

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144806.10 + phase: 0
(407 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
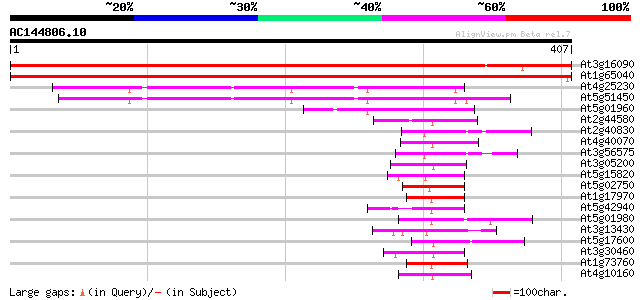
At3g16090 putative zinc finger protein 628 e-180
At1g65040 610 e-175
At4g25230 unknown protein 111 8e-25
At5g51450 unknown protein 101 6e-22
At5g01960 unknown protein 67 1e-11
At2g44580 putative RING zinc finger protein 60 3e-09
At2g40830 unknown protein 58 1e-08
At4g40070 unknown protein 54 1e-07
At3g56575 unknown protein 54 1e-07
At3g05200 RING-H2 zinc finger protein ATL6 (ATL6) 53 3e-07
At5g15820 putative protein 52 4e-07
At5g02750 unknown protein 52 4e-07
At1g17970 unknown protein 52 8e-07
At5g42940 unknown protein 51 1e-06
At5g01980 unknown protein 50 2e-06
At3g13430 unknown protein 50 2e-06
At5g17600 RING-H2 zinc finger protein-like 50 2e-06
At3g30460 RING zinc finger protein 50 2e-06
At1g73760 putative RING zinc finger protein 50 2e-06
At4g10160 unknown protein (At4g10160) 50 3e-06
>At3g16090 putative zinc finger protein
Length = 492
Score = 628 bits (1619), Expect = e-180
Identities = 311/410 (75%), Positives = 355/410 (85%), Gaps = 4/410 (0%)
Query: 1 MMRLQTYAGLSLIATLAITYHAFNSRSQFYPAMVYLATSKISLVLLLNMALVIMCVLWQL 60
M+RL+TYAGLS +ATLA+ YHAF+SR QFYPA VYL+TSKISLVLLLNM LV+M LW L
Sbjct: 1 MIRLRTYAGLSFMATLAVIYHAFSSRGQFYPATVYLSTSKISLVLLLNMCLVLMLSLWHL 60
Query: 61 TKKVFLGSLREAEVERLNEQAWREVMEILFAITIFRQDFSITFLAMVTALLLIKALHWLA 120
K VFLGSLREAEVERLNEQAWRE+MEILFAITIFRQDFS FL +V LLLIKALHWLA
Sbjct: 61 VKFVFLGSLREAEVERLNEQAWRELMEILFAITIFRQDFSSGFLPLVVTLLLIKALHWLA 120
Query: 121 QKRVEYIETTPSVSVLSQVRIVSFLGFLLLLDGAFLYSSVKHLLATKQASVSIFFSFEYM 180
QKRVEYIETTPSVS LS RIVSF+GFLLL+D F+YSS++HL+ ++QASVS+FFSFEYM
Sbjct: 121 QKRVEYIETTPSVSKLSHFRIVSFMGFLLLVDSLFMYSSIRHLIQSRQASVSLFFSFEYM 180
Query: 181 ILATTTVSIFVKYIFYVSDMLMEGQWEKKPVFTFYLELVRDLLHLSMYMCFFLAIFINYG 240
ILATTTV+IFVKY+FYV+DMLM+GQWEKKPV+TFYLEL+RDLLHLSMY+CFF IF+NYG
Sbjct: 181 ILATTTVAIFVKYVFYVTDMLMDGQWEKKPVYTFYLELIRDLLHLSMYICFFFVIFMNYG 240
Query: 241 VPLHLIRELYETFRNFKVRISDYLRYRKITSNMNDRFPDATPEELNSNDLTCIICREEMT 300
VPLHL+RELYETFRNF++R+SDYLRYRKITSNMNDRFPDATPEEL ++D TCIICREEMT
Sbjct: 241 VPLHLLRELYETFRNFQIRVSDYLRYRKITSNMNDRFPDATPEELTASDATCIICREEMT 300
Query: 301 TAKKLICGHLFHVHCLRSWLERQHTCPTCRALVVPSENGPTATGGQQRPQSDANRQGEGT 360
AKKLICGHLFHVHCLRSWLERQ TCPTCRALVVP EN +A G QR ++QG +
Sbjct: 301 NAKKLICGHLFHVHCLRSWLERQQTCPTCRALVVPPENATSAAPG-QRELHQGSQQGTSS 359
Query: 361 GSSAQTEVGD--GVARDNLSRHQARLQAAAAAASIYEKSYVYPSP-TFAW 407
+ +E+ GV+ ++LSRH ARLQAAA+AAS+Y KS VYPS T AW
Sbjct: 360 SGNQGSEISSSAGVSNNSLSRHHARLQAAASAASVYGKSMVYPSANTVAW 409
>At1g65040
Length = 537
Score = 610 bits (1573), Expect = e-175
Identities = 298/410 (72%), Positives = 350/410 (84%), Gaps = 3/410 (0%)
Query: 1 MMRLQTYAGLSLIATLAITYHAFNSRSQFYPAMVYLATSKISLVLLLNMALVIMCVLWQL 60
M++L+ YAGLS +ATL + YHAF+SR QFYPA VYL+TSKI+LV+LLNM LV+M LW L
Sbjct: 1 MIQLKVYAGLSTLATLVVIYHAFSSRGQFYPATVYLSTSKINLVVLLNMGLVLMLSLWNL 60
Query: 61 TKKVFLGSLREAEVERLNEQAWREVMEILFAITIFRQDFSITFLAMVTALLLIKALHWLA 120
K VFLGSLREAEVERLNEQAWRE+MEILFAITIFRQDFS+ F+++V LLLIK LHW+A
Sbjct: 61 VKIVFLGSLREAEVERLNEQAWRELMEILFAITIFRQDFSVGFISLVVTLLLIKGLHWMA 120
Query: 121 QKRVEYIETTPSVSVLSQVRIVSFLGFLLLLDGAFLYSSVKHLLATKQASVSIFFSFEYM 180
QKRVEYIETTPSV++LS VRIVSF+ FLL+LD YSS++ L+ +++AS+S+FF+FEYM
Sbjct: 121 QKRVEYIETTPSVTLLSHVRIVSFMVFLLILDCLLTYSSIQQLIQSRKASMSVFFTFEYM 180
Query: 181 ILATTTVSIFVKYIFYVSDMLMEGQWEKKPVFTFYLELVRDLLHLSMYMCFFLAIFINYG 240
ILATTTVSI VKY FYV+DML EGQWE KPV+TFYLELVRDLLHLSMY+CFFL IF+NYG
Sbjct: 181 ILATTTVSIIVKYAFYVTDMLKEGQWEGKPVYTFYLELVRDLLHLSMYLCFFLMIFMNYG 240
Query: 241 VPLHLIRELYETFRNFKVRISDYLRYRKITSNMNDRFPDATPEELNSNDLTCIICREEMT 300
+PLHLIRELYETFRNFK+R++DYLRYRKITSNMNDRFPDATPEEL+SND TCIICREEMT
Sbjct: 241 LPLHLIRELYETFRNFKIRVTDYLRYRKITSNMNDRFPDATPEELSSNDATCIICREEMT 300
Query: 301 TAKKLICGHLFHVHCLRSWLERQHTCPTCRALVVPSENGPTATGGQQRPQSDANRQGEGT 360
+AKKL+CGHLFHVHCLRSWLERQ+TCPTCRALVVP+EN + G + P ++ +QG GT
Sbjct: 301 SAKKLVCGHLFHVHCLRSWLERQNTCPTCRALVVPAENATSTASGNRGPHQESLQQGTGT 360
Query: 361 GSS-AQTEVGDGVARDNLSRHQARLQAAAAAASIYEKSYVYPSP--TFAW 407
SS Q A +N+SRH+AR QAAA+AASIY +S VYPS T W
Sbjct: 361 SSSDGQGSSVSAAASENMSRHEARFQAAASAASIYGRSIVYPSSANTLVW 410
>At4g25230 unknown protein
Length = 578
Score = 111 bits (277), Expect = 8e-25
Identities = 92/337 (27%), Positives = 153/337 (45%), Gaps = 44/337 (13%)
Query: 32 AMVYLATSKISLVLLLNMALVIMCVLWQLTKKVFLGSLREAEVERLNEQAWREV------ 85
A+ L S ++ LL N L + +L K +F G L + E ++L E+ +
Sbjct: 49 ALELLLGSYFTIALLTNFVLNVYILLVLSLKTLFFGDLYDVETKKLVERLANYIIYKGTF 108
Query: 86 MEILFAITIFRQDFSITFLAMVTALLLIKALHWLAQKRVEYIETTPSVSVLSQVRIVSFL 145
+ ++ TIF+ + + +T L +K LA+ R+E + +PS + + R+ S L
Sbjct: 109 LPLVIPPTIFQ---GVLWTVWLTVLCTLKMFQALARDRLERLNASPSSTPWTYFRVYSVL 165
Query: 146 GFLLLLDGAFLYSSVKHLLATKQASVSIFFSFEYMILATTTVSIFVKYIFYVSDMLME-- 203
+L +D ++ S+ T ++V + FE +A T+ + + F + DM +
Sbjct: 166 FLVLSVDMFWIKLSLM-TYNTIGSAVYLLLLFEPCSIAFETLQALLIHGFQLLDMWINHL 224
Query: 204 -------------------GQWEKKPVFTFYLELVRDLLHLSMYMCFFLAIFINYGVPLH 244
E K + L D+ L M + +L I+ +G+ H
Sbjct: 225 AVKNSDCQRSKFIDSMTAGSLLEWKGLLNRNLGFFLDMATLVMALGHYLHIWWLHGIAFH 284
Query: 245 LIRELYETFRNFKV-------RISDYLRYRKITSNMNDRFPDATPEELNSNDLTCIICRE 297
L+ + F N + RI Y++ R ++ PDAT EEL + D C ICRE
Sbjct: 285 LVDAVL--FLNIRALLSAILKRIKGYIKLRIALGALHAALPDATSEELRAYDDECAICRE 342
Query: 298 EMTTAKKLICGHLFHVHCLRSWLER----QHTCPTCR 330
M AK+L C HLFH+ CLRSWL++ ++CPTCR
Sbjct: 343 PMAKAKRLHCNHLFHLGCLRSWLDQGLNEVYSCPTCR 379
>At5g51450 unknown protein
Length = 577
Score = 101 bits (252), Expect = 6e-22
Identities = 96/368 (26%), Positives = 159/368 (43%), Gaps = 46/368 (12%)
Query: 36 LATSKISLVLLLNMALVIMCVLWQLTKKVFLGSLREAEVERLNEQAWREV------MEIL 89
L +S ++ LL + L I +L K +F G L E +L E+ + + +
Sbjct: 53 LLSSHTTIALLASFVLNIYILLVLSLKTLFFGDLYAIETRKLVERLANYIIYKGTFLPFV 112
Query: 90 FAITIFRQDFSITFLAMVTALLLIKALHWLAQKRVEYIETTPSVSVLSQVRIVSFLGFLL 149
T+F+ + + +T L +K LA+ R++ + +PS + + R+ S L +L
Sbjct: 113 VPRTVFQ---GVLWTIWLTVLCTLKMFQALARDRLDRLNASPSSTPWTYFRVYSALFMVL 169
Query: 150 LLDGAFLYSSVKHLLATKQASVSIFFSFEYMILATTTVSIFVKYIFYVSDMLME------ 203
D ++ S+ + T +SV + FE +A T+ + + F + DM +
Sbjct: 170 STDLCWIKLSLM-IYNTVGSSVYLLLLFEPCGIAFETLQALLIHGFQLLDMWINHLAVKN 228
Query: 204 ---------------GQWEKKPVFTFYLELVRDLLHLSMYMCFFLAIFINYGVPLHLIRE 248
E K + L D+ L M + +L I+ +G+ HL+
Sbjct: 229 SDCQRSKFYDSMTAGSLLEWKGLLNRNLGFFLDMATLVMALGHYLHIWWLHGMAFHLVDA 288
Query: 249 LYETFRNFKV-------RISDYLRYRKITSNMNDRFPDATPEELNSNDLTCIICREEMTT 301
+ F N + RI Y++ R ++ DAT EEL D C ICRE M
Sbjct: 289 VL--FLNIRALLSSILKRIKGYIKLRVALGALHAALLDATSEELRDYDDECAICREPMAK 346
Query: 302 AKKLICGHLFHVHCLRSWLER----QHTCPTCR--ALVVPSENGPTATGGQQRPQSDANR 355
AK+L C HLFH+ CLRSWL++ ++CPTCR V +E+ + G+ R
Sbjct: 347 AKRLHCNHLFHLGCLRSWLDQGLNEVYSCPTCRKPLFVGRTESEANPSRGEVSSDEHLAR 406
Query: 356 QGEGTGSS 363
Q E +S
Sbjct: 407 QFERQNNS 414
>At5g01960 unknown protein
Length = 426
Score = 67.4 bits (163), Expect = 1e-11
Identities = 44/128 (34%), Positives = 66/128 (51%), Gaps = 6/128 (4%)
Query: 214 FYLELVRDLLHLSMYMCFFLAIFINYGVPLH-LIRELYETFRNFKV--RISDYLRYRKIT 270
+++ L R LL ++ FFL +YG L+ LY TF+ V ++ + K
Sbjct: 285 YFMLLYRSLLPTPVWYRFFLNK--DYGSLFSSLMTGLYLTFKLTSVVEKVQSFFTALKAL 342
Query: 271 SNMNDRFPD-ATPEELNSNDLTCIICREEMTTAKKLICGHLFHVHCLRSWLERQHTCPTC 329
S + AT E++N+ C IC+E+M T L C H+F C+ W ER+ TCP C
Sbjct: 343 SRKEVHYGSYATTEQVNAAGDLCAICQEKMHTPILLRCKHMFCEDCVSEWFERERTCPLC 402
Query: 330 RALVVPSE 337
RALV P++
Sbjct: 403 RALVKPAD 410
>At2g44580 putative RING zinc finger protein
Length = 624
Score = 59.7 bits (143), Expect = 3e-09
Identities = 31/79 (39%), Positives = 41/79 (51%), Gaps = 5/79 (6%)
Query: 265 RYRKITSNMNDRFPDATPEELNSNDLTCIICREEMTTAKKL----ICGHLFHVHCLRSWL 320
RYR T + R+ T EE +S+ C IC E T +K+ C H FHV C+ WL
Sbjct: 523 RYRTFTVHHRRRWRKTTAEEKHSSPY-CTICLENATEGEKMRRIAACSHCFHVDCIDPWL 581
Query: 321 ERQHTCPTCRALVVPSENG 339
E++ CP CRA + P G
Sbjct: 582 EKKSMCPLCRAEIPPVPPG 600
Score = 55.8 bits (133), Expect = 4e-08
Identities = 29/79 (36%), Positives = 39/79 (48%), Gaps = 5/79 (6%)
Query: 265 RYRKITSNMNDRFPDATPEELNSNDLTCIICREEMTTAKKL----ICGHLFHVHCLRSWL 320
RYR T R+ EE +S+ C IC E+ +K+ C H FHV C+ WL
Sbjct: 372 RYRTFTVQHRRRWRKTAAEEKHSSPY-CTICLEDAAEGEKMRRITACSHCFHVDCIDPWL 430
Query: 321 ERQHTCPTCRALVVPSENG 339
++ TCP CRA + P G
Sbjct: 431 MKKSTCPLCRAEIPPVPPG 449
>At2g40830 unknown protein
Length = 328
Score = 57.8 bits (138), Expect = 1e-08
Identities = 29/97 (29%), Positives = 52/97 (52%), Gaps = 6/97 (6%)
Query: 285 LNSNDLTCIICREEM---TTAKKLICGHLFHVHCLRSWLERQHTCPTCRALVVPSENGPT 341
L S+D C +C++E + AK++ C H++H C+ WL + ++CP CR +PS +GP+
Sbjct: 183 LRSSDSNCPVCKDEFELGSEAKQMPCNHIYHSDCIVPWLVQHNSCPVCRQ-ELPSASGPS 241
Query: 342 ATGGQQRPQSDANRQGEGTGSSAQTEVGDGVARDNLS 378
++ Q R N + + SS+ + R+ S
Sbjct: 242 SS--QNRTTPTRNYRSSSSSSSSNSRENGNERRNPFS 276
>At4g40070 unknown protein
Length = 322
Score = 54.3 bits (129), Expect = 1e-07
Identities = 23/61 (37%), Positives = 33/61 (53%), Gaps = 4/61 (6%)
Query: 284 ELNSNDLTCIICREEMTTAKKL----ICGHLFHVHCLRSWLERQHTCPTCRALVVPSENG 339
++ S DL C IC E+ + + IC HLFH+ C+ +WL TCP CR+ + N
Sbjct: 115 KIGSKDLECAICLNELEDHETVRLLPICNHLFHIDCIDTWLYSHATCPVCRSNLTAKSNK 174
Query: 340 P 340
P
Sbjct: 175 P 175
>At3g56575 unknown protein
Length = 320
Score = 54.3 bits (129), Expect = 1e-07
Identities = 28/91 (30%), Positives = 48/91 (51%), Gaps = 10/91 (10%)
Query: 281 TPEELNSNDLTCIICREEM---TTAKKLICGHLFHVHCLRSWLERQHTCPTCRALVVPSE 337
T + L S+D C +C++E + AK++ C H++H C+ WL + ++CP CR +PS
Sbjct: 175 TQKHLKSSDSHCPVCKDEFELKSEAKQMPCHHIYHSDCIVPWLVQHNSCPVCRK-ELPSR 233
Query: 338 NGPTATGGQQRPQSDANRQGEGTGSSAQTEV 368
++T QS NR G +S + +
Sbjct: 234 GSSSST------QSSQNRSTNGRENSRRRNI 258
>At3g05200 RING-H2 zinc finger protein ATL6 (ATL6)
Length = 398
Score = 53.1 bits (126), Expect = 3e-07
Identities = 23/59 (38%), Positives = 31/59 (51%), Gaps = 4/59 (6%)
Query: 277 FPDATPEELNSNDLTCIICREEMTTAKKLI----CGHLFHVHCLRSWLERQHTCPTCRA 331
+ D ++L +L C IC E + L C H+FH HC+ +WLE TCP CRA
Sbjct: 113 YSDVKTQKLGKGELECAICLNEFEDDETLRLLPKCDHVFHPHCIDAWLEAHVTCPVCRA 171
>At5g15820 putative protein
Length = 348
Score = 52.4 bits (124), Expect = 4e-07
Identities = 25/61 (40%), Positives = 35/61 (56%), Gaps = 5/61 (8%)
Query: 275 DRFPDA--TPEELNSNDLTCIICREEMT---TAKKLICGHLFHVHCLRSWLERQHTCPTC 329
D PD T EEL+S + C IC++E+ K+L C H +H C+ WL ++TCP C
Sbjct: 272 DGLPDVELTIEELSSVSIVCAICKDEVVFKEKVKRLPCKHYYHGECIIPWLGIRNTCPVC 331
Query: 330 R 330
R
Sbjct: 332 R 332
>At5g02750 unknown protein
Length = 283
Score = 52.4 bits (124), Expect = 4e-07
Identities = 19/48 (39%), Positives = 32/48 (66%), Gaps = 3/48 (6%)
Query: 286 NSNDLTCIICREEMTTAK---KLICGHLFHVHCLRSWLERQHTCPTCR 330
N+ ++ C+IC+EEM+ + ++ C H FH C+ WL +++TCP CR
Sbjct: 208 NAGEVECVICKEEMSEGRDVCEMPCQHFFHWKCILPWLSKKNTCPFCR 255
>At1g17970 unknown protein
Length = 368
Score = 51.6 bits (122), Expect = 8e-07
Identities = 20/45 (44%), Positives = 28/45 (61%), Gaps = 3/45 (6%)
Query: 289 DLTCIICREEMTTAKK---LICGHLFHVHCLRSWLERQHTCPTCR 330
D C IC++E + L CGH FHVHC++ WL R++ CP C+
Sbjct: 317 DRKCSICQDEYEREDEVGELNCGHSFHVHCVKQWLSRKNACPVCK 361
>At5g42940 unknown protein
Length = 691
Score = 50.8 bits (120), Expect = 1e-06
Identities = 27/74 (36%), Positives = 39/74 (52%), Gaps = 13/74 (17%)
Query: 260 ISDYLRYRKITSNMNDRFPDATPEELNSNDLTCIICREEMTTAKK---LICGHLFHVHCL 316
IS+ L+ RK SN DA P C +C+EE T + L CGH FH C+
Sbjct: 615 ISNRLKQRKYKSNTKSP-QDAEP---------CCVCQEEYTEGEDMGTLECGHEFHSQCI 664
Query: 317 RSWLERQHTCPTCR 330
+ WL++++ CP C+
Sbjct: 665 KEWLKQKNLCPICK 678
>At5g01980 unknown protein
Length = 493
Score = 50.4 bits (119), Expect = 2e-06
Identities = 30/102 (29%), Positives = 49/102 (47%), Gaps = 6/102 (5%)
Query: 283 EELNSNDLTCIICREEMTTAKK---LICGHLFHVHCLRSWLERQHTCPTCRALVVPSENG 339
EE L C IC+E + + L C HL+H HC+ WL +++CP CR +P+++
Sbjct: 341 EEHVMKGLVCAICKELFSLRNETTQLPCLHLYHAHCIVPWLSARNSCPLCR-YELPTDDK 399
Query: 340 PTATGGQQ--RPQSDANRQGEGTGSSAQTEVGDGVARDNLSR 379
G + D++ +GT S + V G + +SR
Sbjct: 400 DYEEGKRNVLDVSEDSSSSDDGTESGEEEYVERGESESGVSR 441
>At3g13430 unknown protein
Length = 315
Score = 50.4 bits (119), Expect = 2e-06
Identities = 32/103 (31%), Positives = 49/103 (47%), Gaps = 21/103 (20%)
Query: 264 LRYRKITSNMNDRF--PDATPE--------ELNSNDLTCIICREEMTT---AKKLICGHL 310
L R +++N+R+ P AT E ++ + L C +C ++ AK++ C H
Sbjct: 187 LLQRLAENDLNNRYGTPPATKEAVEALAMVKIEDSLLQCSVCLDDFEIGMEAKEMPCKHK 246
Query: 311 FHVHCLRSWLERQHTCPTCRALVVPSENGPTATGGQQRPQSDA 353
FH CL WLE +CP CR L+ TG P++DA
Sbjct: 247 FHSDCLLPWLELHSSCPVCRYLL--------PTGDDDEPKTDA 281
>At5g17600 RING-H2 zinc finger protein-like
Length = 362
Score = 50.1 bits (118), Expect = 2e-06
Identities = 27/86 (31%), Positives = 40/86 (46%), Gaps = 5/86 (5%)
Query: 292 CIICREEMTTAKKLI----CGHLFHVHCLRSWLERQHTCPTCRALVVPSENGPTATGGQQ 347
C +C E + L C H FH+ C+ +WL+ CP CRA V N PTA+ GQ
Sbjct: 142 CSVCLSEFEENESLRLLPKCNHAFHLPCIDTWLKSHSNCPLCRAFVT-GVNNPTASVGQN 200
Query: 348 RPQSDANRQGEGTGSSAQTEVGDGVA 373
AN+ + + +E+ +A
Sbjct: 201 VSVVVANQSNSAHQTGSVSEINLNLA 226
>At3g30460 RING zinc finger protein
Length = 147
Score = 50.1 bits (118), Expect = 2e-06
Identities = 24/65 (36%), Positives = 35/65 (52%), Gaps = 6/65 (9%)
Query: 272 NMNDRFP--DATPEELNSNDLT-CIICREEMTTAKKLI---CGHLFHVHCLRSWLERQHT 325
N + P + T EE+ L C ICREE+ ++L C H +H C+ +WL ++T
Sbjct: 74 NAGEELPVVEFTAEEMMERGLVVCAICREELAANERLSELPCRHYYHKECISNWLSNRNT 133
Query: 326 CPTCR 330
CP CR
Sbjct: 134 CPLCR 138
>At1g73760 putative RING zinc finger protein
Length = 367
Score = 50.1 bits (118), Expect = 2e-06
Identities = 19/47 (40%), Positives = 29/47 (61%), Gaps = 3/47 (6%)
Query: 289 DLTCIICREEMTTAKK---LICGHLFHVHCLRSWLERQHTCPTCRAL 332
D CIIC++E + L CGH FH+ C+ WL R+++CP C+ +
Sbjct: 316 DRKCIICQDEYEAKDEVGELRCGHRFHIDCVNQWLVRKNSCPVCKTM 362
>At4g10160 unknown protein (At4g10160)
Length = 225
Score = 49.7 bits (117), Expect = 3e-06
Identities = 21/57 (36%), Positives = 28/57 (48%), Gaps = 4/57 (7%)
Query: 283 EELNSNDLTCIICREEMTTAKKL----ICGHLFHVHCLRSWLERQHTCPTCRALVVP 335
E ND C +C + +KL CGH FH+ C+ WL TCP CR ++P
Sbjct: 88 ESFTVNDTQCSVCLGDYQAEEKLQQMPSCGHTFHMECIDLWLTSHTTCPLCRLSLIP 144
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.137 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,316,868
Number of Sequences: 26719
Number of extensions: 336136
Number of successful extensions: 1553
Number of sequences better than 10.0: 311
Number of HSP's better than 10.0 without gapping: 235
Number of HSP's successfully gapped in prelim test: 76
Number of HSP's that attempted gapping in prelim test: 1250
Number of HSP's gapped (non-prelim): 337
length of query: 407
length of database: 11,318,596
effective HSP length: 102
effective length of query: 305
effective length of database: 8,593,258
effective search space: 2620943690
effective search space used: 2620943690
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144806.10


