

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144806.12 - phase: 1 /pseudo
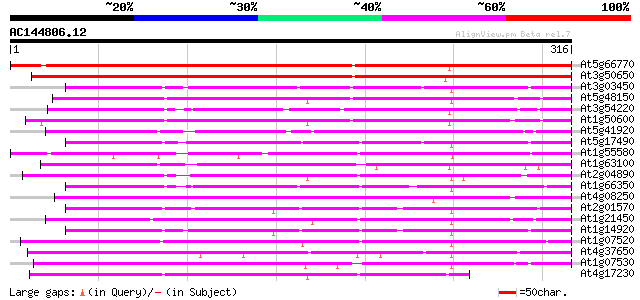
(316 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g66770 SCARECROW gene regulator 431 e-121
At3g50650 scarecrow-like 7 (SCL7) 397 e-111
At3g03450 RGA1-like protein 179 2e-45
At5g48150 SCARECROW gene regulator-like 175 3e-44
At3g54220 SCARECROW1 174 4e-44
At1g50600 scarecrow-like 5 (SCL5) 169 1e-42
At5g41920 SCARECROW gene regulator-like protein 164 6e-41
At5g17490 RGA-like protein 164 8e-41
At1g55580 unknown protein 161 5e-40
At1g63100 transcription factor SCARECROW, putative 157 9e-39
At2g04890 putative SCARECROW gene regulator 156 2e-38
At1g66350 gibberellin regulatory protein like 154 6e-38
At4g08250 putative protein 149 3e-36
At2g01570 putative RGA1, giberellin repsonse modulation protein 147 7e-36
At1g21450 scarecrow-like 1 (SCL1) 146 1e-35
At1g14920 signal response protein (GAI) 143 1e-34
At1g07520 transcription factor scarecrow-like 14, putative 133 1e-31
At4g37650 SHORT-ROOT (SHR) 128 5e-30
At1g07530 transcription factor scarecrow-like 14, putative 128 5e-30
At4g17230 scarecrow-like 13 (SCL13) 124 5e-29
>At5g66770 SCARECROW gene regulator
Length = 584
Score = 431 bits (1107), Expect = e-121
Identities = 222/322 (68%), Positives = 259/322 (79%), Gaps = 9/322 (2%)
Query: 1 TNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHI 60
+N+++ S SS+SSST E+L LSYK LNDACPYSKFAHLTANQAILEATE SN IHI
Sbjct: 265 SNRLSPNSPATSSSSSST--EDLILSYKTLNDACPYSKFAHLTANQAILEATEKSNKIHI 322
Query: 61 VDFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKL 120
VDFGIVQGIQW ALLQA ATR+SGKP +R+SGIPA +LG SP S+ ATGNRL +FAK+
Sbjct: 323 VDFGIVQGIQWPALLQALATRTSGKPTQIRVSGIPAPSLGESPEPSLIATGNRLRDFAKV 382
Query: 121 LGLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLN 180
L LNF+F PILTPI LL+ SSF + PDE LAVNFMLQLY LLDE V+ ALRLAKSLN
Sbjct: 383 LDLNFDFIPILTPIHLLNGSSFRVDPDEVLAVNFMLQLYKLLDETPTIVDTALRLAKSLN 442
Query: 181 PKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRI 240
P++VTLGEYE SL RVGF R + A +++A FESLEPN+ DS ER +VE L GRRI
Sbjct: 443 PRVVTLGEYEVSL-NRVGFANRVKNALQFYSAVFESLEPNLGRDSEERVRVERELFGRRI 501
Query: 241 DGVIGV------RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLV 294
G+IG RERME+KEQW+VLMEN GFESV LS+YA+SQAKILLWNY+YS+LYS+V
Sbjct: 502 SGLIGPEKTGIHRERMEEKEQWRVLMENAGFESVKLSNYAVSQAKILLWNYNYSNLYSIV 561
Query: 295 ESQPAFLSLAWKDVPLLTVSSW 316
ES+P F+SLAW D+PLLT+SSW
Sbjct: 562 ESKPGFISLAWNDLPLLTLSSW 583
>At3g50650 scarecrow-like 7 (SCL7)
Length = 542
Score = 397 bits (1021), Expect = e-111
Identities = 207/314 (65%), Positives = 245/314 (77%), Gaps = 11/314 (3%)
Query: 13 SNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWA 72
S+SSS++ E+ LSYK LNDACPYSKFAHLTANQAILEAT SNNIHIVDFGI QGIQW+
Sbjct: 229 SSSSSSSLEDFILSYKTLNDACPYSKFAHLTANQAILEATNQSNNIHIVDFGIFQGIQWS 288
Query: 73 ALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILT 132
ALLQA ATRSSGKP +RISGIPA +LG SP S+ ATGNRL +FA +L LNFEF P+LT
Sbjct: 289 ALLQALATRSSGKPTRIRISGIPAPSLGDSPGPSLIATGNRLRDFAAILDLNFEFYPVLT 348
Query: 133 PIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEAS 192
PI+LL+ SSF + PDE L VNFML+LY LLDE +V ALRLA+SLNP+IVTLGEYE S
Sbjct: 349 PIQLLNGSSFRVDPDEVLVVNFMLELYKLLDETATTVGTALRLARSLNPRIVTLGEYEVS 408
Query: 193 LTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVI-------- 244
L RV F R + + +++A FESLEPN+ DS ER +VE +L GRRI ++
Sbjct: 409 L-NRVEFANRVKNSLRFYSAVFESLEPNLDRDSKERLRVERVLFGRRIMDLVRSDDDNNK 467
Query: 245 -GVR-ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLS 302
G R ME+KEQW+VLME GFE V S+YA+SQAK+LLWNY+YS+LYSLVES+P F+S
Sbjct: 468 PGTRFGLMEEKEQWRVLMEKAGFEPVKPSNYAVSQAKLLLWNYNYSTLYSLVESEPGFIS 527
Query: 303 LAWKDVPLLTVSSW 316
LAW +VPLLTVSSW
Sbjct: 528 LAWNNVPLLTVSSW 541
>At3g03450 RGA1-like protein
Length = 547
Score = 179 bits (453), Expect = 2e-45
Identities = 110/292 (37%), Positives = 168/292 (56%), Gaps = 14/292 (4%)
Query: 32 DACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRI 91
++CPY KFAH TANQAILEA + +H++D G+ QG+QW AL+QA A R G P S R+
Sbjct: 260 ESCPYLKFAHFTANQAILEAVTTARRVHVIDLGLNQGMQWPALMQALALRPGGPP-SFRL 318
Query: 92 SGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILT-PIELLDESSFCIQPD-EA 149
+GI T S+ G +L++FA+ +G+ FEF + + L+ F +P+ E
Sbjct: 319 TGIGPPQ--TENSDSLQQLGWKLAQFAQNMGVEFEFKGLAAESLSDLEPEMFETRPESET 376
Query: 150 LAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNY 209
L VN + +L+ LL + S+EK L K++ P IVT+ E EA+ + F++RF A +Y
Sbjct: 377 LVVNSVFELHRLL-ARSGSIEKLLNTVKAIKPSIVTVVEQEAN-HNGIVFLDRFNEALHY 434
Query: 210 FAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCG 264
+++ F+SLE + +L S +R E + LGR+I V+ ER E QW++ M++ G
Sbjct: 435 YSSLFDSLEDSYSLPSQDRVMSE-VYLGRQILNVVAAEGSDRVERHETAAQWRIRMKSAG 493
Query: 265 FESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
F+ + L A QA +LL Y+ Y VE L + W+ PL+T S+W
Sbjct: 494 FDPIHLGSSAFKQASMLLSLYATGDGYR-VEENDGCLMIGWQTRPLITTSAW 544
>At5g48150 SCARECROW gene regulator-like
Length = 490
Score = 175 bits (443), Expect = 3e-44
Identities = 114/301 (37%), Positives = 157/301 (51%), Gaps = 14/301 (4%)
Query: 25 LSYK-ALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSS 83
LSY L + CPY KF +++AN AI EA + N +HI+DF I QG QW L+QAFA R
Sbjct: 194 LSYMHILYEVCPYFKFGYMSANGAIAEAMKEENRVHIIDFQIGQGSQWVTLIQAFAARPG 253
Query: 84 GKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFC 143
G P +RI+GI M + +S GNRL++ AK + FEF + + + +
Sbjct: 254 GPPR-IRITGIDDMTSAYARGGGLSIVGNRLAKLAKQFNVPFEFNSVSVSVSEVKPKNLG 312
Query: 144 IQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFV 200
++P EALAVNF L+++ DE+ N ++ LR+ KSL+PK+VTL E E++ T F
Sbjct: 313 VRPGEALAVNFAFVLHHMPDESVSTENHRDRLLRMVKSLSPKVVTLVEQESN-TNTAAFF 371
Query: 201 ERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQ 255
RF NY+AA FES++ + D +R VE L R + +I ER E +
Sbjct: 372 PRFMETMNYYAAMFESIDVTLPRDHKQRINVEQHCLARDVVNIIACEGADRVERHELLGK 431
Query: 256 WKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSS 315
W+ GF LS S K LL N YS Y L E A L L W L+ +
Sbjct: 432 WRSRFGMAGFTPYPLSPLVNSTIKSLLRN--YSDKYRLEERDGA-LYLGWMHRDLVASCA 488
Query: 316 W 316
W
Sbjct: 489 W 489
>At3g54220 SCARECROW1
Length = 653
Score = 174 bits (442), Expect = 4e-44
Identities = 114/299 (38%), Positives = 168/299 (56%), Gaps = 18/299 (6%)
Query: 22 ELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATR 81
++ +++ N P KF+H TANQAI EA E +++HI+D I+QG+QW L A+R
Sbjct: 365 KMVSAFQVFNGISPLVKFSHFTANQAIQEAFEKEDSVHIIDLDIMQGLQWPGLFHILASR 424
Query: 82 SSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESS 141
G P+ VR++G LGTS + ++ ATG RLS+FA LGL FEF P+ + LD
Sbjct: 425 PGGPPH-VRLTG-----LGTS-MEALQATGKRLSDFADKLGLPFEFCPLAEKVGNLDTER 477
Query: 142 FCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVE 201
++ EA+AV++ L + L + T S L L + L PK+VT+ E L+ F+
Sbjct: 478 LNVRKREAVAVHW---LQHSLYDVTGSDAHTLWLLQRLAPKVVTV--VEQDLSHAGSFLG 532
Query: 202 RFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV----RERMEDKEQWK 257
RF A +Y++A F+SL + +S ER VE LL + I V+ V R E W+
Sbjct: 533 RFVEAIHYYSALFDSLGASYGEESEERHVVEQQLLSKEIRNVLAVGGPSRSGEVKFESWR 592
Query: 258 VLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
M+ CGF+ + L+ A +QA +LL + S Y+LV+ L L WKD+ LLT S+W
Sbjct: 593 EKMQQCGFKGISLAGNAATQATLLLGMFP-SDGYTLVDDN-GTLKLGWKDLSLLTASAW 649
>At1g50600 scarecrow-like 5 (SCL5)
Length = 597
Score = 169 bits (429), Expect = 1e-42
Identities = 118/323 (36%), Positives = 165/323 (50%), Gaps = 21/323 (6%)
Query: 10 IASSNSS--------STTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIV 61
+ASS SS T EL L +ACPY KF + +AN AI EA + + +HI+
Sbjct: 279 LASSGSSIYKALRCKDPTGPELLTYMHILYEACPYFKFGYESANGAIAEAVKNESFVHII 338
Query: 62 DFGIVQGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLL 121
DF I QG QW +L++A R G PN VRI+GI + + G RL + A++
Sbjct: 339 DFQISQGGQWVSLIRALGARPGGPPN-VRITGIDDPRSSFARQGGLELVGQRLGKLAEMC 397
Query: 122 GLNFEFTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKS 178
G+ FEF ++ ++ EALAVNF L L+++ DE+ N ++ LRL K
Sbjct: 398 GVPFEFHGAALCCTEVEIEKLGVRNGEALAVNFPLVLHHMPDESVTVENHRDRLLRLVKH 457
Query: 179 LNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGR 238
L+P +VTL E EA+ T F+ RF N++ A FES++ +A D ER VE L R
Sbjct: 458 LSPNVVTLVEQEAN-TNTAPFLPRFVETMNHYLAVFESIDVKLARDHKERINVEQHCLAR 516
Query: 239 RIDGVIGV-----RERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSL 293
+ +I ER E +W+ GF+ LS Y + K LL SYS Y+L
Sbjct: 517 EVVNLIACEGVEREERHEPLGKWRSRFHMAGFKPYPLSSYVNATIKGLL--ESYSEKYTL 574
Query: 294 VESQPAFLSLAWKDVPLLTVSSW 316
E A L L WK+ PL+T +W
Sbjct: 575 EERDGA-LYLGWKNQPLITSCAW 596
>At5g41920 SCARECROW gene regulator-like protein
Length = 405
Score = 164 bits (415), Expect = 6e-41
Identities = 102/297 (34%), Positives = 163/297 (54%), Gaps = 13/297 (4%)
Query: 21 EELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFAT 80
+++ + + N P KF+H TANQAI +A +G +++HI+D ++QG+QW AL A+
Sbjct: 115 QKIFSALQTYNSVSPLIKFSHFTANQAIFQALDGEDSVHIIDLDVMQGLQWPALFHILAS 174
Query: 81 RSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPI-ELLDE 139
R K S+RI+G + + +++TG RL++FA L L FEF PI I L+D
Sbjct: 175 RPR-KLRSIRITGFGSSS------DLLASTGRRLADFASSLNLPFEFHPIEGIIGNLIDP 227
Query: 140 SSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGF 199
S + EA+ V++M + L D N++E L + + L P ++T+ E E S F
Sbjct: 228 SQLATRQGEAVVVHWMQ--HRLYDVTGNNLE-TLEILRRLKPNLITVVEQELSYDDGGSF 284
Query: 200 VERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVRERMEDKEQWKVL 259
+ RF A +Y++A F++L + +S ERF VE ++LG I ++ + +WK
Sbjct: 285 LGRFVEALHYYSALFDALGDGLGEESGERFTVEQIVLGTEIRNIVAHGGGRRKRMKWKEE 344
Query: 260 MENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
+ GF V L +QA +LL ++ Y+LVE + L L WKD+ LLT S+W
Sbjct: 345 LSRVGFRPVSLRGNPATQAGLLLGMLPWNG-YTLVE-ENGTLRLGWKDLSLLTASAW 399
>At5g17490 RGA-like protein
Length = 523
Score = 164 bits (414), Expect = 8e-41
Identities = 107/292 (36%), Positives = 161/292 (54%), Gaps = 16/292 (5%)
Query: 32 DACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRI 91
D+CPY KFAH TANQAILEA S +H++D G+ QG+QW AL+QA A R G P S R+
Sbjct: 233 DSCPYLKFAHFTANQAILEAVTTSRVVHVIDLGLNQGMQWPALMQALALRPGGPP-SFRL 291
Query: 92 SGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTP-IELLDESSFCIQPD-EA 149
+G+ S I G +L++ A+ +G+ F+F + T + L+ F + + E
Sbjct: 292 TGVG----NPSNREGIQELGWKLAQLAQAIGVEFKFNGLTTERLSDLEPDMFETRTESET 347
Query: 150 LAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNY 209
L VN + +L+ +L + S+EK L K++ P +VT+ E EA+ V F++RF A +Y
Sbjct: 348 LVVNSVFELHPVLSQ-PGSIEKLLATVKAVKPGLVTVVEQEANHNGDV-FLDRFNEALHY 405
Query: 210 FAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCG 264
+++ F+SLE + + S +R E + LGR+I ++ ER E QW+ M + G
Sbjct: 406 YSSLFDSLEDGVVIPSQDRVMSE-VYLGRQILNLVATEGSDRIERHETLAQWRKRMGSAG 464
Query: 265 FESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
F+ V L A QA +LL Y VE L LAW+ PL+ S+W
Sbjct: 465 FDPVNLGSDAFKQASLLLALSGGGDGYR-VEENDGSLMLAWQTKPLIAASAW 515
>At1g55580 unknown protein
Length = 445
Score = 161 bits (407), Expect = 5e-40
Identities = 116/336 (34%), Positives = 182/336 (53%), Gaps = 33/336 (9%)
Query: 1 TNKITAQSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNN--I 58
T+ + + + + ++++ +E + Y LN P+ +F HLTANQAIL+ATE ++N +
Sbjct: 122 TSSVCKEQFLFRTKNNNSDFE--SCYYLWLNQLTPFIRFGHLTANQAILDATETNDNGAL 179
Query: 59 HIVDFGIVQGIQWAALLQAFATRS---SGKPNSVRISGIPAMALGTSPVSSISATGNRLS 115
HI+D I QG+QW L+QA A RS S P S+RI+G V+ ++ TG+RL+
Sbjct: 180 HILDLDISQGLQWPPLMQALAERSSNPSSPPPSLRITGC------GRDVTGLNRTGDRLT 233
Query: 116 EFAKLLGLNFEF----------TPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDEN 165
FA LGL F+F +L I LL S+ E +AVN + L+ + +++
Sbjct: 234 RFADSLGLQFQFHTLVIVEEDLAGLLLQIRLLALSAV---QGETIAVNCVHFLHKIFNDD 290
Query: 166 TNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDS 225
+ + L KSLN +IVT+ E EA+ F+ RF A +++ A F+SLE + +S
Sbjct: 291 GDMIGHFLSAIKSLNSRIVTMAEREANHGDH-SFLNRFSEAVDHYMAIFDSLEATLPPNS 349
Query: 226 PERFQVESLLLGRRIDGVIGVRE-----RMEDKEQWKVLMENCGFESVGLSHYAISQAKI 280
ER +E G+ I V+ E R E W+ +M+ GF +V + +A+SQAK+
Sbjct: 350 RERLTLEQRWFGKEILDVVAAEETERKQRHRRFEIWEEMMKRFGFVNVPIGSFALSQAKL 409
Query: 281 LLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
LL + S Y+L + L L W++ PL +VSSW
Sbjct: 410 LLRLHYPSEGYNL-QFLNNSLFLGWQNRPLFSVSSW 444
>At1g63100 transcription factor SCARECROW, putative
Length = 658
Score = 157 bits (396), Expect = 9e-39
Identities = 101/320 (31%), Positives = 169/320 (52%), Gaps = 33/320 (10%)
Query: 18 TTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQA 77
T +E + + LN P KF H TAN+ +L A EG +HI+DF I QG+QW + Q+
Sbjct: 346 TVEDESGNALRFLNQVTPIPKFIHFTANEMLLRAFEGKERVHIIDFDIKQGLQWPSFFQS 405
Query: 78 FATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELL 137
A+R + P+ VRI+GI L ++ TG+RL FA+ + L FEF P++ +E +
Sbjct: 406 LASRIN-PPHHVRITGIGESKL------ELNETGDRLHGFAEAMNLQFEFHPVVDRLEDV 458
Query: 138 DESSFCIQPDEALAVNFMLQLYNLLDENTN-SVEKALRLAKSLNPKIVTLGEYEASLTTR 196
++ E++AVN ++Q++ L + T ++ L L +S NP + L E EA +
Sbjct: 459 RLWMLHVKEGESVAVNCVMQMHKTLYDGTGAAIRDFLGLIRSTNPIALVLAEQEAEHNS- 517
Query: 197 VGFVERFET----AFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV-----R 247
E+ ET + Y++A F+++ N+A DS R +VE +L GR I ++ +
Sbjct: 518 ----EQLETRVCNSLKYYSAMFDAIHTNLATDSLMRVKVEEMLFGREIRNIVACEGSHRQ 573
Query: 248 ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSS--LYSLVES--------- 296
ER W+ ++E GF S+G+S + Q+K+LL Y + +++ S
Sbjct: 574 ERHVGFRHWRRMLEQLGFRSLGVSEREVLQSKMLLRMYGSDNEGFFNVERSDEDNGGEGG 633
Query: 297 QPAFLSLAWKDVPLLTVSSW 316
+ ++L W + PL T+S+W
Sbjct: 634 RGGGVTLRWSEQPLYTISAW 653
>At2g04890 putative SCARECROW gene regulator
Length = 413
Score = 156 bits (394), Expect = 2e-38
Identities = 110/318 (34%), Positives = 160/318 (49%), Gaps = 21/318 (6%)
Query: 8 SSIASSNSSSTTWEELTLSYK-ALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIV 66
SSI S S LSY L++ CPY KF +++AN AI EA + IHI+DF I
Sbjct: 107 SSIYKSLQSREPESYEFLSYVYVLHEVCPYFKFGYMSANGAIAEAMKDEERIHIIDFQIG 166
Query: 67 QGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFE 126
QG QW AL+QAFA R G PN +RI+G+ S + RL + AK + F
Sbjct: 167 QGSQWIALIQAFAARPGGAPN-IRITGV-------GDGSVLVTVKKRLEKLAKKFDVPFR 218
Query: 127 FTPILTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPKI 183
F + P ++ + ++ EAL VNF L++L DE+ N ++ LR+ KSL+PK+
Sbjct: 219 FNAVSRPSCEVEVENLDVRDGEALGVNFAYMLHHLPDESVSMENHRDRLLRMVKSLSPKV 278
Query: 184 VTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGV 243
VTL E E + T F+ RF +Y+ A FES++ + + ER +E + R + +
Sbjct: 279 VTLVEQECNTNTS-PFLPRFLETLSYYTAMFESIDVMLPRNHKERINIEQHCMARDVVNI 337
Query: 244 IGVR--ERMEDKE---QWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQP 298
I ER+E E +WK GFE LS + + LL +YS +E +
Sbjct: 338 IACEGAERIERHELLGKWKSRFSMAGFEPYPLSSIISATIRALLRDYSNG---YAIEERD 394
Query: 299 AFLSLAWKDVPLLTVSSW 316
L L W D L++ +W
Sbjct: 395 GALYLGWMDRILVSSCAW 412
>At1g66350 gibberellin regulatory protein like
Length = 511
Score = 154 bits (389), Expect = 6e-38
Identities = 104/292 (35%), Positives = 163/292 (55%), Gaps = 21/292 (7%)
Query: 32 DACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRI 91
++CPY KFAH TANQAILE + +H++D G+ G+QW AL+QA A R +G P+ R+
Sbjct: 228 ESCPYLKFAHFTANQAILEVFATAEKVHVIDLGLNHGLQWPALIQALALRPNGPPD-FRL 286
Query: 92 SGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPI-LTPIELLDESSFCIQPD-EA 149
+GI G S ++ I G +L + A +G+NFEF I L + L I+P E+
Sbjct: 287 TGI-----GYS-LTDIQEVGWKLGQLASTIGVNFEFKSIALNNLSDLKPEMLDIRPGLES 340
Query: 150 LAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFNY 209
+AVN + +L+ LL + S++K L KS+ P I+T+ E EA+ V F++RF + +Y
Sbjct: 341 VAVNSVFELHRLL-AHPGSIDKFLSTIKSIRPDIMTVVEQEANHNGTV-FLDRFTESLHY 398
Query: 210 FAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENCG 264
+++ F+SLE + D + L LGR+I ++ ER E QW+ G
Sbjct: 399 YSSLFDSLEGPPSQDR----VMSELFLGRQILNLVACEGEDRVERHETLNQWRNRFGLGG 454
Query: 265 FESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
F+ V + A QA +LL Y+ + Y++ E++ L L W+ PL+ S+W
Sbjct: 455 FKPVSIGSNAYKQASMLLALYAGADGYNVEENEGCLL-LGWQTRPLIATSAW 505
>At4g08250 putative protein
Length = 483
Score = 149 bits (375), Expect = 3e-36
Identities = 96/296 (32%), Positives = 150/296 (50%), Gaps = 7/296 (2%)
Query: 26 SYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSG- 84
+++ L + PY F +LTA QAILEA + IHIVD+ I +G+QWA+L+QA +R++G
Sbjct: 183 AFELLQNMSPYVNFGYLTATQAILEAVKYERRIHIVDYDINEGVQWASLMQALVSRNTGP 242
Query: 85 KPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDESSFCI 144
+RI+ + G V+++ TG RL+ FA +G F + SS +
Sbjct: 243 SAQHLRITALSRATNGKKSVAAVQETGRRLTAFADSIGQPFSYQHCKLDTNAFSTSSLKL 302
Query: 145 QPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFE 204
EA+ +N ML L + +SV L AK+LNPK+VTL E L GF+ RF
Sbjct: 303 VRGEAVVINCMLHLPRFSHQTPSSVISFLSEAKTLNPKLVTLVHEEVGLMGNQGFLYRFM 362
Query: 205 TAFNYFAAFFESLEPNMALDSPERFQVESLLLG----RRIDGVIGVRERMEDKEQWKVLM 260
+ F+A F+SLE +++ +P R VE + +G + + +E W +
Sbjct: 363 DLLHQFSAIFDSLEAGLSIANPARGFVERVFIGPWVANWLTRITANDAEVESFASWPQWL 422
Query: 261 ENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
E GF+ + +S QAK+LL ++ + + E L L WK L++ S W
Sbjct: 423 ETNGFKPLEVSFTNRCQAKLLL--SLFNDGFRVEELGQNGLVLGWKSRRLVSASFW 476
>At2g01570 putative RGA1, giberellin repsonse modulation protein
Length = 587
Score = 147 bits (371), Expect = 7e-36
Identities = 99/293 (33%), Positives = 154/293 (51%), Gaps = 17/293 (5%)
Query: 32 DACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRI 91
+ CPY KFAH TANQAILEA EG +H++DF + QG+QW AL+QA A R G P + R+
Sbjct: 297 ETCPYLKFAHFTANQAILEAFEGKKRVHVIDFSMNQGLQWPALMQALALREGGPP-TFRL 355
Query: 92 SGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTP-ILTPIELLDESSFCIQPD--E 148
+GI A S + G +L++ A+ + + FE+ + + LD S ++P E
Sbjct: 356 TGIGPPAPDNS--DHLHEVGCKLAQLAEAIHVEFEYRGFVANSLADLDASMLELRPSDTE 413
Query: 149 ALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFN 208
A+AVN + +L+ LL +EK L + K + P I T+ E E++ V F++RF + +
Sbjct: 414 AVAVNSVFELHKLLG-RPGGIEKVLGVVKQIKPVIFTVVEQESNHNGPV-FLDRFTESLH 471
Query: 209 YFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENC 263
Y++ F+SLE + + + + + LG++I ++ ER E QW +
Sbjct: 472 YYSTLFDSLE---GVPNSQDKVMSEVYLGKQICNLVACEGPDRVERHETLSQWGNRFGSS 528
Query: 264 GFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
G L A QA +LL ++ Y + ES L L W PL+T S+W
Sbjct: 529 GLAPAHLGSNAFKQASMLLSVFNSGQGYRVEESN-GCLMLGWHTRPLITTSAW 580
>At1g21450 scarecrow-like 1 (SCL1)
Length = 593
Score = 146 bits (369), Expect = 1e-35
Identities = 96/304 (31%), Positives = 155/304 (50%), Gaps = 12/304 (3%)
Query: 21 EELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFAT 80
+E + + L + CP KF L AN AILEA +G +HI+DF I QG Q+ L+++ A
Sbjct: 293 DERLAAMQVLFEVCPCFKFGFLAANGAILEAIKGEEEVHIIDFDINQGNQYMTLIRSIA- 351
Query: 81 RSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTPIELLDES 140
GK +R++GI + + G RL + A+ G++F+F + + ++ S
Sbjct: 352 ELPGKRPRLRLTGIDDPESVQRSIGGLRIIGLRLEQLAEDNGVSFKFKAMPSKTSIVSPS 411
Query: 141 SFCIQPDEALAVNFMLQLYNLLDENTNSV---EKALRLAKSLNPKIVTLGEYEASLTTRV 197
+ +P E L VNF QL+++ DE+ +V ++ L + KSLNPK+VT+ E + + T
Sbjct: 412 TLGCKPGETLIVNFAFQLHHMPDESVTTVNQRDELLHMVKSLNPKLVTVVEQDVNTNTS- 470
Query: 198 GFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMED 252
F RF A+ Y++A FESL+ + +S ER VE L R I ++ ER E
Sbjct: 471 PFFPRFIEAYEYYSAVFESLDMTLPRESQERMNVERQCLARDIVNIVACEGEERIERYEA 530
Query: 253 KEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLT 312
+W+ M GF +S + + L+ Y + Y L E + L W++ L+
Sbjct: 531 AGKWRARMMMAGFNPKPMSAKVTNNIQNLI-KQQYCNKYKLKE-EMGELHFCWEEKSLIV 588
Query: 313 VSSW 316
S+W
Sbjct: 589 ASAW 592
>At1g14920 signal response protein (GAI)
Length = 533
Score = 143 bits (361), Expect = 1e-34
Identities = 94/293 (32%), Positives = 154/293 (52%), Gaps = 17/293 (5%)
Query: 32 DACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAALLQAFATRSSGKPNSVRI 91
+ CPY KFAH TANQAILEA +G +H++DF + QG+QW AL+QA A R G P R+
Sbjct: 245 ETCPYLKFAHFTANQAILEAFQGKKRVHVIDFSMSQGLQWPALMQALALRPGGPP-VFRL 303
Query: 92 SGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTP-ILTPIELLDESSFCIQPD--E 148
+GI A + G +L+ A+ + + FE+ + + LD S ++P E
Sbjct: 304 TGIGPPA--PDNFDYLHEVGCKLAHLAEAIHVEFEYRGFVANTLADLDASMLELRPSEIE 361
Query: 149 ALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVTLGEYEASLTTRVGFVERFETAFN 208
++AVN + +L+ LL +++K L + + P+I T+ E E++ + + F++RF + +
Sbjct: 362 SVAVNSVFELHKLLG-RPGAIDKVLGVVNQIKPEIFTVVEQESNHNSPI-FLDRFTESLH 419
Query: 209 YFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVR-----ERMEDKEQWKVLMENC 263
Y++ F+SLE + S + + + LG++I V+ ER E QW+ +
Sbjct: 420 YYSTLFDSLE---GVPSGQDKVMSEVYLGKQICNVVACDGPDRVERHETLSQWRNRFGSA 476
Query: 264 GFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFLSLAWKDVPLLTVSSW 316
GF + + A QA +LL ++ Y VE L L W PL+ S+W
Sbjct: 477 GFAAAHIGSNAFKQASMLLALFNGGEGYR-VEESDGCLMLGWHTRPLIATSAW 528
>At1g07520 transcription factor scarecrow-like 14, putative
Length = 695
Score = 133 bits (334), Expect = 1e-31
Identities = 94/320 (29%), Positives = 147/320 (45%), Gaps = 13/320 (4%)
Query: 7 QSSIASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIV 66
QS S +S T ++ SY A P+ + +N+ IL+A + ++ +HIVDFGI+
Sbjct: 376 QSYYDSISSKKRTAAQILKSYSVFLSASPFMTLIYFFSNKMILDAAKDASVLHIVDFGIL 435
Query: 67 QGIQWAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFE 126
G QW +Q + + G +RI+GI G P I TG RL+E+ K G+ FE
Sbjct: 436 YGFQWPMFIQHLSKSNPGL-RKLRITGIEIPQHGLRPTERIQDTGRRLTEYCKRFGVPFE 494
Query: 127 FTPILTP-IELLDESSFCIQPDEALAVNFMLQLYNLLD----ENTNSVEKALRLAKSLNP 181
+ I + E + F I+P+E LAVN +L+ NL D E + L+L + +NP
Sbjct: 495 YNAIASKNWETIKMEEFKIRPNEVLAVNAVLRFKNLRDVIPGEEDCPRDGFLKLIRDMNP 554
Query: 182 KIVTLGEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRID 241
+ S F RF+ A +++A F+ ++ ++PER E GR +
Sbjct: 555 NVFLSSTVNGSFNAPF-FTTRFKEALFHYSALFDLFGATLSKENPERIHFEGEFYGREVM 613
Query: 242 GVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVES 296
VI ER E +QW+V M GF+ + + + + + Y + L E
Sbjct: 614 NVIACEGVDRVERPETYKQWQVRMIRAGFKQKPVEAELVQLFREKMKKWGYHKDFVLDED 673
Query: 297 QPAFLSLAWKDVPLLTVSSW 316
FL WK L + S W
Sbjct: 674 SNWFLQ-GWKGRILFSSSCW 692
>At4g37650 SHORT-ROOT (SHR)
Length = 531
Score = 128 bits (321), Expect = 5e-30
Identities = 95/323 (29%), Positives = 152/323 (46%), Gaps = 20/323 (6%)
Query: 11 ASSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQ 70
A++ + ++E + + P++ F H+ AN AILEA +G IHIVD Q
Sbjct: 209 AAATEKTCSFESTRKTVLKFQEVSPWATFGHVAANGAILEAVDGEAKIHIVDISSTFCTQ 268
Query: 71 WAALLQAFATRSSGKPNSVRISGIPAMALGTSPVSS---ISATGNRLSEFAKLLGLNFEF 127
W LL+A ATRS P+ + + A +S + GNR+ +FA+L+G+ F+F
Sbjct: 269 WPTLLEALATRSDDTPHLRLTTVVVANKFVNDQTASHRMMKEIGNRMEKFARLMGVPFKF 328
Query: 128 TPI--LTPIELLDESSFCIQPDEALAVNFMLQLYNLLDENTNSVEKALRLAKSLNPKIVT 185
I + + D + ++PDE LA+N + ++ + + + + + L P+IVT
Sbjct: 329 NIIHHVGDLSEFDLNELDVKPDEVLAINCVGAMHGIASRGSPR-DAVISSFRRLRPRIVT 387
Query: 186 LGEYEASLT--TRVGFVERFETAF----NYFAAFFESLEPNMALDSPERFQVESLLLGRR 239
+ E EA L GF + F F +F FES E + S ER +E GR
Sbjct: 388 VVEEEADLVGEEEGGFDDEFLRGFGECLRWFRVCFESWEESFPRTSNERLMLER-AAGRA 446
Query: 240 IDGVIGVR-----ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLV 294
I ++ ER E +W M N GF +VG S + LL Y ++S+V
Sbjct: 447 IVDLVACEPSDSTERRETARKWSRRMRNSGFGAVGYSDEVADDVRALLRRYK-EGVWSMV 505
Query: 295 E-SQPAFLSLAWKDVPLLTVSSW 316
+ A + L W+D P++ S+W
Sbjct: 506 QCPDAAGIFLCWRDQPVVWASAW 528
>At1g07530 transcription factor scarecrow-like 14, putative
Length = 769
Score = 128 bits (321), Expect = 5e-30
Identities = 87/315 (27%), Positives = 141/315 (44%), Gaps = 19/315 (6%)
Query: 14 NSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQWAA 73
+S T+ ++ +Y+ CP+ K A + AN +++ T +N IHI+DFGI G QW A
Sbjct: 457 SSKKTSAADMLKAYQTYMSVCPFKKAAIIFANHSMMRFTANANTIHIIDFGISYGFQWPA 516
Query: 74 LLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPILTP 133
L+ + G +RI+GI G P + TG+RL+ + + + FE+ I
Sbjct: 517 LIHRLSLSRPGGSPKLRITGIELPQRGFRPAEGVQETGHRLARYCQRHNVPFEYNAIAQK 576
Query: 134 IELLDESSFCIQPDEALAVNFMLQLYNLLDEN--TNSVEKA-LRLAKSLNPKI----VTL 186
E + ++ E + VN + + NLLDE NS A L+L + +NP + +
Sbjct: 577 WETIQVEDLKLRQGEYVVVNSLFRFRNLLDETVLVNSPRDAVLKLIRKINPNVFIPAILS 636
Query: 187 GEYEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGV 246
G Y A FV RF A +++A F+ + +A + R E GR I V+
Sbjct: 637 GNYNAPF-----FVTRFREALFHYSAVFDMCDSKLAREDEMRLMYEKEFYGREIVNVVAC 691
Query: 247 R-----ERMEDKEQWKVLMENCGFESVGLSHYAISQAKILLWNYSYSSLYSLVESQPAFL 301
ER E +QW+ + GF + L + K+ + N Y + V+ +L
Sbjct: 692 EGTERVERPETYKQWQARLIRAGFRQLPLEKELMQNLKLKIEN-GYDKNFD-VDQNGNWL 749
Query: 302 SLAWKDVPLLTVSSW 316
WK + S W
Sbjct: 750 LQGWKGRIVYASSLW 764
>At4g17230 scarecrow-like 13 (SCL13)
Length = 287
Score = 124 bits (312), Expect = 5e-29
Identities = 84/251 (33%), Positives = 125/251 (49%), Gaps = 6/251 (2%)
Query: 12 SSNSSSTTWEELTLSYKALNDACPYSKFAHLTANQAILEATEGSNNIHIVDFGIVQGIQW 71
S + T EL L + CPY KFA+ TAN ILEA G +HI+DF I QG Q+
Sbjct: 34 SLKCNEPTGRELMSYMSVLYEICPYWKFAYTTANVEILEAIAGETRVHIIDFQIAQGSQY 93
Query: 72 AALLQAFATRSSGKPNSVRISGIPAMALGTSPVSSISATGNRLSEFAKLLGLNFEFTPIL 131
L+Q A G P +R++G+ + +S G RL+ A+ G+ FEF +
Sbjct: 94 MFLIQELAKHPGGPP-LLRVTGVDDSQSTYARGGGLSLVGERLATLAQSCGVPFEFHDAI 152
Query: 132 TPIELLDESSFCIQPDEALAVNFMLQLYNLLDENT---NSVEKALRLAKSLNPKIVTLGE 188
+ ++P A+ VNF L+++ DE+ N ++ L L KSL+PK+VTL E
Sbjct: 153 MSGCKVQREHLGLEPGFAVVVNFPYVLHHMPDESVSVENHRDRLLHLIKSLSPKLVTLVE 212
Query: 189 YEASLTTRVGFVERFETAFNYFAAFFESLEPNMALDSPERFQVESLLLGRRIDGVIGVRE 248
E++ T F+ RF +Y+ A FES++ D +R E + R I +I E
Sbjct: 213 QESNTNTS-PFLSRFVETLDYYTAMFESIDAARPRDDKQRISAEQHCVARDIVNMIAC-E 270
Query: 249 RMEDKEQWKVL 259
E E+ +VL
Sbjct: 271 ESERVERHEVL 281
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.131 0.372
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,498,688
Number of Sequences: 26719
Number of extensions: 258078
Number of successful extensions: 810
Number of sequences better than 10.0: 37
Number of HSP's better than 10.0 without gapping: 31
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 673
Number of HSP's gapped (non-prelim): 40
length of query: 316
length of database: 11,318,596
effective HSP length: 99
effective length of query: 217
effective length of database: 8,673,415
effective search space: 1882131055
effective search space used: 1882131055
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144806.12


