

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC145061.22 - phase: 0 /pseudo
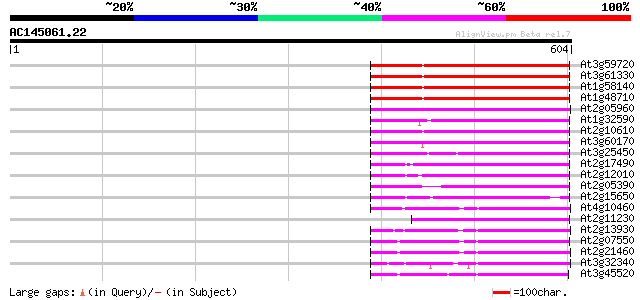
(604 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g59720 copia-type reverse transcriptase-like protein 190 2e-48
At3g61330 copia-type polyprotein 189 3e-48
At1g58140 hypothetical protein 189 3e-48
At1g48710 hypothetical protein 189 3e-48
At2g05960 putative retroelement pol polyprotein 154 1e-37
At1g32590 hypothetical protein, 5' partial 143 3e-34
At2g10610 pseudogene 139 6e-33
At3g60170 putative protein 137 1e-32
At3g25450 hypothetical protein 134 1e-31
At2g17490 putative retroelement pol polyprotein 126 3e-29
At2g12010 pseudogene 125 7e-29
At2g05390 putative retroelement pol polyprotein 119 4e-27
At2g15650 putative retroelement pol polyprotein 116 3e-26
At4g10460 putative retrotransposon 112 5e-25
At2g11230 putative retroelement pol polyprotein 107 2e-23
At2g13930 putative retroelement pol polyprotein 106 3e-23
At2g07550 putative retroelement pol polyprotein 105 9e-23
At2g21460 putative retroelement pol polyprotein 103 2e-22
At3g32340 hypothetical protein 103 3e-22
At3g45520 copia-like polyprotein 100 3e-21
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 190 bits (482), Expect = 2e-48
Identities = 90/214 (42%), Positives = 144/214 (67%), Gaps = 1/214 (0%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N+LS+GQL++KGY I +KDN+L + DK L+ K P++KN+ F +++ CL +
Sbjct: 403 NILSLGQLLEKGYDIRLKDNNLSIRDKESNLITKVPMSKNRMFVLNIRNDIAQCLK-MCY 461
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
E+SWLWH FGHLN+ L L KEM+ +P ++ P++ + L+G Q + +F ++
Sbjct: 462 KEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEGCLLGNQFKMSFPKESSS 521
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R+++ L +++ DV GP++ S+G+ YF+ F+D+FS+ WV +K KS F+IF+KFK
Sbjct: 522 RAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAH 581
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VEK+SG I ++R+D GGE+T K F K+CE+NG+
Sbjct: 582 VEKESGLVIKTMRSDSGGEFTSKEFLKYCEDNGI 615
Score = 40.8 bits (94), Expect = 0.002
Identities = 21/65 (32%), Positives = 39/65 (59%), Gaps = 4/65 (6%)
Query: 6 LPTLESGNWESWSVVMKNLFGAKDLFNIVQNGYDELGENP---TEMHRNAYKELKKKDCK 62
+P L N+++WS+ MK + GA D++ IV+ G+ E EN ++ ++ ++ +K+D K
Sbjct: 10 VPVLTKSNYDNWSLRMKAILGAHDVWEIVEKGFIE-PENEGSLSQTQKDGLRDSRKRDKK 68
Query: 63 TLFFI 67
L I
Sbjct: 69 ALCLI 73
>At3g61330 copia-type polyprotein
Length = 1352
Score = 189 bits (481), Expect = 3e-48
Identities = 90/214 (42%), Positives = 145/214 (67%), Gaps = 1/214 (0%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N+LS+GQL++KGY I +KDN+L + D+ L+ K P++KN+ F +++ CL +
Sbjct: 403 NILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLK-MCY 461
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
E+SWLWH FGHLN+ L L KEM+ +P ++ P++ + L+GKQ + +F ++
Sbjct: 462 KEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSS 521
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R+++ L +++ DV GP++ S+G+ YF+ F+D+FS+ WV +K KS F+IF+KFK
Sbjct: 522 RAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAH 581
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VEK+SG I ++R+D GGE+T K F K+CE+NG+
Sbjct: 582 VEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGI 615
Score = 40.8 bits (94), Expect = 0.002
Identities = 21/65 (32%), Positives = 39/65 (59%), Gaps = 4/65 (6%)
Query: 6 LPTLESGNWESWSVVMKNLFGAKDLFNIVQNGYDELGENP---TEMHRNAYKELKKKDCK 62
+P L N+++WS+ MK + GA D++ IV+ G+ E EN ++ ++ ++ +K+D K
Sbjct: 10 VPVLTKSNYDNWSLRMKAILGAHDVWEIVEKGFIE-PENEGSLSQTQKDGLRDSRKRDKK 68
Query: 63 TLFFI 67
L I
Sbjct: 69 ALCLI 73
>At1g58140 hypothetical protein
Length = 1320
Score = 189 bits (481), Expect = 3e-48
Identities = 90/214 (42%), Positives = 145/214 (67%), Gaps = 1/214 (0%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N+LS+GQL++KGY I +KDN+L + D+ L+ K P++KN+ F +++ CL +
Sbjct: 403 NILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLK-MCY 461
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
E+SWLWH FGHLN+ L L KEM+ +P ++ P++ + L+GKQ + +F ++
Sbjct: 462 KEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSS 521
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R+++ L +++ DV GP++ S+G+ YF+ F+D+FS+ WV +K KS F+IF+KFK
Sbjct: 522 RAQKPLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAH 581
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VEK+SG I ++R+D GGE+T K F K+CE+NG+
Sbjct: 582 VEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGI 615
Score = 40.8 bits (94), Expect = 0.002
Identities = 21/65 (32%), Positives = 39/65 (59%), Gaps = 4/65 (6%)
Query: 6 LPTLESGNWESWSVVMKNLFGAKDLFNIVQNGYDELGENP---TEMHRNAYKELKKKDCK 62
+P L N+++WS+ MK + GA D++ IV+ G+ E EN ++ ++ ++ +K+D K
Sbjct: 10 VPVLTKSNYDNWSLRMKAILGAHDVWEIVEKGFIE-PENEGSLSQTQKDGLRDSRKRDKK 68
Query: 63 TLFFI 67
L I
Sbjct: 69 ALCLI 73
>At1g48710 hypothetical protein
Length = 1352
Score = 189 bits (481), Expect = 3e-48
Identities = 90/214 (42%), Positives = 145/214 (67%), Gaps = 1/214 (0%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N+LS+GQL++KGY I +KDN+L + D+ L+ K P++KN+ F +++ CL +
Sbjct: 403 NILSLGQLLEKGYDIRLKDNNLSIRDQESNLITKVPMSKNRMFVLNIRNDIAQCLK-MCY 461
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
E+SWLWH FGHLN+ L L KEM+ +P ++ P++ + L+GKQ + +F ++
Sbjct: 462 KEESWLWHLRFGHLNFGGLELLSRKEMVRGLPCINHPNQVCEGCLLGKQFKMSFPKESSS 521
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R+++ L +++ DV GP++ S+G+ YF+ F+D+FS+ WV +K KS F+IF+KFK
Sbjct: 522 RAQKSLELIHTDVCGPIKPKSLGKSNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAH 581
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VEK+SG I ++R+D GGE+T K F K+CE+NG+
Sbjct: 582 VEKESGLVIKTMRSDRGGEFTSKEFLKYCEDNGI 615
Score = 40.8 bits (94), Expect = 0.002
Identities = 21/65 (32%), Positives = 39/65 (59%), Gaps = 4/65 (6%)
Query: 6 LPTLESGNWESWSVVMKNLFGAKDLFNIVQNGYDELGENP---TEMHRNAYKELKKKDCK 62
+P L N+++WS+ MK + GA D++ IV+ G+ E EN ++ ++ ++ +K+D K
Sbjct: 10 VPVLTKSNYDNWSLRMKAILGAHDVWEIVEKGFIE-PENEGSLSQTQKDGLRDSRKRDKK 68
Query: 63 TLFFI 67
L I
Sbjct: 69 ALCLI 73
>At2g05960 putative retroelement pol polyprotein
Length = 1200
Score = 154 bits (389), Expect = 1e-37
Identities = 73/215 (33%), Positives = 128/215 (58%), Gaps = 1/215 (0%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N++S+GQ + G + +KDN L L D++ L++K ++N+ + ++KV CL
Sbjct: 325 NIISLGQATEAGCDVRLKDNYLTLHDRDGNLLVKATRSRNRLYRVELKVKNTKCLQLAAL 384
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
N D WH GH+N ++ +V KE ++ +P + + ++GKQ R F TT+
Sbjct: 385 N-DLTKWHARLGHINLETIKAMVTKEFVIGIPSAPKEKEICASCMLGKQARQVFPKATTY 443
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R+ ++L +++ D+ GP+ P+ + +Y + +D+ S+ +W L+K KS AF F+ FK
Sbjct: 444 RASQILELIHGDLCGPISPPTAAKRRYILVLIDDHSRFMWSFLLKEKSEAFGKFKTFKAT 503
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGVI 603
VE+++G+ I +LRTD GGE+ + F+ FCEE G+I
Sbjct: 504 VEQETGEKIKTLRTDRGGEFLSQEFQTFCEEEGII 538
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 143 bits (360), Expect = 3e-34
Identities = 82/225 (36%), Positives = 127/225 (56%), Gaps = 15/225 (6%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQ-RLVMKTPLAKNKTF*TDMKVAK------LN 441
NL SVGQL QKG ++ + +++ K + R+VM + + KN+ F V K
Sbjct: 325 NLFSVGQLQQKGLRFIIEGDVCEVWHKTEKRMVMHSTMTKNRMFVVFAAVKKSKETEETR 384
Query: 442 CLSAI--VNNEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHK--FYDTRLIGKQ 497
CL I NN +WH FGHLN+ L L +KEM+ +PK D+ + D L GKQ
Sbjct: 385 CLQVIGKANN----MWHKRFGHLNHQGLRSLAEKEMVKGLPKFDLGEEEAVCDICLKGKQ 440
Query: 498 PRTAFISTTTHRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSG 557
R + + +S +VL +V+ D+ GP+ S +Y ++F+D+FS+ W L+ KS
Sbjct: 441 IRESIPKESAWKSTQVLQLVHTDICGPINPASTSGKRYILNFIDDFSRKCWTYLLSEKSE 500
Query: 558 AFDIFQKFKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
F F++FK VE++SGK + LR+D GGEY + F+++C+E G+
Sbjct: 501 TFQFFKEFKAEVERESGKKLVCLRSDRGGEYNSREFDEYCKEFGI 545
>At2g10610 pseudogene
Length = 531
Score = 139 bits (349), Expect = 6e-33
Identities = 72/214 (33%), Positives = 119/214 (54%), Gaps = 1/214 (0%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N++S+GQ + G I +KD L + D+ +L++K ++N + M + K CL + +
Sbjct: 240 NIISLGQATESGCEIKLKDGYLTMHDQEGKLLVKAERSRNSLYKVKMGLRKEACLY-LTS 298
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
S LWH GH++ I+L ++DKE+I P L I + + L+GKQ R +F T
Sbjct: 299 TSMSSLWHARMGHVSLITLKSMIDKELIQGAPSLVIESEVCGSCLLGKQTRQSFPQATVF 358
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R+ + L +++ D+ GP+ + +Y +D++S+ +W L+K K AF F+ FK
Sbjct: 359 RATKKLELIHGDLCGPITPKTSVGNRYIFVLIDDYSRYMWTVLLKEKGDAFFKFKNFKAL 418
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VEK+SG+ I + TD GGE+ F FCE G+
Sbjct: 419 VEKESGEKIQTFITDRGGEFVSGEFNSFCERTGI 452
>At3g60170 putative protein
Length = 1339
Score = 137 bits (346), Expect = 1e-32
Identities = 76/217 (35%), Positives = 124/217 (57%), Gaps = 3/217 (1%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCL---SA 445
NLLS+GQL ++G +I ++D + K++ ++ +M+T ++ N+ F + N L +
Sbjct: 367 NLLSLGQLQERGLAILIRDGTCKVYHPSKGAIMETNMSGNRMFFLLASKPQKNSLCLQTE 426
Query: 446 IVNNEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFIST 505
V ++++ LWH FGHLN L L K+M++ +P L + L GKQ R +
Sbjct: 427 EVMDKENHLWHCRFGHLNQEGLKLLAHKKMVIGLPILKATKEICAICLTGKQHRESMSKK 486
Query: 506 TTHRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKF 565
T+ +S L +V+ D+ GP+ S +Y +SF+D+F++ WV + KS AF F+ F
Sbjct: 487 TSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKTWVYFLHEKSEAFATFKIF 546
Query: 566 KFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
K VEK+ G +T LRTD GGE+T F +FC +G+
Sbjct: 547 KASVEKEIGAFLTCLRTDRGGEFTSNEFGEFCRSHGI 583
Score = 36.2 bits (82), Expect = 0.054
Identities = 19/59 (32%), Positives = 34/59 (57%), Gaps = 3/59 (5%)
Query: 12 GNWESWSVVMKNLFGAKDLFNIVQNGYDEL--GENP-TEMHRNAYKELKKKDCKTLFFI 67
G ++ WS+ M+N +++L+ +V+ G + G P +E R+A +E K KD K F+
Sbjct: 17 GYYDFWSMTMENFLRSRELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKLKDLKVKNFL 75
>At3g25450 hypothetical protein
Length = 1343
Score = 134 bits (338), Expect = 1e-31
Identities = 67/216 (31%), Positives = 125/216 (57%), Gaps = 4/216 (1%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N+LS+GQ + G I M+++ L L D+ L++K ++N+ + ++V CL
Sbjct: 360 NILSLGQATESGCDIRMREDYLTLHDREGNLLIKAQRSRNRLYKVSLEVENSKCLQLTTT 419
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHK--FYDTRLIGKQPRTAFISTT 506
NE S +WH GH+++ ++ ++ KE+++ + +P + + L GKQ R +F T
Sbjct: 420 NE-STIWHARLGHISFETIKAMIKKELVIGISS-SVPQEKETCGSCLFGKQARHSFPKAT 477
Query: 507 THRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFK 566
++R+ +VL +++ D+ GP+ + + +Y +D+ S+ +W L+K KS AF F++FK
Sbjct: 478 SYRAAQVLELIHGDLCGPISPSTAAKKRYVFVLIDDHSRYMWSILLKEKSEAFGKFKEFK 537
Query: 567 FFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VE++ G I + RTD GGE+ F++FC + G+
Sbjct: 538 ALVEQECGAIIKTFRTDRGGEFLSHEFQEFCAKEGI 573
>At2g17490 putative retroelement pol polyprotein
Length = 822
Score = 126 bits (317), Expect = 3e-29
Identities = 68/214 (31%), Positives = 117/214 (53%), Gaps = 3/214 (1%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NLLSV Q++ GY +T+K+N + D ++ + + + NK+F ++ + +
Sbjct: 97 NLLSVPQMIINGYQVTLKNNYCTIHDSARKKIGEVEMV-NKSF--HLRWLSNEETAMVAK 153
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
+E + LWH GH + +L L KEM+ +PK ++ ++ ++ K R F +
Sbjct: 154 DEATELWHKRLGHTGHSNLKILQSKEMVTGLPKFNVEEGKCESCILSKHSRDPFPKESET 213
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R+K L +++ DV GP++ S+ +Y ++F+D+ ++ +WV +KAKS F F+KFK
Sbjct: 214 RAKHKLELIHSDVCGPMQNSSINGSRYILTFIDDATRMVWVYFLKAKSEVFQTFKKFKNL 273
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VE + I LR D G EY K F +F E NG+
Sbjct: 274 VENNANCRIKKLRIDRGTEYLSKEFSEFLEGNGI 307
>At2g12010 pseudogene
Length = 616
Score = 125 bits (314), Expect = 7e-29
Identities = 69/216 (31%), Positives = 118/216 (53%), Gaps = 6/216 (2%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NLLSV Q++ GYS+ ++ D+ + +M+ NK+F ++ S +V
Sbjct: 180 NLLSVPQIISVGYSVLFENKCCTNLDRTGKKIMEIQRV-NKSFPIRWNSSQE---SELVT 235
Query: 449 NEDSW--LWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTT 506
+E+ LWH GH+ + + +K+M+ P + + +GKQ R AF + +
Sbjct: 236 SEEEGMELWHRRLGHVGNSRMEQMHNKKMVDGFPNFHVNKEICGVCKLGKQVREAFPTES 295
Query: 507 THRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFK 566
++KE L +V+ +V GP++ S+ YF+ +D+++ W+ +K KS F +F+K K
Sbjct: 296 QTKTKEKLEIVHTNVCGPMQTESLNGSIYFLLLVDDYTHMAWIYFLKQKSEVFSVFKKSK 355
Query: 567 FFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VE QSGK I LR+DGGGE+T F +FC++ G+
Sbjct: 356 ALVETQSGKKIKRLRSDGGGEFTSSGFNEFCDQEGI 391
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 119 bits (299), Expect = 4e-27
Identities = 62/214 (28%), Positives = 115/214 (52%), Gaps = 20/214 (9%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N++S+GQ + G + MKD+ L L D+ L+++ ++N+ + D+ V + CL
Sbjct: 348 NIISLGQATEAGCDVRMKDDQLTLHDREGCLLLRATRSRNRLYKVDLNVENVKCLQ---- 403
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
+ +V KE+++ + + + + L+GKQ R F TT+
Sbjct: 404 ----------------LEAATMVRKELVIGISNIPKEKETCGSCLLGKQARQPFPKATTY 447
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
R+ +VL +V+ D+ GP+ + + +Y + +D+ ++ +W L+K KS AF+ F+ FK
Sbjct: 448 RASQVLELVHGDLCGPITQSTTAKKRYILVLIDDHTRYMWSMLLKEKSEAFEKFRDFKTK 507
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VE++SG I + RTD GGE+ + F+ FC + G+
Sbjct: 508 VEQESGVKIKTFRTDKGGEFVSQEFQDFCAKEGI 541
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 116 bits (291), Expect = 3e-26
Identities = 67/214 (31%), Positives = 114/214 (52%), Gaps = 13/214 (6%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NLLSV Q++ GY + +D + D N + +M + +K+F + + ++A V
Sbjct: 393 NLLSVPQIISSGYWVRFQDKRCIIQDANGKEIMNIEMT-DKSFKIKLSSVEEEAMTANVQ 451
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
E++W H GH++ L + DKE++ +P+ + + +GKQ R +F +
Sbjct: 452 TEETW--HKRLGHVSNKRLQQMQDKELVNGLPRFKVTKETCKACNLGKQSRKSFPKESQT 509
Query: 509 RSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKFF 568
+++E L +V+ DV GP++ S+ +Y++ FLD+++ WV +K KS F F+KFK
Sbjct: 510 KTREKLEIVHTDVCGPMQHQSIDGSRYYVLFLDDYTHMCWVYFLKQKSETFATFKKFKAL 569
Query: 569 VEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VEKQS SI +LR E FCE+ G+
Sbjct: 570 VEKQSNCSIKTLRP----------MEVFCEDEGI 593
>At4g10460 putative retrotransposon
Length = 1230
Score = 112 bits (281), Expect = 5e-25
Identities = 71/218 (32%), Positives = 119/218 (54%), Gaps = 10/218 (4%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NLLS+G L + GYS K+ L + + + L++ + K + K + ++
Sbjct: 361 NLLSMGTLEEHGYSFESKNGVLVVKEGTRTLLIGS--RHEKLYLLQGKPEVSHSMTVERR 418
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMI--LVVPKLDIPHKFYDTRLIGKQPRTAFISTT 506
N+D+ LWH GH++ +++ LV K + V KL++ + + GK R +F+
Sbjct: 419 NDDTVLWHRRLGHISQKNMDILVKKGYLDGKKVSKLEL----CEDCIYGKARRLSFV-VA 473
Query: 507 THRSKEVLNVVYFDVNGPLEVP-SMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKF 565
TH +++ LN V+ D+ G VP S+G+ +YFISF+D +S+ WV +K K AF F ++
Sbjct: 474 THNTEDKLNYVHSDLWGAPSVPLSLGKCQYFISFIDVYSRKTWVYFLKHKDEAFGTFAEW 533
Query: 566 KFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGVI 603
VE Q+G+ I LR D G E+ + F FC+E G++
Sbjct: 534 SVMVENQTGRKIKILRIDNGLEFCNQQFNDFCKEKGIV 571
>At2g11230 putative retroelement pol polyprotein
Length = 962
Score = 107 bits (266), Expect = 2e-23
Identities = 52/170 (30%), Positives = 94/170 (54%)
Query: 433 TDMKVAKLNCLSAIVNNEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTR 492
T + N + + +S WH H N ++ ++ KE++ +P + I + D
Sbjct: 731 TSLLFTSSNACLHLTSLSESNRWHSRLRHTNLDTIKSMIQKELVRGIPHVKIEKEVCDLC 790
Query: 493 LIGKQPRTAFISTTTHRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLI 552
L GKQ R F T++R+ ++L +V+ D+ GP+ + KY +D+ S+ +W L+
Sbjct: 791 LFGKQSRQVFPQGTSYRAAKILELVHGDLCGPITPSTHAGNKYIFVIIDDKSRYMWTALL 850
Query: 553 KAKSGAFDIFQKFKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
K KS +F+ F++F+ VE+++ I + RT+ GGE+ + F KFCEE+G+
Sbjct: 851 KEKSDSFNKFKQFRALVEQETRIKIQTFRTNRGGEFILHEFTKFCEESGI 900
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 106 bits (265), Expect = 3e-23
Identities = 70/218 (32%), Positives = 117/218 (53%), Gaps = 10/218 (4%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NL+S+G L +G +D LK+ K ++K ++ + D + S+
Sbjct: 353 NLISLGTLEDRGCWFKSQDGILKIV-KGCSTILKGQ-KRDTLYILDGVTEEGESHSSAEV 410
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMIL--VVPKLDIPHKFYDTRLIGKQPRTAFISTT 506
+++ LWH GH++ + LV K + V+ +L+ F + + GKQ R +F +
Sbjct: 411 KDETALWHSRLGHMSQKGMEILVKKGCLRREVIKELE----FCEDCVYGKQHRVSF-APA 465
Query: 507 THRSKEVLNVVYFDVNG-PLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKF 565
H +KE L V+ D+ G P S+G +YFISF+D++S+ +W+ ++ K AF+ F ++
Sbjct: 466 QHVTKEKLAYVHSDLWGSPHNPASLGNSQYFISFVDDYSRKVWIYFLRKKDEAFEKFVEW 525
Query: 566 KFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGVI 603
K VE QS + + LRTD G EY FEKFC+E G++
Sbjct: 526 KKMVENQSDRKVKKLRTDNGLEYCNHYFEKFCKEEGIV 563
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 105 bits (261), Expect = 9e-23
Identities = 68/217 (31%), Positives = 115/217 (52%), Gaps = 10/217 (4%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NLLS+G + GY +D L++ NQ V+ T + + + K L+ +
Sbjct: 377 NLLSLGTFEKAGYKFESEDGILRIKAGNQ--VLLTGRRYDTLYLLNWKPVASESLAVVKR 434
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMI--LVVPKLDIPHKFYDTRLIGKQPRTAFISTT 506
+D+ LWH H++ ++ LV K + V LD+ + + GK R +F S
Sbjct: 435 ADDTVLWHQRLCHMSQKNMEILVRKGFLDKKKVSSLDV----CEDCIYGKAKRKSF-SLA 489
Query: 507 THRSKEVLNVVYFDVNGPLEVP-SMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKF 565
H +KE L ++ D+ G VP S+G+ +YF+S +D+F++ +WV +K K AF+ F ++
Sbjct: 490 HHDTKEKLEYIHSDLWGAPFVPLSLGKCQYFMSIIDDFTRKVWVYFMKTKDEAFEKFVEW 549
Query: 566 KFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGV 602
VE Q+ + + +LRTD G E+ K+F+ FCE G+
Sbjct: 550 VNLVENQTDRRVKTLRTDNGLEFCNKLFDGFCESIGI 586
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 103 bits (258), Expect = 2e-22
Identities = 65/218 (29%), Positives = 117/218 (52%), Gaps = 10/218 (4%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NLLS+G + GYS +++ +L + + V+ T + + LS +
Sbjct: 355 NLLSLGTFEKSGYSFKLENGTLSIIAGDS--VLLTVRRCYTLYLLQWRPVTEESLSVVKR 412
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMI--LVVPKLDIPHKFYDTRLIGKQPRTAFISTT 506
+D+ LWH GH++ +++ L+ K ++ V KL+ + + GK R F +
Sbjct: 413 QDDTILWHRRLGHMSQKNMDLLLKKGLLDKKKVSKLET----CEDCIYGKAKRIGF-NLA 467
Query: 507 THRSKEVLNVVYFDVNGPLEVP-SMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKF 565
H ++E L V+ D+ G VP S+G+ +YFISF+D++++ + + +K K AFD F ++
Sbjct: 468 QHDTREKLEYVHSDLWGAPSVPFSLGKCQYFISFIDDYTRKVRIYFLKTKDEAFDKFVEW 527
Query: 566 KFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGVI 603
VE Q+ K I +LRTD G E+ + F++FC + G++
Sbjct: 528 ANLVENQTDKRIKTLRTDNGLEFCNRSFDEFCSQKGIL 565
>At3g32340 hypothetical protein
Length = 871
Score = 103 bits (257), Expect = 3e-22
Identities = 68/221 (30%), Positives = 119/221 (53%), Gaps = 14/221 (6%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
N++SV L +G+ ++K N FD++ PL +N + + + N + I
Sbjct: 221 NIVSVSCLDMEGFHFSIK-NKCCSFDRDDMFYGSAPL-ENGLYVLNQSMPVYNIRTKIFK 278
Query: 449 NED---SWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTR---LIGKQPRTAF 502
+ D ++LWH GH+N + L ++ ++ Y+T L+GK + F
Sbjct: 279 SNDLNPTFLWHCRLGHINEKRIQKLHSDGLLN-----SFDYESYETCESCLLGKMTKAPF 333
Query: 503 ISTTTHRSKEVLNVVYFDVNGPLEVPSMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIF 562
S + R+ ++L +++ DV GP+ + G +YFI+F D+FS+ +V L+K KS +F+ F
Sbjct: 334 -SGHSERAGDLLGLIHTDVCGPMSTSARGNYQYFITFTDDFSRYSYVYLMKHKSESFEKF 392
Query: 563 QKFKFFVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENGVI 603
++F+ V+ Q GKSI +LR+D GGEY +VF E G++
Sbjct: 393 KEFQNEVQNQLGKSIKALRSDRGGEYLSQVFSGHLRECGIV 433
>At3g45520 copia-like polyprotein
Length = 1363
Score = 100 bits (248), Expect = 3e-21
Identities = 66/214 (30%), Positives = 114/214 (52%), Gaps = 6/214 (2%)
Query: 389 NLLSVGQLVQKGYSITMKDNSLKLFDKNQRLVMKTPLAKNKTF*TDMKVAKLNCLSAIVN 448
NLLS+G + G+ ++ L++ NQ V+ + + K A L+
Sbjct: 384 NLLSLGTFEKAGHKFESENGMLRIKSGNQ--VLLEGRRYDTLYILHGKPATDESLAVARA 441
Query: 449 NEDSWLWHYMFGHLNYISLNHLVDKEMILVVPKLDIPHKFYDTRLIGKQPRTAFISTTTH 508
N+D+ LWH H++ +++ L+ K+ L K+ + D + G+ + F + H
Sbjct: 442 NDDTVLWHRRLCHMSQKNMSLLI-KKGFLDKKKVSMLDTCEDC-IYGRAKKIGF-NLAQH 498
Query: 509 RSKEVLNVVYFDVNGPLEVP-SMGRIKYFISFLDEFSKNIWVCLIKAKSGAFDIFQKFKF 567
+K+ L V+ D+ G VP S+G +YFISF+D++++ +WV +K K AF+ F +
Sbjct: 499 DTKKKLEYVHSDLWGAPTVPMSLGNCQYFISFIDDYTRKVWVYFLKTKDEAFEKFVSWIS 558
Query: 568 FVEKQSGKSITSLRTDGGGEYTIKVFEKFCEENG 601
VE QSG+ + +LRTD G E+ ++F+ FCEE G
Sbjct: 559 LVENQSGERVKTLRTDNGLEFCNRMFDGFCEEKG 592
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.349 0.155 0.516
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 11,761,009
Number of Sequences: 26719
Number of extensions: 452049
Number of successful extensions: 1895
Number of sequences better than 10.0: 79
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 1732
Number of HSP's gapped (non-prelim): 108
length of query: 604
length of database: 11,318,596
effective HSP length: 105
effective length of query: 499
effective length of database: 8,513,101
effective search space: 4248037399
effective search space used: 4248037399
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC145061.22


