

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144806.1 + phase: 0
(260 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
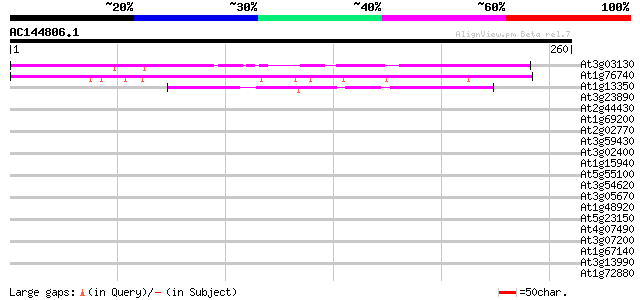
Score E
Sequences producing significant alignments: (bits) Value
At3g03130 hypothetical protein 109 2e-24
At1g76740 hypothetical protein 48 5e-06
At1g13350 Unknown protein 41 6e-04
At3g23890 topoisomerase II 39 0.002
At2g44430 unknown protein 37 0.008
At1g69200 putative fructokinase 35 0.040
At2g02770 unknown protein 35 0.053
At3g59430 hypothetical protein 34 0.090
At3g02400 hypothetical protein 33 0.12
At1g15940 T24D18.4 33 0.12
At5g55100 unknown protein 33 0.15
At3g54620 bZIP transcription factor-like protein 33 0.20
At3g05670 unknown protein 33 0.20
At1g48920 unknown protein 32 0.26
At5g23150 transcription factor-like protein (gb|AAD31171.1) 32 0.34
At4g07490 putative transposon protein 32 0.34
At3g07200 putative RING zinc finger protein 32 0.34
At1g67140 hypothetical protein 32 0.34
At3g13990 unknown protein 31 0.58
At1g72880 unknown protein 31 0.58
>At3g03130 hypothetical protein
Length = 520
Score = 109 bits (272), Expect = 2e-24
Identities = 84/245 (34%), Positives = 127/245 (51%), Gaps = 32/245 (13%)
Query: 1 MDFHTLSRKELQALSKMNKIPANITNVAMADALSALPHVEGLDEILN-QREGGDIGTPAV 59
MDFH+L R++LQ K NKIPAN+TN+AMADAL L VEG+DE ++ R+
Sbjct: 1 MDFHSLPRRDLQFFCKRNKIPANMTNIAMADALRDLEIVEGMDEFMDPSRDQSPTSVARN 60
Query: 60 QP---RTARRTTTQRKPVKEAESTKVSTRVNRGGRGGVAEGEVEQENLDANVDAGTPAVV 116
P RTA RTT ++ E +S+++ TR +A GE++QEN D N+ P+ V
Sbjct: 61 LPSAARTAARTTRRKSTKDETQSSELVTRSCYVVSKSLA-GEMDQENKDMNM-LQNPS-V 117
Query: 117 PTSRRRVPAVSTRRKKEVIVIEDEDDVVSEVQGKATDVAKTPAAAPSSRTRAGRSVRNKT 176
P SR V D +D++ E +V+KTPAA + R +A S +
Sbjct: 118 PQSR--------------AVKLDVNDIMPE-----ANVSKTPAARSTRRAQAAASSKK-- 156
Query: 177 EISDGTSVQKAYSTRRSVRLVGKSLSKMSLADTEDMESTKNDDVSEEMSVSQNEGGSIET 236
SVQ+ YSTRRSVRL+ +S++ +SL ++ ++ + +E
Sbjct: 157 ----DESVQRVYSTRRSVRLLEESMADLSLKTNVPVKKHEDSPAGSKFQAKSDENSENTD 212
Query: 237 ENGML 241
+ G++
Sbjct: 213 KGGVM 217
>At1g76740 hypothetical protein
Length = 1532
Score = 48.1 bits (113), Expect = 5e-06
Identities = 61/288 (21%), Positives = 120/288 (41%), Gaps = 46/288 (15%)
Query: 1 MDFHTLSRKELQALSKMNKIPANITNVAMADALSAL--PHVEG-LDEILNQREGG----- 52
M+FH + RK+LQAL K N IPAN+ N+ MA L+++ EG L + ++ E
Sbjct: 1 MNFHGMKRKDLQALCKKNGIPANLKNIEMATRLASVLQKETEGTLTDTISLSEDSVDVLE 60
Query: 53 -DIGTPAVQ--------------PRTARRTTTQRKPVKEAESTKVSTRVNRGGRGGVAEG 97
D+ V+ R R+ ++RK + + S R G+
Sbjct: 61 DDVAVKKVRFSPENEVFEFTRSLKRCQRKYVSKRKVQESGIELRRSKRSVAKGKAIAGSS 120
Query: 98 EVEQENLDANVDAGTPAV-VPTSRRRVPAVSTRRK----KEVIVIE-------------- 138
E E N T A+ VP +R + + ++ E++ +E
Sbjct: 121 ENESNTRRGNRSDVTKAIEVPRRSKRSGSTGSSQEDCKVTELMALEQFEFENISEGLSGS 180
Query: 139 DEDDVVSEVQGKATD--VAKTPAAAPSSRTRAGRSVR-NKTEISDGTSVQKAYSTRRSVR 195
D+D E G+ + ++KT P ++++ ++ N+ E+ + + RRS+R
Sbjct: 181 DQDGFKFEKIGRRSTQLISKTGNLLPVNKSKRLVDIKDNEDELVKNNVNEVSMDQRRSIR 240
Query: 196 LVGKSLSKMSLADTED-MESTKNDDVSEEMSVSQNEGGSIETENGMLI 242
L ++ ++ ++ D + EE+ + Q++ ++ + L+
Sbjct: 241 LKARAANQSMGGESSDGRPLCSAKETKEEIKIKQSKRSTVVSSQKELV 288
Score = 27.7 bits (60), Expect = 6.4
Identities = 33/182 (18%), Positives = 76/182 (41%), Gaps = 7/182 (3%)
Query: 62 RTARRTTTQRKPVKEAESTKVSTRVNRGGRGGVAEGEVEQENLDANVDAGTPAVVPTSRR 121
R+++ T + + ++ V G R GV+ G+V++ N++ + +SR
Sbjct: 374 RSSKHTASNNMLGQALSNSLKRCSVIDGERAGVSVGDVKKSNVNNEMQIRVRVTRNSSRH 433
Query: 122 RVPAVSTRRKKEVIVIEDEDDVVSEVQGKATDVAKTPAAAPSSRTRAGRSVRNKTEISDG 181
++ +I + ++ V + + + +A P ++ +K +++
Sbjct: 434 HSFKAPIEFERNLIEKKSDETFVKSSKRGTRRMKRGRSAEPGKDIETQSNLTSKKLLNEY 493
Query: 182 TSVQKAYSTRRSVRLVGKSLSKMSLADTEDMESTKN-DDVSEEMSVSQNEGGSIETENGM 240
T ++ + +V + G S A T + E K+ V E S S +E +I E+ M
Sbjct: 494 TQFEQEEACDANVEVRGSS----KKAKTMNQECVKDIPQVMTENSPSSSE--TIAAESVM 547
Query: 241 LI 242
++
Sbjct: 548 IV 549
>At1g13350 Unknown protein
Length = 761
Score = 41.2 bits (95), Expect = 6e-04
Identities = 41/164 (25%), Positives = 69/164 (42%), Gaps = 27/164 (16%)
Query: 74 VKEAESTKVSTRVNRGGRGGVAEGEVEQENLDANVDAGTPAVVPTSRRRVPAVSTRRKK- 132
VKE TR++ +G V EVEQE+ + N ++ SRRR A+ + KK
Sbjct: 237 VKETRGNVERTRIDNDDKGDVVVWEVEQEDEELN-------LIEESRRRTQAIMEKYKKK 289
Query: 133 ------------EVIVIEDEDDVVSEVQGKATDVAKTPAAAPSSRTRAGRSVRNKTEISD 180
E+ I + V++V G T P + ++ +AG + + D
Sbjct: 290 LEQQNGFSSHDLELANIPKQSSTVADVLGSGT---LGPVTSAVNQAKAGLDI----DAVD 342
Query: 181 GTSVQKAYSTRRSVRLVGKSLSKMSLADTEDMESTKNDDVSEEM 224
G + + + S + S S +LA T E + D +S++M
Sbjct: 343 GEVAKLSSAVGESPAQLVISDSDRTLASTGLGEGSPKDKISDDM 386
>At3g23890 topoisomerase II
Length = 1473
Score = 39.3 bits (90), Expect = 0.002
Identities = 39/117 (33%), Positives = 56/117 (47%), Gaps = 16/117 (13%)
Query: 128 TRRKKEVIVI-EDEDDVVSEV--------QGKATDVAKTPAAAPSSRTRAGRSVRNKTEI 178
T + I + +D+DDVV EV + AT AK PAA R R ++V + +
Sbjct: 1322 TAETSKAIAVDDDDDDVVVEVAPVKKGGRKPAATKAAKPPAA---PRKRGKQTVASTEVL 1378
Query: 179 SDGTSVQKAYSTRRSVRLVGKSLSKMS-LADTEDMESTKNDDVSEEMSVSQNEGGSI 234
+ G S +K RS KS S MS LAD ++ ES++N S S+ GG +
Sbjct: 1379 AIGVSPEKKVRKMRSSPFNKKSSSVMSRLADNKEEESSEN---VAGNSSSEKSGGDV 1432
>At2g44430 unknown protein
Length = 646
Score = 37.4 bits (85), Expect = 0.008
Identities = 38/164 (23%), Positives = 69/164 (41%), Gaps = 20/164 (12%)
Query: 89 GGRGGVAEGEVEQENLDANVDAGTPAVVPTSRRRVPAVSTRRK---------KEVIVIED 139
G R G A+ E +L +G P VV RR V A ++ KE + E+
Sbjct: 423 GMRSGKADAETSDSSLSRQKSSG-PLVVCKKRRSVSAKASPSSSSFSQKDDTKEETLSEE 481
Query: 140 EDDVVSEVQGKATDVAKTPAAAPSSRTRAGRSVRNKTE-----ISDGTSVQKAYSTRRSV 194
+D++ + V+ A +++T GR+ + +TE +D +S Q T +
Sbjct: 482 KDNIATGVRSSRRANKVAAVVANNTKTGKGRNKQKQTESKTNSSNDNSSKQDTGKTEKKT 541
Query: 195 RLVGKSLSKMSLADTEDMESTKNDDVSEEMSVSQNEGGSIETEN 238
K K S+AD ++ K + +E GG+++ ++
Sbjct: 542 VSADK---KKSVADF--LKRLKKNSPQKEAKDQNKSGGNVKKDS 580
>At1g69200 putative fructokinase
Length = 614
Score = 35.0 bits (79), Expect = 0.040
Identities = 39/168 (23%), Positives = 68/168 (40%), Gaps = 34/168 (20%)
Query: 100 EQENLDANVDAGTPAVVPTSRRRVPAVSTRRKKEVIV-----------------IEDEDD 142
E L+ + G AVV ++ +V KK+V+V + D++
Sbjct: 53 ESAPLEEEGNDGNGAVV--GKKPSKSVKRTTKKKVVVKDEPLEEISEFLVDNDDVLDKES 110
Query: 143 VVSEVQGKATDVAKTPAAAPSS--RTRAGRSVRNKTEISDGTSVQKAYSTRRSVRLVGKS 200
+VS ++ K T K AAA S + + VR K + V+ +T
Sbjct: 111 IVSALKPKKTRTRKKAAAASSDVEEVKTEKKVRRKRTVKKDKDVEDDLAT---------- 160
Query: 201 LSKMSLADTED---MESTKNDDVSEEMSVSQNEGGSIETENGMLISLC 245
+ ++D E+ +EST + EE+ +S++EG I G +C
Sbjct: 161 IMDAEVSDVEEALAVESTDTESEEEEIDLSKHEGEDISHTYGWPPLVC 208
>At2g02770 unknown protein
Length = 569
Score = 34.7 bits (78), Expect = 0.053
Identities = 43/197 (21%), Positives = 77/197 (38%), Gaps = 19/197 (9%)
Query: 46 LNQREGGDIGTPAVQPRTARRTTTQRKPVKEAESTKVSTRVNRGGRGGVAEGEVEQENLD 105
L +RE D+ A T+++ +K +STK S + G +GEVE
Sbjct: 108 LLERETNDLTIKEAPVTQAGDDKTKKRKLKSLDSTKRSHKKRSLGDQSSGKGEVESL--- 164
Query: 106 ANVDAGTPAVVPTSRRRVPAVSTRRKKEVIVIEDEDDVVSEVQGKATDVAKTPAAAPSSR 165
A +P + A + ++ ++ +++DE++ E + + + P+A+ +
Sbjct: 165 ----AMSPTGKDLEKGEKSAATIDQENDLALVDDENE-SHEEDVRVGAIVEAPSASKNLL 219
Query: 166 T------RAGRSVRNKTEISDGTSVQKAYSTRRSVRLVGKSLSKMSLA-----DTEDMES 214
T V N+ DG + A SV+ K +K A DTE S
Sbjct: 220 TAEFLDINQSADVENEGLNKDGVDGKYADEATTSVKAAKKGRTKKDKAAPTKEDTEVASS 279
Query: 215 TKNDDVSEEMSVSQNEG 231
++N D + Q G
Sbjct: 280 SRNVDHDVVIREPQENG 296
>At3g59430 hypothetical protein
Length = 451
Score = 33.9 bits (76), Expect = 0.090
Identities = 27/89 (30%), Positives = 43/89 (47%), Gaps = 11/89 (12%)
Query: 1 MDFHT-LSRKELQALSKMNKIPANITNVAMADALSALPHVEGLDEILNQREGGDIGTPAV 59
+DF+ SRKELQ+L K +PAN ++ MA++L++ L+ + G P
Sbjct: 7 VDFYCGFSRKELQSLCKKYNLPANRSSSDMAESLASYFEKNNLNPV-------SFGVPGN 59
Query: 60 QPRTARRTTTQRKPVKEAESTKVSTRVNR 88
Q +A TT R P + K + N+
Sbjct: 60 QDSSA---TTSRAPAIRTWNVKRDSYGNK 85
>At3g02400 hypothetical protein
Length = 585
Score = 33.5 bits (75), Expect = 0.12
Identities = 48/216 (22%), Positives = 77/216 (35%), Gaps = 61/216 (28%)
Query: 68 TTQRKPVKEAESTKV-------STRVNRGGRGGVAEGEVEQENLDANVDAGTPAVVPTSR 120
+ Q KP + S+K STR +R + + + ++E LD ++ A V R
Sbjct: 140 SVQEKPKRTRGSSKQEENELPKSTRASR--KKNLDDIADKEEELDVEIEKVVKARVGRPR 197
Query: 121 RRVPA--------------VSTRRKKEVIVIEDEDDVVSEVQGKAT-------------- 152
+ + V RK I +E++VV E +G +
Sbjct: 198 KNAGSAIAKEEEVVEEKKRVGRPRKNASSAITEEEEVVEEKKGNSRARRGKNSEIVQKSI 257
Query: 153 --DVAKTPAAAPSSRTRAGRSVRNKTEISD---GTSVQKAYSTRRSVRL----------- 196
+V TP A S ++ + V +I + G V+ T RS R
Sbjct: 258 KLEVEDTPKAVEISEVKSRKRVTRSKQIENECFGLEVKDEKRTTRSTRSKTTEIGGESFL 317
Query: 197 --------VGKSLSKMSLADTEDMESTKNDDVSEEM 224
KS +K D E + T+NDD EE+
Sbjct: 318 ELEMVLNQARKSRAKRKKMDEEPSKETRNDDAGEEV 353
>At1g15940 T24D18.4
Length = 1012
Score = 33.5 bits (75), Expect = 0.12
Identities = 49/207 (23%), Positives = 87/207 (41%), Gaps = 17/207 (8%)
Query: 41 GLDEILNQREGGDIGTPAVQPRTARRTTTQR--KPV-----KEAESTKVST--RVNRGGR 91
G ++++ EG T A TA +T +R KP +E S K S+ +V
Sbjct: 341 GDEKVITANEGLSESTDA---ETASGSTRKRGWKPKSLMNPEEGYSFKTSSSKKVQEKEL 397
Query: 92 GGVAEGEVEQEN--LDANVDAGTPAVVPTSRRRVPAVSTRRKKEVIVIEDEDDVVSEVQG 149
G + G+V + L + V +VV + A + RK+ +E+ D VS V
Sbjct: 398 GDSSLGKVAAKKVPLPSKVGQTNQSVVISLSSSGRARTGSRKRSRTKMEETDHDVSSV-- 455
Query: 150 KATDVAKTPAAAPSSRTRAGRSVRNKTEISDGTSVQKAYSTRRSVRLVGKSLSKMSLADT 209
AT AK ++ + + N + DG K+ ++ + K+ +K LA+T
Sbjct: 456 -ATQPAKKQTVKKTNPAKEDLTKSNVKKHEDGIKTGKSSKKEKADNGLAKTSAKKPLAET 514
Query: 210 EDMESTKNDDVSEEMSVSQNEGGSIET 236
++ + V + +EG S++T
Sbjct: 515 MMVKPSGKKLVHSDAKKKNSEGASMDT 541
>At5g55100 unknown protein
Length = 844
Score = 33.1 bits (74), Expect = 0.15
Identities = 31/133 (23%), Positives = 55/133 (41%), Gaps = 7/133 (5%)
Query: 101 QENLDANVDAGTPAVVPTSRRRVPAVSTRRKKEVIVIEDEDDVVSEVQGKATDVAKTPAA 160
Q + NVDA T A + + RR R + I+ D S+ G DV+ +
Sbjct: 454 QPQMRVNVDANTAAAILQAARR----GIRNPQLGILTGKPMDETSQTLGN--DVSYPSSK 507
Query: 161 APSSRTRAGRSVRNKTEISDGTSVQKAYSTRRSVRLVGKSLSKMSLADTE-DMESTKNDD 219
+P G+S+ T S+ S + S + ++ +KM +A + D + + +
Sbjct: 508 SPDLAKSTGQSLSGSTAASEADSSEAGLSKEQKLKAERLKRAKMFVAKLKPDAQPVQQAE 567
Query: 220 VSEEMSVSQNEGG 232
S +SV + G
Sbjct: 568 PSRSISVEPLDSG 580
>At3g54620 bZIP transcription factor-like protein
Length = 403
Score = 32.7 bits (73), Expect = 0.20
Identities = 23/76 (30%), Positives = 37/76 (48%), Gaps = 15/76 (19%)
Query: 62 RTARRTTTQRKPVKEAESTKVSTRVNRGGRGGVAEGEVEQENLDANVDAGTPAVVPTSRR 121
R + T+TQ+KP A T +S+R + + ++LD + D G P V +RR
Sbjct: 187 RFSPTTSTQKKPDVPARQTSISSR-----------DDSDDDDLDGDADNGDPTDVKRARR 235
Query: 122 ----RVPAVSTRRKKE 133
R A +RR+K+
Sbjct: 236 MLSNRESARRSRRRKQ 251
>At3g05670 unknown protein
Length = 883
Score = 32.7 bits (73), Expect = 0.20
Identities = 36/153 (23%), Positives = 61/153 (39%), Gaps = 14/153 (9%)
Query: 98 EVEQENLDA-NVDAGTPAVVPTSRRRVPAVSTRRKKEVIV-------IEDEDDVVSEVQG 149
EVE+E++ NV+ P R++ +R+ +V+V ++D DD E++
Sbjct: 74 EVEEEDVMLRNVEWPKVKTGPRGNRKITGCKSRKTNQVVVSDNEDVDLDDADDEDDEIRN 133
Query: 150 KATDVAKTPAAAPSSRTRAGRSVRNKT--EISDGTSVQKAYSTRRSVRLVGKSLSKMSLA 207
V K+ + R G R + EI D +++ SL
Sbjct: 134 SRNAVGKSGSLDGEKHQRIGLGKRRRVFYEIEDEDGDYPEEDGDEEEERDVENVDSNSLH 193
Query: 208 DTED--MESTKNDDVSEEMSVSQNEGGSIETEN 238
D ED M + D+VS E + + G E E+
Sbjct: 194 DGEDGKMALEEQDNVSHE--TEKEDDGDYEDED 224
>At1g48920 unknown protein
Length = 557
Score = 32.3 bits (72), Expect = 0.26
Identities = 37/169 (21%), Positives = 62/169 (35%), Gaps = 13/169 (7%)
Query: 78 ESTKVSTRVNRGGRGGVAEGEVEQENLDANVDAGTPAVVPTSRRRVPAVSTRRKKEVIVI 137
E V+ + + G + + E + D + PA P ++ PA +
Sbjct: 133 EEVAVTKKPAAAAKNGSVKAKKESSSEDDSSSEDEPAKKPAAKIAKPAAKDSSSSDDDSD 192
Query: 138 EDEDDVVSEVQGKATDVAKTPAAAPSSRTRAGRSVRNKTEISDGTSVQKAYSTRRSVRLV 197
ED +D + A AK +++ SS + E D QK T+ S +
Sbjct: 193 EDSEDEKPATKKAAPAAAKAASSSDSS------DEDSDEESEDEKPAQKKADTKASKKSS 246
Query: 198 GKSLSKMSLADTEDMEST---KNDDV----SEEMSVSQNEGGSIETENG 239
S+ ++ED E T K+ DV +E+ S Q + S G
Sbjct: 247 SDESSESEEDESEDEEETPKKKSSDVEMVDAEKSSAKQPKTPSTPAAGG 295
>At5g23150 transcription factor-like protein (gb|AAD31171.1)
Length = 1392
Score = 32.0 bits (71), Expect = 0.34
Identities = 30/122 (24%), Positives = 50/122 (40%), Gaps = 20/122 (16%)
Query: 67 TTTQRKPVKEAESTKVSTRVNRGGRGGVAEGEVEQENLDANVDAGTPAVVPTSRRRVPAV 126
T QR+ ++ A S S G R +G +EQ++ + P + PA
Sbjct: 383 TKRQRQTMEHATSPSFS-----GSRDKSGKGHLEQKDRSS----------PVRNVKAPAA 427
Query: 127 STRRKKEVIVIEDED---DVVSEVQGKATDVAKTPAAAPSSRTRAG--RSVRNKTEISDG 181
+ +K+ + I DED D + + GK V + + RA S K +IS G
Sbjct: 428 QSLKKRRAVCIYDEDDDEDPKTPLHGKPAIVPQAASVLTDGPKRANVCHSTSTKAKISAG 487
Query: 182 TS 183
++
Sbjct: 488 ST 489
>At4g07490 putative transposon protein
Length = 559
Score = 32.0 bits (71), Expect = 0.34
Identities = 27/104 (25%), Positives = 44/104 (41%), Gaps = 16/104 (15%)
Query: 124 PAVSTRRKKEVIVIEDEDDVVSEVQGKATDVAKTPA----AAPSSRTRAGRSVRNKTEIS 179
P S+RR + I+ + E + + +PA + SS++ A RNK +I+
Sbjct: 38 PLGSSRRDQTSFEIDHSESSQEEEVMQEEETFSSPAGNTRSQKSSKSSAREFARNKADIT 97
Query: 180 DGTSVQKAYSTRRSVRLVGKSLSKMSLADTEDMESTKNDDVSEE 223
G + VG+ L + EDM S K DD +E+
Sbjct: 98 QGK------------KPVGRRLELVDEDSEEDMASPKEDDQTEK 129
>At3g07200 putative RING zinc finger protein
Length = 182
Score = 32.0 bits (71), Expect = 0.34
Identities = 31/112 (27%), Positives = 47/112 (41%), Gaps = 23/112 (20%)
Query: 81 KVSTRVNRGGRGGVAEGEVEQENLDANVDAGTPAVVPTSRRRVPAVSTRRKKEVIVIEDE 140
+ S R+ R R GV + E + + A+ P +V T R V+ IED+
Sbjct: 7 RTSMRLRR--RDGVTQNEHQGDQQVADQAPTVPQIVATPPRI----------NVVAIEDD 54
Query: 141 DDVVSEVQGKATDVAKTPAAAPSSRTRAGRSVRNKTEISDGTSVQKAYSTRR 192
DDVV ++ A+A + RS R +++ D S STRR
Sbjct: 55 DDVV-----------ESTASAFAQAKNKSRSARRGSQVVDVESAGANRSTRR 95
>At1g67140 hypothetical protein
Length = 2149
Score = 32.0 bits (71), Expect = 0.34
Identities = 52/214 (24%), Positives = 87/214 (40%), Gaps = 23/214 (10%)
Query: 53 DIGTPAVQPRTARRTTTQRKPVKEAESTKVSTRVNRGG--RGGVAEGEVEQENLDANVDA 110
DI PA T + T+ VKE + + T N+ G E E E+E+ D + D
Sbjct: 1895 DIKLPAPVVATPEKVTSTANMVKEEALSTMPTSFNQVSTVESGTDEEEEEEED-DDDDDW 1953
Query: 111 GTPAVVPTSRRRVPAVSTRRKKEVIVIEDEDDVV-------SEVQGKATDVAKTPAAAPS 163
T P S + K V E+E D+ E + TD + +
Sbjct: 1954 DTFQSFPASTN----LEGSESKTESVAEEEPDLPGRSSIQDDESNAEETDDQHLASDHAT 2009
Query: 164 SRTRAGRSVRNKTEISDGTSVQKAYSTRRSVRLVGKSLSKMSLAD--TEDMESTKNDDVS 221
TR + ++K E+ + +V+ ++TR K + + ++ ED +++ND S
Sbjct: 2010 DITREDSNDKSK-EVVEEETVEPCFTTREDSVDKSKEVEEETVKPCLIEDALTSQNDKTS 2068
Query: 222 E-----EMSVSQNEGGSIETEN-GMLISLCLTIV 249
E++ E ++E+EN G I L T V
Sbjct: 2069 SGDHPVEINEQSVESKNLESENIGTDIKLASTEV 2102
>At3g13990 unknown protein
Length = 848
Score = 31.2 bits (69), Expect = 0.58
Identities = 52/226 (23%), Positives = 83/226 (36%), Gaps = 25/226 (11%)
Query: 26 NVAMADALSALPHVEGLDEILNQREGGDIGTPAVQPRTARRTTTQRKPVKEAES-TKVST 84
N+ +A+ L H + E+ +R+ T V+P + KP++ S KV T
Sbjct: 46 NMDANEAVEKLIHQDPFHEVKRKRDRKKEDTVFVEPANIK------KPLENVTSEVKVRT 99
Query: 85 RVNRG-GRGGVAEGEVEQENLDANVDAGTPAVVPTSRRRVPAVSTRRKKEVIVIEDEDDV 143
+ RGG + + N PA RV V R + DE+
Sbjct: 100 QPEHNVRRGGYSRNFFPRNAAQRNASPRNPATGSNKEFRV--VRDNRSNPNV---DEELK 154
Query: 144 VSEVQGKATDVAKTPAAAPSSRTRAGRSVRNKTEISDGTSVQKAYSTRRSVRL------- 196
S Q +++ K A +R G RN + D T A + R +
Sbjct: 155 HSSAQSSGSNINKVVATVNKLGSRGGLGNRNSSGAQDSTDECNAPADARLRQAEIRPLHC 214
Query: 197 -VGKSLSKMSLADTED---MESTKNDDVSEEMSVSQNEG-GSIETE 237
V K L ++L T + S+ D V VS++ G+I+ E
Sbjct: 215 PVNKELGGVTLPSTNSVLGVYSSSKDPVHVPSPVSRSSPVGAIKRE 260
>At1g72880 unknown protein
Length = 385
Score = 31.2 bits (69), Expect = 0.58
Identities = 33/99 (33%), Positives = 44/99 (44%), Gaps = 11/99 (11%)
Query: 32 ALSALPHVEGLDEILNQREGG----DIGTPAVQPRTARRTTTQRKPVKEAEST----KVS 83
A+SA H + + NQ+ G +G A ARR TTQ+K + E ES K
Sbjct: 262 AVSANRHPGAGNFMSNQQSLGAQLAQLGRDASAAGAARRFTTQKKSIVEIESVGVAGKTD 321
Query: 84 TRVNRGGRGGVAEGEVEQENLDANVDA---GTPAVVPTS 119
TRV + R E E + D +V A G +V P S
Sbjct: 322 TRVKKFFRLEFLAKEQEHTDEDLDVKALEDGFVSVTPFS 360
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.310 0.126 0.332
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,178,013
Number of Sequences: 26719
Number of extensions: 209139
Number of successful extensions: 810
Number of sequences better than 10.0: 67
Number of HSP's better than 10.0 without gapping: 11
Number of HSP's successfully gapped in prelim test: 56
Number of HSP's that attempted gapping in prelim test: 784
Number of HSP's gapped (non-prelim): 77
length of query: 260
length of database: 11,318,596
effective HSP length: 97
effective length of query: 163
effective length of database: 8,726,853
effective search space: 1422477039
effective search space used: 1422477039
T: 11
A: 40
X1: 16 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144806.1


