 268
S++E QKLV LGL + D C++ CM+F+ ++ L C+FC PRY+ +
Sbjct: 135 SYFEIQKLVLSLGLPSEMIDVCIDYCMIFWGDDVN-----LQECRFCGKPRYQT-----T 184
Query: 269 QKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQ 328
+ RV K M+YLPI RL+RL+ S TA KM WH + G ++HP D AWKHF
Sbjct: 185 SGRTRVPYKRMWYLPITDRLKRLYQSERTAAKMRWHAQHTTADGEIKHPSDTKAWKHFQT 244
Query: 329 VHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCI 388
V+ DFA + NV LGLC+DGF P++ S YS W I+LTPYN+P +M M +LFLS +
Sbjct: 245 VYPDFASECRNVYLGLCTDGFSPFV-LSGRQYSLWHIILTPYNLPPDMYMQSEFLFLSIL 303
Query: 389 VPGPTSSLDGIDVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTINDFPAYGML 447
VPGP +DV++QPLI +L +LW GV T+D + K+ F MRA LMWTI+DFPAYGML
Sbjct: 304 VPGPKHPKRALDVFLQPLIHELKKLWYEGVHTFDYSSKQNFNMRAVLMWTISDFPAYGML 363
Query: 448 S*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLFTKGKTEKD 507
S W+THGRL+CP+CM +T A GRK FD HR FLP H +R+ K LF K K +
Sbjct: 364 SGWTTHGRLSCPYCMGRTDAFQLKKGRKPCWFDYHRRFLPLHHPYRRNKKLFRKNKIVRL 423
Query: 508 GPPPRTTSEEVYRRVNELGLLKFTDVG----KRVRHP-KYGNEHHWTKRSIFWDLPYWKD 562
PP + +++++++ G + G P +YG+ H+W K+SIFW LPYWKD
Sbjct: 424 PPPSYVSGTDLFKQIDYYGAQETCKRGGNWLTTANMPDEYGSTHNWHKQSIFWQLPYWKD 483
Query: 563 NLLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNEKARMDLPLHC 607
LRH DVMHIEKNF DNI++ ++NV KTKD+ K+R+DL C
Sbjct: 484 LFLRHCLDVMHIEKNFFDNIMNTLLNVQGKTKDSLKSRLDLEEIC 528
Score = 41.2 bits (95), Expect = 0.002
Identities = 22/65 (33%), Positives = 32/65 (48%)
Query: 20 RKLKAAFVDGVRDFVAYAMAQDAFQLEGGIRCSCVKCTCRLIRSPKDVLNHLKDLGFMEN 79
RK+ + DG+R F+ A+ + + G I C C KC V HL + GFM N
Sbjct: 62 RKVTREYYDGLRGFMYQAINEPFARENGTIFCPCRKCLNEKYLEFNVVKKHLYNRGFMPN 121
Query: 80 YYVWI 84
YY ++
Sbjct: 122 YYEYL 126
>At4g08000 putative transposon protein
Length = 609
Score = 369 bits (946), Expect = e-102
Identities = 184/425 (43%), Positives = 260/425 (60%), Gaps = 16/425 (3%)
Query: 189 SECYTDIMRDTTPVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDGA 248
S+ + D++ + P + S Y+ +KL+ LGL T D C N CMLF+ K D
Sbjct: 68 SKKFYDLLEEFLPEGNQATSSHYQTEKLMRNLGLPYHTIDVCSNNCMLFW-----KEDEQ 122
Query: 249 LVACKFCNAPRYEVCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYNK 308
C+FC A R++ DD +++ +V M+YLPI RL+R++ S TA M WH ++
Sbjct: 123 EDHCRFCGAQRWKPKDD---RRRTKVPYSRMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQ 179
Query: 309 PNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILLT 368
G M HP D W++F ++H FAE+P NV LGLC+DGF P+ + +S WP++LT
Sbjct: 180 SKEGEMNHPSDAAEWRYFQELHPRFAEEPRNVYLGLCTDGFNPF--GMSRNHSLWPVILT 237
Query: 369 PYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLWN-GVSTYDIARKKI 427
PYN+P MCM+ YLFL+ + P +DV++QPLI++L LW+ GV YD++ +
Sbjct: 238 PYNLPPSMCMNTKYLFLTILNVEPNHPRASLDVFLQPLIEELKELWSTGVDAYDVSLSQN 297
Query: 428 FRMRAALMWTINDFPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFLP 487
F ++A L+WTI+DFPAY MLS W THG+ +CP CME TK+ + +GRK FDCHR FLP
Sbjct: 298 FNLKAVLLWTISDFPAYSMLSGWITHGKFSCPICMESTKSFYLPNGRKTCWFDCHRRFLP 357
Query: 488 ATHEFRKKKNLFTKGK-TEKDGPPPRTTSEEV-YRRVNELGLLKFTDV---GKRVRHPKY 542
H R+ K F KG+ + + PP T E+V Y R+ + K DV G + P Y
Sbjct: 358 HGHPSRRNKKDFLKGRDSSSEYPPKSLTGEQVYYERLASVNPTKTKDVGGNGHEKKMPGY 417
Query: 543 GNEHHWTKRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNEKARMD 602
G EH+W K SI W+L YW D LRHN DVMH EKNF D+I++ +M+V K+KDN +R+D
Sbjct: 418 GKEHNWHKESILWELSYWNDLNLRHNIDVMHTEKNFLDDIMNTLMSVKDKSKDNIMSRLD 477
Query: 603 LPLHC 607
+ + C
Sbjct: 478 IEIFC 482
>At1g36470 hypothetical protein
Length = 753
Score = 362 bits (928), Expect = e-100
Identities = 183/417 (43%), Positives = 251/417 (59%), Gaps = 16/417 (3%)
Query: 197 RDTTPVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCN 256
RD P + S Y+ +KL+ LGL T D C N CML + K D C+FC
Sbjct: 54 RDFLPEGNQATTSHYQTEKLMRNLGLPYHTIDVCQNNCMLSW-----KEDEKEDQCRFCG 108
Query: 257 APRYEVCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRH 316
A R++ DD +++ +V M+YLPI RL+R++ S TA M WH ++ G M H
Sbjct: 109 AKRWKPKDD---RRRTKVPYSRMWYLPIGDRLKRMYQSHKTAAAMRWHAEHQSKEGEMNH 165
Query: 317 PCDGVAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEM 376
D W++F +H +FAE+P NV LGLC+DGF P+ + +S WP++LTPYN+P M
Sbjct: 166 LSDAAEWRYFQGLHPEFAEEPRNVYLGLCTDGFNPF--GMSRNHSLWPVILTPYNLPPGM 223
Query: 377 CMSKPYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLWN-GVSTYDIARKKIFRMRAALM 435
CM+ YLFL+ + GP +DV++QPLI++L W+ GV YD++ + F ++A L+
Sbjct: 224 CMNTEYLFLTILNSGPNHPRASLDVFLQPLIEELKEFWSTGVDAYDVSLSQNFNLKAVLL 283
Query: 436 WTINDFPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKK 495
WTI+DFPAY ML W+THG+L+CP CME TK+ + +GRK FDCHR FLP H R+
Sbjct: 284 WTISDFPAYNMLLGWTTHGKLSCPVCMESTKSFYLPNGRKTCWFDCHRRFLPHGHPSRRN 343
Query: 496 KNLFTKGK-TEKDGPPPRTTSEEV-YRRVNELGLLKFTDVGKRVRHPK---YGNEHHWTK 550
+ F KG+ + PP T E+V Y R+ + K DVG K YG EH+W K
Sbjct: 344 RKDFLKGRDASSEYPPESLTGEQVYYERLASVNPPKTKDVGGNGHEKKMRGYGKEHNWHK 403
Query: 551 RSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNEKARMDLPLHC 607
SI W+L YWKD LRHN DVMH EKNF DNI++ +M V K+KDN +R+D+ C
Sbjct: 404 ESILWELSYWKDLNLRHNIDVMHTEKNFLDNIMNTLMRVKGKSKDNIMSRLDIEKFC 460
Score = 31.2 bits (69), Expect = 1.8
Identities = 15/39 (38%), Positives = 20/39 (50%)
Query: 6 FHRSWMYDRKYPGKRKLKAAFVDGVRDFVAYAMAQDAFQ 44
F+R WMY R L A +V GV F+ +A +Q Q
Sbjct: 13 FYRDWMYKRFDEVTGNLSAEYVAGVEQFMTFANSQPIVQ 51
>At1g34660 hypothetical protein
Length = 501
Score = 360 bits (925), Expect = e-99
Identities = 195/491 (39%), Positives = 276/491 (55%), Gaps = 24/491 (4%)
Query: 11 MYDRKYPGKRKLKAAFVDGVRDFVAYAMAQDAFQLEGGIRCSCVKCTCRLIRSPKDVLNH 70
MY+ G +L F +G+ DF+ +A Q+ G + C C C + H
Sbjct: 1 MYNYIDEGTGELTKEFKEGLVDFMRFAANQEIPLECGKMYCPCSTCGNEKFLPVDSIWKH 60
Query: 71 LKDLGFMENYYVWIYHGEQ-EPTNNTEFDVNMHASSSEARMECENFGVMEDMVGDAVGVN 129
L GFM YY+W HG+ E N+ D +H E E E+++ D GV
Sbjct: 61 LYSRGFMPGYYIWFSHGKDFETLANSNEDDEIHGLEPEDNNSNEE---EENIIRDPTGVI 117
Query: 130 LSYNEGGEEETI-----PNEKALKFYKMMQEVNKPLFEGSSD--SKLSMSVRLLAAASDW 182
E T PN +A KF+ M+ KP+++G + S+LS + RL+ +++
Sbjct: 118 QMVTNAFRETTESGIEEPNLEAKKFFDMLDAAKKPIYDGCKEGHSRLSAATRLMNIKTEY 177
Query: 183 SVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEF 242
+++E+ + D ++D P + P S+YE QKLV LGL + D CV+ CM+F+ +
Sbjct: 178 NLSEDCVDAIADFVKDLLPEPNLAPGSYYEIQKLVSSLGLPYQMIDVCVDNCMIFWRDTV 237
Query: 243 GKNDGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMT 302
L AC FC PR++V + K RV K M+YLPI RL+RL+ S TA M
Sbjct: 238 N-----LDACTFCQKPRFQV-----TSGKSRVPFKRMWYLPITDRLKRLYQSERTAGPMR 287
Query: 303 WHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSC 362
WH + + G + HP D AWKHF+ +++DFA + NV LGLC+DGF P+ + S YS
Sbjct: 288 WHAEHS-SDGNISHPSDAEAWKHFNSMYKDFANEHRNVYLGLCTDGFNPFGK-SGRKYSL 345
Query: 363 WPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLWN-GVSTYD 421
WP++LTPYN+P +CM + +LFLS +VPGP +DV++QPLI +L LW+ GV YD
Sbjct: 346 WPVILTPYNLPPSLCMKREFLFLSILVPGPEHPRRSLDVFLQPLIHELQLLWSEGVVAYD 405
Query: 422 IARKKIFRMRAALMWTINDFPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDC 481
++ K F MRA LMWTI+DFPAYGMLS W+THGRL+CP+C + A HG+K FDC
Sbjct: 406 VSLKNNFVMRAVLMWTISDFPAYGMLSGWTTHGRLSCPYCQDTIDAFQLKHGKKSCWFDC 465
Query: 482 HRPFLPATHEF 492
HR FLP H +
Sbjct: 466 HRRFLPKNHSY 476
>At2g12210 putative TNP2-like transposon protein
Length = 889
Score = 301 bits (771), Expect = 7e-82
Identities = 199/586 (33%), Positives = 284/586 (47%), Gaps = 56/586 (9%)
Query: 49 IRCSCVKCTCRLIRSPKDVLNHLKDLGFMENYYV---WIYHGEQEPTNNTEFDVNMHASS 105
I C C C +S DV++HL G E Y + W +HG+ + + E S
Sbjct: 37 IVCPCKDCRNVERKSGSDVVDHLVRRGMDEAYKLRADWYHHGDVDSVADCE--------S 88
Query: 106 SEARMECENFGVMEDMVGDAVGVNLSYNEGGEEETIPNEKALKFYKMMQEVNKPLFEGS- 164
+ +R E + ++ A G + E E+E I ++ F + + PL+
Sbjct: 89 NFSRWNAE----ILELYQAAQGFDADLGEIAEDEDIREDE---FLAKLADAEIPLYPSCL 141
Query: 165 SDSKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEKLGLEV 224
+ SKLS V L + +++ + + P + L S YE +K + +
Sbjct: 142 NHSKLSAIVTLFRIKTKNGWSDKSFNELLGTLPEMLPADNVLHTSLYEVKKFLRSFDMGY 201
Query: 225 KTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSMFYLPI 284
+ CVN C LF K L C CNA R++ G + KK V K + Y PI
Sbjct: 202 EKIHACVNDCCLFR-----KRYKNLENCPKCNASRWKSNMQTG-EAKKGVPQKVLRYFPI 255
Query: 285 IPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGL 344
IPR +R+F S A + WHY NK G +RHP D V W + + +FA + N+RLGL
Sbjct: 256 IPRFKRMFRSEEMAKDLRWHYSNKSTDGKLRHPVDSVTWDKMNDKYPEFAIEERNLRLGL 315
Query: 345 CSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQ 404
+DGF P+ T YSCWP+LL YN+P ++CM K + LS ++PGP + IDVY++
Sbjct: 316 STDGFNPF-NMKNTRYSCWPVLLVNYNLPPDLCMKKENIMLSLLIPGPQQPGNSIDVYLE 374
Query: 405 PLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTINDFPAYGMLS*WSTHGRLACPHCME 463
PLI+DLN LW G TYD K F ++A L+WTI+DFPAYG L+ G++ CP C +
Sbjct: 375 PLIEDLNHLWKKGELTYDAFSKTTFTLKAMLLWTISDFPAYGNLAGCKVKGKMGCPLCGK 434
Query: 464 QTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLFTKGKTEKDGPPPRTTSEEVYRRV- 522
T ++W + RKH F CHR L TH +R KK F GK E T EV + +
Sbjct: 435 GTDSMWLKYSRKHV-FMCHRKGLAPTHSYRNKKAWF-DGKIEHGRKARILTGREVSQTLK 492
Query: 523 ---NELGLLK--------------FTDVGKRVRHPKYGNE--------HHWTKRSIFWDL 557
N+ G K TD + E W KRSIF+ L
Sbjct: 493 NFKNDFGNAKESGRKRKRNDCIESVTDSDDESSESEEDEELEVDEDELSRWKKRSIFFKL 552
Query: 558 PYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNEKARMDL 603
YW+D +RHN DVMH+E+N +I+ +++ K+KD AR DL
Sbjct: 553 SYWEDLPVRHNLDVMHVERNVGASIVSTLLH-CGKSKDGLNARKDL 597
>At2g10630 En/Spm transposon hypothetical protein 1
Length = 515
Score = 301 bits (770), Expect = 9e-82
Identities = 174/467 (37%), Positives = 256/467 (54%), Gaps = 37/467 (7%)
Query: 6 FHRSWMYDRKYPGKRKLKAAFVDGVRDFVAYAMAQDAFQLEGG-IRCSCVKCTC-RLIRS 63
F+R W+Y R L A +V GV F+ +A +Q Q G C C C + I S
Sbjct: 13 FYRDWVYKRFDEVTGNLSAEYVAGVEQFMTFANSQPIVQSSRGKFHCPCSVCKNEKHIIS 72
Query: 64 PKDVLNHLKDLGFMENYYVWIYHGEQ-------EPTNNTEFDVNMHASSSEARMECENFG 116
+ V +HL GFM +YYVW HGE+ TN T F N E E+
Sbjct: 73 GRRVSSHLFSHGFMHDYYVWYKHGEEMNMDLGTSYTNRTYFSEN----HEEVGNIVEDPY 128
Query: 117 VMEDMVGDAVGVNLSYNEG--------GEEETIPNEKALKFYKMMQEVNKPLFEGSSD-- 166
V DMV DA N+ Y++ EE + N KFY +++ N PL++G +
Sbjct: 129 V--DMVNDAFNFNVGYDDNYRHDDSYQNVEEPVRNHSN-KFYDLLEGANNPLYDGCRERQ 185
Query: 167 SKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEKLGLEVKT 226
S+LS++ RL+ +++++ E+ + + D P ++ S Y+ +KL+ LGL T
Sbjct: 186 SQLSLASRLMHNKAEYNMNEKFVDSVCETFTDFLPEENQATTSHYQTEKLMRNLGLPYHT 245
Query: 227 TDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSMFYLPIIP 286
D C N CMLF+ E ++ C+FC A R++ DD +++ +V M+YLPI
Sbjct: 246 IDVCQNNCMLFWKEEEKEDQ-----CRFCGAKRWKPKDD---RRRTKVPYSCMWYLPIGD 297
Query: 287 RLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGLCS 346
RL+R++ S TA M WH ++ G M HP D W++F +H FAEDP NV LGLC+
Sbjct: 298 RLKRMYQSHKTAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQGLHPQFAEDPRNVYLGLCT 357
Query: 347 DGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQPL 406
DGF P+ + +S WP++LTPYN+P MCM+ YLFL+ + GP +DV++QPL
Sbjct: 358 DGFNPF--GMSRNHSLWPVILTPYNLPPGMCMNTEYLFLTILNSGPNHPRASLDVFLQPL 415
Query: 407 IDDLNRLWN-GVSTYDIARKKIFRMRAALMWTINDFPAYGMLS*WST 452
I++L LW+ GV YD++ + F ++A L+WTI+DFPAY MLS W+T
Sbjct: 416 IEELKELWSTGVDAYDVSLSQNFNLKAVLLWTISDFPAYSMLSGWTT 462
>At1g35280 hypothetical protein
Length = 557
Score = 281 bits (719), Expect = 8e-76
Identities = 165/430 (38%), Positives = 223/430 (51%), Gaps = 37/430 (8%)
Query: 201 PVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRY 260
P + L S YE +K ++ + + CVN C LF K L C CNA R+
Sbjct: 3 PADNVLHTSLYEVKKFLKSFDMGYEKIHACVNDCCLFR-----KRYKNLENCPKCNASRW 57
Query: 261 EVCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDG 320
+ G + KK V K + Y PIIPR +R+F S A + WHY NK G +RHP D
Sbjct: 58 KSNMQTG-EAKKGVPQKVLRYFPIIPRFKRMFRSEEMAKDLRWHYSNKSIDGKLRHPVDS 116
Query: 321 VAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSK 380
V W + + +FA + N+RLGL +DGF P+ T YSCWP+LL YN+PL++CM K
Sbjct: 117 VTWDKMNDKYPEFAAEERNLRLGLSTDGFNPF-NMKNTRYSCWPVLLVNYNLPLDLCMKK 175
Query: 381 PYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTIN 439
+ LS ++PGP + IDVY++PLI+DLN LW G TYD K F ++A L+WTI+
Sbjct: 176 ENIMLSLLIPGPQQPGNSIDVYLEPLIEDLNHLWKKGELTYDAFSKTTFTLKAMLLWTIS 235
Query: 440 DFPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLF 499
DF AYG L+ G++ CP C + T ++W + RKH F CHR L TH +R KK F
Sbjct: 236 DFSAYGNLAGCKVKGKMGCPLCGKGTDSMWLKYSRKHV-FMCHRKGLAPTHSYRNKKAWF 294
Query: 500 TKGKTEKDGPPPRTTSEEV----------YRRVNELG--------LLKFTDVGKRVRHPK 541
GK E T EV +R E G + TD +
Sbjct: 295 -DGKIEHGRKARILTGREVSQTLKNFKNDFRNAKESGRKRKRNDCIESVTDSDDESSESE 353
Query: 542 YGNE--------HHWTKRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKT 593
E W KRSIF+ L YW+D +RHN DVMH+E+N +I+ +++ K+
Sbjct: 354 EDEELEVDKDELSRWKKRSIFFKLSYWEDLPVRHNLDVMHVERNVGASIVSTLLH-CGKS 412
Query: 594 KDNEKARMDL 603
KD AR DL
Sbjct: 413 KDGLNARKDL 422
>At4g03730 putative transposon protein
Length = 696
Score = 280 bits (715), Expect = 2e-75
Identities = 139/308 (45%), Positives = 187/308 (60%), Gaps = 14/308 (4%)
Query: 301 MTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPY 360
M WH + G M +P DG AWK+F++V+ +FA + N+ LGL +DGF + + +
Sbjct: 3 MRWHAEHMSPEGEMHNPSDGAAWKNFNEVYPNFAAESRNIYLGLSTDGF-NLVSMNGEAH 61
Query: 361 SCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLW-NGVST 419
S W +++TPYN+P MC+ + + FLS +VPGP +D+Y+QPLI +L LW +G
Sbjct: 62 SVWHVIVTPYNLPSGMCIKREFFFLSVLVPGPKHPKKNLDIYLQPLIKELVSLWIDGEEA 121
Query: 420 YDIARKKIFRMRAALMWTINDFPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*F 479
YD ++KK F MR AL W I+DF YGMLS W THG L+CP+C++ T + W + RKHS F
Sbjct: 122 YDKSKKKKFTMRVALTWAISDFSTYGMLSVWMTHGLLSCPYCLDNTISFWLPNERKHSWF 181
Query: 480 DCHRPFLPATHEFRKKKNLFTKGKTEKDGPPPRTTSEEVYRRVNELGLLKFTDVGKRVRH 539
DCH FLP H + + F GK +D PP V NE T G
Sbjct: 182 DCHGMFLPLGHMYMQNVQAFRGGKEVRDDPP-----FTVDCGGNEHEKFAQTIDG----- 231
Query: 540 PKYGNEHHWTKRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNEKA 599
YG H+W KRSIFW+LPYW++ +LRHN D MHI+KNF DN+I+ V+ V KTKD K+
Sbjct: 232 --YGIHHNWVKRSIFWELPYWENLILRHNLDFMHIKKNFFDNLINTVLKVLEKTKDKIKS 289
Query: 600 RMDLPLHC 607
RMDL C
Sbjct: 290 RMDLATLC 297
>At4g07750 putative transposon protein
Length = 569
Score = 265 bits (678), Expect = 4e-71
Identities = 177/550 (32%), Positives = 265/550 (48%), Gaps = 61/550 (11%)
Query: 83 WIYHGEQEPTNNTEFDVNMHASSSEARMECENFGVMEDMVGDAVGVNLSYNEGGEEETIP 142
W +HGE + E D + + E + F +++ VG++ E
Sbjct: 11 WYHHGESN--SRAESDTKVTQWNDEVLYQAAEF--LDEEFAYKVGLS-------EIAECD 59
Query: 143 NEKALKFYKMMQEVNKPLFEGS-SDSKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTP 201
++K +F + + PL+ + SKLS V L + +++ +I+ + P
Sbjct: 60 DKKEDEFLAKLADAETPLYPSCVNHSKLSAIVSLFRLNTKNGWSDKSFNDLLEILPEMLP 119
Query: 202 VKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYE 261
+ L S Y+ +K ++ + + CVN C LF KN L +C CNA R++
Sbjct: 120 KDNVLHTSLYKVKKFLKSFDMGYQKIHACVNDCCLFR-----KNYKKLDSCPKCNASRWK 174
Query: 262 VCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGV 321
G + KK V K + Y PIIPRL+ +F S + WH+ NK + + HP D V
Sbjct: 175 TNLHIG-EIKKGVPQKVLRYFPIIPRLKGMFRSKEMDKDLRWHFSNKSSDEKLCHPVDLV 233
Query: 322 AWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKP 381
W + + FA + N+R GL +DGF P+ T YSCWP ++CM K
Sbjct: 234 TWDQMNAKYPSFAGEERNLRHGLSTDGFNPF-NMKNTRYSCWP----------DLCMKKE 282
Query: 382 YLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTIND 440
+ L+ ++PGP + IDVY++PLI+DL LW NG TYD K F ++A L+WTI+D
Sbjct: 283 NIMLTLLIPGPQQPGNNIDVYLEPLIEDLCHLWDNGELTYDAYSKSTFDLKAMLLWTISD 342
Query: 441 FPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLFT 500
FPAYG L+ G++ CP C + T ++W RKH + CH LP TH FR KK+ F
Sbjct: 343 FPAYGNLAGCKVKGKMGCPLCGKNTDSMWLKFSRKHV-YMCHIKGLPPTHHFRMKKSWF- 400
Query: 501 KGKTEKDGPPPRTTSEEVYRRV----NELGLLKFTDVGKRVRHPKYGNE----------- 545
GK E + +E+Y+ + N G LK KR R +
Sbjct: 401 DGKAENGRKRRILSGQEIYQNLKNFKNNFGTLK-QSASKRKRTVNIEADSDSDDQSSESE 459
Query: 546 ------------HHWTKRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKT 593
W K+SIF+ LPYW++ L+RHN DVMH+E+N +II +++ K+
Sbjct: 460 EDEQDQVDEEELSRWKKKSIFFKLPYWQELLVRHNLDVMHVERNVAASIISTLLH-CWKS 518
Query: 594 KDNEKARMDL 603
KD AR DL
Sbjct: 519 KDGLNARKDL 528
>At1g31560 putative protein
Length = 1431
Score = 265 bits (678), Expect = 4e-71
Identities = 158/430 (36%), Positives = 216/430 (49%), Gaps = 37/430 (8%)
Query: 201 PVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRY 260
P + L S YE +K ++ + + CVN C LF K L C CNA R+
Sbjct: 109 PADNVLRTSLYEVKKFLKSFDMGYEKIHACVNDCCLFR-----KRYKNLENCPKCNASRW 163
Query: 261 EVCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDG 320
+ G + KK V K + Y IIPR +++F S A + WHY NK G RHP D
Sbjct: 164 KNYMQTG-EAKKGVPQKVLRYFLIIPRFKKMFRSKEMAKDLRWHYSNKSTDGKHRHPVDS 222
Query: 321 VAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSK 380
V W + +FA + N+RLGL +DGF P+ T YSCWP+LL YN+P + CM
Sbjct: 223 VTWDKMIDKYPEFAAEERNMRLGLSTDGFNPF-NMKNTRYSCWPVLLVNYNLPPDQCMKN 281
Query: 381 PYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAALMWTIN 439
+ LS ++PGP + IDVY++P I+DLN LW G TYD K F ++A L WTI+
Sbjct: 282 ENIMLSLLIPGPQQPGNSIDVYLEPFIEDLNHLWKKGELTYDAFSKTTFTLKAMLFWTIS 341
Query: 440 DFPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLF 499
DFPAYG L+ G++ CP C + T ++W + RKH F CHR L TH ++ KK F
Sbjct: 342 DFPAYGNLAGCKVKGKMGCPLCGKGTDSMWLKYSRKHV-FMCHRKGLAPTHSYKNKKAWF 400
Query: 500 TKGKTEKDGPPPRTTSEEVYRRV----NELGLLK--------------FTDVGKRVRHPK 541
GK E T EV + N+ G K TD +
Sbjct: 401 -DGKVEHGRKARILTGREVSHNLKNFKNDFGNAKESGRKRKRNDCIESVTDSDNESSESE 459
Query: 542 YGNE--------HHWTKRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKT 593
E W KRSIF+ L YW++ +RHN DVMH+E+N +I+ +++ K+
Sbjct: 460 EDEEVEVDEDELSRWKKRSIFFKLSYWENLPVRHNLDVMHVERNVGASIVSTLLH-CGKS 518
Query: 594 KDNEKARMDL 603
KD +R DL
Sbjct: 519 KDGLNSRKDL 528
>At3g31920 hypothetical protein
Length = 725
Score = 254 bits (648), Expect = 1e-67
Identities = 175/561 (31%), Positives = 259/561 (45%), Gaps = 52/561 (9%)
Query: 51 CSCVKCTCRLIRSPKDVLNHLKDLGFMENYY---VWIYHGEQEPTNNTEFDVNMHASSSE 107
C C+ C + + +L HL G Y W HGE + S
Sbjct: 38 CPCIDCRNLCHQPIETILEHLVMKGMDHKYKRNRCWSKHGEIRADK-------LEVEPSS 90
Query: 108 ARMECENFGVMEDMV--GDAVGVNLSYNEGGEEETIPNEKALKFYKMMQEVNKPLFEGSS 165
ECE + ++ GD + + NE +E E+A F + ++E PL+ G
Sbjct: 91 TTSECEAYELIRTAYFDGDVHSDSQNQNEDDSKEPETKEEA-DFREKLKEAETPLYHGCP 149
Query: 166 D-SKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEKLGLEV 224
+K+S + L ++E + ++ D P ++ LP S E +K ++ G
Sbjct: 150 KYTKVSAIMGLYRIKVKSGMSENYFDQLLSLVHDILPGENVLPTSTNEIKKFLKMFGFGY 209
Query: 225 KTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSMFYLPI 284
C N C+L Y E+ D C CNA R+E G ++KK + K + Y PI
Sbjct: 210 DIIHACQNDCIL-YRKEYELRD----TCPRCNASRWERDKHTG-EEKKGIPAKVLGYFPI 263
Query: 285 IPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGL 344
R +R+F S A+ WH+ N + G MRH D +AW + +FA +P N+RLGL
Sbjct: 264 KDRFKRMFRSKKMAEDQRWHFSNASDDGTMRHHVDSLAWAQVNDKWPEFAAEPRNLRLGL 323
Query: 345 CSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQ 404
+DG P+ T YS WPILL YN+ MCM + L+ ++ GPT+ + IDVY+
Sbjct: 324 SADGMNPF-SIQNTKYSTWPILLVNYNMAPTMCMKAENIMLTLLIHGPTAPSNNIDVYLA 382
Query: 405 PLIDDLNRLWN-GVSTYDIARKKIFRMRAALMWTINDFPAYGMLS*WSTHGRLACPHCME 463
PLIDDL LW+ G+ YD +K+ F +RA +W+I+D+PA G L+ G+ AC C +
Sbjct: 383 PLIDDLKDLWSEGIHVYDSFKKESFTLRAMFLWSISDYPALGTLAGCKVKGKQACNVCGK 442
Query: 464 QTKAIWFHHGRKHS*FDCHRPFLPATHEFRKKKNLFTKGKTEKDGPPPR-TTSEEVYRRV 522
T W RKH R P H +R+++ F T ++G R T EE++ +
Sbjct: 443 DTPFRWLKFSRKHVYLRNRRRLRPG-HPYRRRRGWF--DNTVEEGTTSRIQTGEEIFEAL 499
Query: 523 NELGLLKFTDVGKRVRHPKYGNEHHWTKRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNI 582
L + I DL D +RHN DVMH+EKN D I
Sbjct: 500 KAL-------------------------KMILGDLWRETDMPVRHNIDVMHVEKNVSDAI 534
Query: 583 IHIVMNVSAKTKDNEKARMDL 603
+ ++M S K+KD AR DL
Sbjct: 535 VSMLMQ-SVKSKDGLNARKDL 554
>At2g14980 pseudogene
Length = 665
Score = 250 bits (639), Expect = 1e-66
Identities = 161/474 (33%), Positives = 243/474 (50%), Gaps = 37/474 (7%)
Query: 136 GEEETIPNEKALKFYKMMQEVNKPLFEGS-SDSKLSMSVRLLAAASDWSVAEEGSECYTD 194
GE ++K + + + PL+ + SKLS V L + +++ +
Sbjct: 5 GEIAERDDKKEDELLAKLVDAETPLYPSCVNHSKLSAIVSLFRLKTKNGWSDKSFNDLLE 64
Query: 195 IMRDTTPVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKF 254
+ + P + L S YE +K ++ + + CVN C LF K L C
Sbjct: 65 TLPEMLPEDNVLHTSLYEVKKFLKSFDMGYQKIHACVNDCCLFR-----KKYKKLQNCLK 119
Query: 255 CNAPRYEVCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVM 314
CNA R++ G + KK V K + Y I+PRL+R+F S A + WH++NK G +
Sbjct: 120 CNASRWKTNMHTG-EVKKGVPQKVLRYFSIVPRLKRMFRSEEMARDLRWHFHNKSTDGKL 178
Query: 315 RHPCDGVAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPILLTPYNVPL 374
HP D V W + + FA + N+RLGL ++GF P+ T YSCWP+LL YN+P
Sbjct: 179 CHPVDSVTWDQMNAKYPLFASEERNLRLGLSTNGFNPF-NMKNTRYSCWPVLLVNYNLPP 237
Query: 375 EMCMSKPYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRLW-NGVSTYDIARKKIFRMRAA 433
++CM K + L+ ++PG + IDVY++PLIDDL LW NG TYD K F ++A
Sbjct: 238 DLCMKKENIMLTLLIPGSQQPGNSIDVYLEPLIDDLCHLWDNGEWTYDAFSKSSFTLKAM 297
Query: 434 LMWTINDFPAYGMLS*WSTHGRLACPHCMEQTKAIWFHHGRKHS*FDCHRPFLPATHEFR 493
L+WTI+DFPAYG L+ G++AC C + T+++W RKH + HR LP +H FR
Sbjct: 298 LLWTISDFPAYGNLAGCKVKGKMACLLCGKHTESMWLKFSRKHV-YMGHRMGLPPSHRFR 356
Query: 494 KKKNLFTKGKTEKDGPPPRTTSEEVYRRVNELGLLKFTDVGKRVRH--PKYGNEHHWT-- 549
KK+ F GK E +RR + +L ++ +++ +GN T
Sbjct: 357 NKKSWF-DGKAE-------------HRRKSR--ILSGLEISHNLKNFQNNFGNFKQSTRK 400
Query: 550 KRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNEKARMDL 603
++ DLP +RHN DVMH+E+N +I+ +M+ K+KD AR DL
Sbjct: 401 RKQPESDLP------VRHNLDVMHVERNVVASIVSTLMH-CGKSKDGVNARKDL 447
>At2g10140 putative protein
Length = 663
Score = 248 bits (632), Expect = 9e-66
Identities = 183/569 (32%), Positives = 269/569 (47%), Gaps = 95/569 (16%)
Query: 6 FHRSWMYDRKYPGKRKLKAAFVDGVRDFVAYAMAQDAFQLEGGIRCSCVKCTCRLIRSPK 65
F+R WMY R L A +V GV +F+ +A +Q Q S V + S
Sbjct: 13 FYRDWMYKRFDEVTENLSAEYVAGVEEFMTFANSQPIVQ------SSRVYARMKNTSSQV 66
Query: 66 D---VLNHLKDLGFMENYYVWIYHGEQ-------EPTNNTEFDVNMHASSSEARMECENF 115
+ V+ +KDL + + + + HGE+ TN T F N E E+
Sbjct: 67 EELVVICLVKDLCLIIMFGISM-HGEELNMDIGTSYTNRTYFREN----HEEVGNIVEDP 121
Query: 116 GVMEDMVGDAVGVNLSYNEG-GEEETIPNEKALKFYKMMQEVNKPLFEGSSDSKLSMSVR 174
V DMV DA N+ Y++ G ++ ++ + YK ++E
Sbjct: 122 YV--DMVNDAFNFNVGYDDNVGYDDNYHHDGS---YKNVEE------------------- 157
Query: 175 LLAAASDWSVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNG- 233
PV+ N FY+ L+E G D C G
Sbjct: 158 --------------------------PVR-NHSNKFYD---LLE--GANNTLYDYCREGQ 185
Query: 234 CMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGSQK--KKRVSIKSMFYLPIIPRLQRL 291
L + N + C F + ++ D + K R +++ Y ++ + +
Sbjct: 186 SQLSLASRLMHNKAGTIVCYFGKKTKKKINVDFVAHKDGSLRTTVEEPKYHIVVCGIYK- 244
Query: 292 FASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGLCSDGFIP 351
TA M WH ++ G M HP D W++F ++H FA++P NV LGLC DGF P
Sbjct: 245 -----TAAAMRWHAEHQSKEGEMNHPSDAAEWRYFQELHTWFAKEPRNVYLGLCIDGFNP 299
Query: 352 YIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQPLIDDLN 411
+ + +S WP++LT YN+P MCM+ YLFL+ + GP +DV++QPLI++L
Sbjct: 300 F--GISRNHSLWPVILTSYNLPPGMCMNTKYLFLTILNSGPNHPRASLDVFLQPLIEELK 357
Query: 412 RLW-NGVSTYDIARKKIFRMRAALMWTINDFPAYGMLS*WSTHGRLACPHCMEQTKAIWF 470
LW GV TYD++ + F ++A L+W I+DFPAY MLS W+THG+L+CP CME TK+ +
Sbjct: 358 ELWCTGVDTYDVSLSQNFNLKAVLLWAISDFPAYSMLSGWTTHGKLSCPVCMESTKSFYL 417
Query: 471 HHGRKHS*FDCHRPFLPATHEFRKKKNLFTKGK-TEKDGPPPRTTSEEV-YRRVNELGLL 528
+GRK FDCHR FLP +H R+ K F KG+ ++ PP T E+V Y R+ +
Sbjct: 418 PNGRKTCWFDCHRRFLPHSHPSRRNKKDFLKGRDASREYPPGSLTGEQVYYERLASVNPP 477
Query: 529 KFTDVGKRVRHPK---YGNEHHWTKRSIF 554
K DVG K YG EH+W K SIF
Sbjct: 478 KTKDVGGNGHEKKMRGYGKEHNWDKESIF 506
>At1g33580 En/Spm-like transposon protein, putative
Length = 428
Score = 176 bits (446), Expect = 3e-44
Identities = 125/392 (31%), Positives = 184/392 (46%), Gaps = 33/392 (8%)
Query: 51 CSCVKCTCRLIRSPKD-VLNHLKDLGFMENYY---VWIYHGEQEPTNNTEFDVNMHASSS 106
C C C L P D VL HL G Y W HGE + A
Sbjct: 48 CPCTDCR-NLCHQPIDTVLEHLVIKGMNHKYKRNGCWSKHGE------------IRADKL 94
Query: 107 EARMECENFGVMEDMV-----GDAVGVNLSYNEGGEEETIPNEKALKFYKMMQEVNKPLF 161
EA E FG E + G+ + + NE +E E++ F + +++ PL+
Sbjct: 95 EAESSSE-FGAYELIRTAYFDGEDHSDSQNQNENDSKEPSTKEES-DFREKLKDAETPLY 152
Query: 162 EGSSD-SKLSMSVRLLAAASDWSVAEEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEKL 220
G +K+S +RL ++E + ++ D P ++ L S E +K ++
Sbjct: 153 YGCPKYTKVSAIMRLYRIKVKSGMSENYFDQLLSLVHDILPGENVLLTSTNEIKKFLKMF 212
Query: 221 GLEVKTTDCCVNGCMLFYDNEFGKNDGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSMF 280
G C N C+L Y E+ D C C+A R+E G ++KK + K +
Sbjct: 213 GFGYDIIHACPNDCIL-YRKEYELRD----TCPRCSAFRWERDKHTG-EEKKGIPAKVLR 266
Query: 281 YLPIIPRLQRLFASTHTADKMTWHYYNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNV 340
Y PI R +R+F S A+ + WH+ N + G MRHP D +AW + FA +P N+
Sbjct: 267 YFPIKDRFKRMFRSKKMAEDLRWHFSNASDDGTMRHPVDSLAWAQVNYKWPQFAAEPRNL 326
Query: 341 RLGLCSDGFIPYIQASATPYSCWPILLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGID 400
RLGL +DG P+ T YS WP+LL YN+ MCM + L+ ++PGPT+ + ID
Sbjct: 327 RLGLSTDGMNPF-SIQNTKYSTWPVLLVNYNMAPTMCMKAENIMLTLLIPGPTTPNNNID 385
Query: 401 VYIQPLIDDLNRLW-NGVSTYDIARKKIFRMR 431
VY+ PLIDDL LW +G+ YD K+ F +R
Sbjct: 386 VYLAPLIDDLKDLWSDGIQVYDSFLKESFTLR 417
>At2g15070 En/Spm-like transposon protein
Length = 672
Score = 171 bits (432), Expect = 1e-42
Identities = 102/324 (31%), Positives = 165/324 (50%), Gaps = 10/324 (3%)
Query: 128 VNLSYNEGGEEETIPNEKALKFYK-MMQEVNKPLFEGSSD-SKLSMSVRLLAAASDWSVA 185
+ +Y +G E+ P K +++ +++V PL+ G +K+S + L ++
Sbjct: 36 IRTAYFDGEEDSKEPVTKEESYFREKLKDVETPLYYGCPKYTKVSAIMGLYRIKVKSGMS 95
Query: 186 EEGSECYTDIMRDTTPVKDNLPLSFYEAQKLVEKLGLEVKTTDCCVNGCMLFYDNEFGKN 245
E + ++ D P ++ LP S E +K ++ G C N C+L Y E+
Sbjct: 96 ENYFDQLLSLVHDILPGENVLPTSTNEIKKFLKMFGFGYDIIHACPNDCIL-YKKEYELR 154
Query: 246 DGALVACKFCNAPRYEVCDDAGSQKKKRVSIKSMFYLPIIPRLQRLFASTHTADKMTWHY 305
D C C+A R++ G ++KK + K ++Y I R +R+F S + + WH+
Sbjct: 155 D----TCLGCSASRWKRDKHTG-EEKKGIPAKVLWYFSIKDRFKRMFRSKKMVEDLQWHF 209
Query: 306 YNKPNSGVMRHPCDGVAWKHFDQVHRDFAEDPHNVRLGLCSDGFIPYIQASATPYSCWPI 365
N G MRHP D +AW + FA + N+RLGL +D P+ T YS WP+
Sbjct: 210 SNASVDGTMRHPIDSLAWAQVNDKWPQFAAESRNLRLGLSTDKMNPF-SIQNTKYSTWPV 268
Query: 366 LLTPYNVPLEMCMSKPYLFLSCIVPGPTSSLDGIDVYIQPLIDDLNRL-WNGVSTYDIAR 424
LL YN+ MCM + L+ ++ GP + + IDVY+ PLI+DL L +G+ YD
Sbjct: 269 LLVNYNMAPTMCMKAKNIMLTLLIHGPKAPSNNIDVYLAPLINDLKDLSSDGIQVYDSFF 328
Query: 425 KKIFRMRAALMWTINDFPAYGMLS 448
K+ F +RA L+W+IND+P G L+
Sbjct: 329 KENFTLRAMLLWSINDYPTLGTLA 352
Score = 70.1 bits (170), Expect = 3e-12
Identities = 35/66 (53%), Positives = 45/66 (68%), Gaps = 4/66 (6%)
Query: 541 KYGNEHH---WTKRSIFWDLPYWKDNLLRHNFDVMHIEKNFCDNIIHIVMNVSAKTKDNE 597
+YG + H W KRSI ++LPYWKD +RHN DVMH+EKN D I+ +M SAK+KD
Sbjct: 381 EYGEDTHQWRWKKRSILFELPYWKDLPVRHNIDVMHVEKNVSDVIVSTLMQ-SAKSKDGL 439
Query: 598 KARMDL 603
AR +L
Sbjct: 440 NARKNL 445
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.324 0.138 0.450
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,389,917
Number of Sequences: 26719
Number of extensions: 703817
Number of successful extensions: 1674
Number of sequences better than 10.0: 32
Number of HSP's better than 10.0 without gapping: 28
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 1495
Number of HSP's gapped (non-prelim): 53
length of query: 615
length of database: 11,318,596
effective HSP length: 105
effective length of query: 510
effective length of database: 8,513,101
effective search space: 4341681510
effective search space used: 4341681510
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC145061.27


