

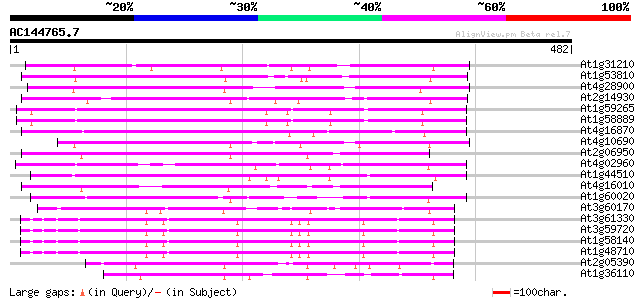
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.7 - phase: 0
(482 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g31210 putative reverse transcriptase 186 2e-47
At1g53810 175 6e-44
At4g28900 putative protein 171 1e-42
At2g14930 pseudogene 154 8e-38
At1g59265 polyprotein, putative 150 2e-36
At1g58889 polyprotein, putative 150 2e-36
At4g16870 retrotransposon like protein 145 4e-35
At4g10690 retrotransposon like protein 145 5e-35
At2g06950 putative retroelement pol polyprotein 144 8e-35
At4g02960 putative polyprotein of LTR transposon 135 5e-32
At1g44510 polyprotein, putative 129 3e-30
At4g16010 reverse transcriptase like protein 127 1e-29
At1g60020 hypothetical protein 127 1e-29
At3g60170 putative protein 111 1e-24
At3g61330 copia-type polyprotein 90 2e-18
At3g59720 copia-type reverse transcriptase-like protein 89 7e-18
At1g58140 hypothetical protein 89 7e-18
At1g48710 hypothetical protein 88 9e-18
At2g05390 putative retroelement pol polyprotein 77 3e-14
At1g36110 hypothetical protein 75 1e-13
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 186 bits (473), Expect = 2e-47
Identities = 129/411 (31%), Positives = 202/411 (48%), Gaps = 44/411 (10%)
Query: 14 SSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEE---FITSSDSSKSNNPAFE 70
SSV++KL +NY LWK+ ++ KL G++ G P + + +S+ NP +E
Sbjct: 15 SSVTLKLTDSNYLLWKTQFESLLSSQKLIGFVNGAVNAPSQSRLVVNGEVTSEEPNPLYE 74
Query: 71 EWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQI---IYLKSEF 127
W DQ + W+ +++ E+ + + TS+Q+W SLA +S + L+
Sbjct: 75 SWFCTDQLVRSWLFGTLSEEVLGHVHNLSTSRQIW---VSLAENFNKSSVAREFSLRQNL 131
Query: 128 HSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVV----KLS 183
+ K E Y + K + D L G P+ S I LNGL +Y+PI LS
Sbjct: 132 QLLSKKEKPFSVYCREFKTICDALSSIGKPVDESMKIFGFLNGLGRDYDPITTVIQSSLS 191
Query: 184 DHTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRG--- 240
T ++ D+ +++ F+S+++ ++ + N+ + + ++N N RG
Sbjct: 192 KLPTPTFNDVVSEVQGFDSKLQSYEEAASVTPHLAFNI-ERSESGSPQYNPNQKGRGRSG 250
Query: 241 -SNFRGWRGGRGRGRSS----------KAPCQVCGKTNHTAINCFHRFDKNYSRSNYSAD 289
+ RG RGRG S + CQ+CG+T HTA+ C++RFD NY
Sbjct: 251 QNKGRGGYSTRGRGFSQHQSSPQVSGPRPVCQICGRTGHTALKCYNRFDNNY-------- 302
Query: 290 SDKQGSHNAFIASQNSVED-YDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKL 348
Q AF + S + +W+ DS A+ HVT TN Q TE+ G ++++VG+G L
Sbjct: 303 ---QAEIQAFSTLRVSDDTGKEWHPDSAATAHVTSSTNGLQSATEYEGDDAVLVGDGTYL 359
Query: 349 EIVATCSSKLKSLN----LDDVLYVPNITKNLLSVSKLAADNNIFVEFDKN 395
I T S+ +KS N L++VL VPNI K+LLSVSKL D V FD N
Sbjct: 360 PITHTGSTTIKSSNGKIPLNEVLVVPNIQKSLLSVSKLCDDYPCGVYFDAN 410
>At1g53810
Length = 1522
Score = 175 bits (443), Expect = 6e-44
Identities = 121/413 (29%), Positives = 200/413 (48%), Gaps = 40/413 (9%)
Query: 11 DLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEF--ITSSD-SSKSNNP 67
++ + V+V L++ NY LWKS + G L G++ G+ P + +T ++ +S+ NP
Sbjct: 10 NISNCVTVTLNQQNYILWKSQFESFLSGQGLLGFVTGSISAPAQTRSVTHNNVTSEEPNP 69
Query: 68 AFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEF 127
F W DQ + W+L S A ++ + +++C TS Q+W + + S++ L+
Sbjct: 70 EFYTWHQTDQVVKSWLLGSFAEDILSVVVNCFTSHQVWLTLANHFNRVSSSRLFELQRRL 129
Query: 128 HSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSD--- 184
++ K + ME +L +K++ D+L G+P+ I LNGL EY PI + +
Sbjct: 130 QTLEKKDNTMEVFLKDLKHICDQLASVGSPVPEKMKIFSALNGLGREYEPIKTTIENSVD 189
Query: 185 -HTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNF 243
+ +LS ++ ++L ++ R++ ++ + NV H D+ + NNN RG
Sbjct: 190 SNPSLSLDEVASKLRGYDDRLQSYVTEPTISPHVAFNVT----HSDSGYYHNNN-RGKGR 244
Query: 244 RGWRGG------RGRG-------------RSSKAPCQVCGKTNHTAINCFHRFDKNYSRS 284
G RGRG +S CQ+CGK H A+ C+HRFD +Y
Sbjct: 245 SNSGSGKSSFSTRGRGFHQQISPTSGSQAGNSGLVCQICGKAGHHALKCWHRFDNSYQHE 304
Query: 285 NYSADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGN 344
+ I ++W DS AS HVT+ + Q +HG +S++V +
Sbjct: 305 DL-----PMALATMRITDVTDHHGHEWIPDSAASAHVTNNRHVLQQSQPYHGSDSIMVAD 359
Query: 345 GDKLEIVATCSSKLKS----LNLDDVLYVPNITKNLLSVSKLAADNNIFVEFD 393
G+ L I T S + S + L +VL P+I K+LLSVSKL +D VEFD
Sbjct: 360 GNFLPITHTGSGSIASSSGKIPLKEVLVCPDIVKSLLSVSKLTSDYPCSVEFD 412
>At4g28900 putative protein
Length = 1415
Score = 171 bits (432), Expect = 1e-42
Identities = 114/392 (29%), Positives = 190/392 (48%), Gaps = 39/392 (9%)
Query: 16 VSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPE--EFITSSDS-SKSNNPAFEEW 72
V++KL NY LWK + +L G++ G CP I + D +++ NP F W
Sbjct: 17 VTLKLSTANYLLWKIQFETWLNNQRLLGFVTGANPCPNATRSIRNGDQVTEATNPDFLTW 76
Query: 73 QANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRK 132
NDQ+++GW+L S++ + + TS+++W + S+ L+ + + K
Sbjct: 77 VQNDQKIMGWLLGSLSEDALRSVYGLHTSREVWFSLAKKYNRVSASRKSDLQRRLNPVSK 136
Query: 133 GEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLS---DHTTLS 189
E M +YL +K + D+L G P+ ++ I LNGL EY + + D +S
Sbjct: 137 NEKSMLEYLNCVKQICDQLDSIGCPVPENEKIFGVLNGLGQEYMLVSTMIKGSMDTYPMS 196
Query: 190 WVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNF-RGWRG 248
+ D+ +L+ F+ +++ + NR +N +G F +
Sbjct: 197 FEDVVFKLINFDDKLQNGQS------------------GGNRGRNNYTTKGRGFPQQISS 238
Query: 249 GRGRGRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSVED 308
G ++ CQ+C K H+A C+ RFD + ++S AF A + S +
Sbjct: 239 GSPSDSGTRPTCQICNKYGHSAYKCWKRFDHAFQSEDFS---------KAFAAMRVSDQK 289
Query: 309 YD-WYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIV----ATCSSKLKSLNL 363
+ W DSGA++H+T+ T++ Q + G++S++VGN D L I A +S +L L
Sbjct: 290 SNPWVTDSGATSHITNSTSQLQSAQPYSGEDSVIVGNSDFLPITHIGSAVLTSNQGNLPL 349
Query: 364 DDVLYVPNITKNLLSVSKLAADNNIFVEFDKN 395
DVL PNITK+LLSVSKL +D +EFD +
Sbjct: 350 RDVLVCPNITKSLLSVSKLTSDYPCVIEFDSD 381
>At2g14930 pseudogene
Length = 1149
Score = 154 bits (390), Expect = 8e-38
Identities = 114/401 (28%), Positives = 191/401 (47%), Gaps = 32/401 (7%)
Query: 11 DLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSN----N 66
++ + V+VKL NY LWKS + G L G++ G P ++ + N
Sbjct: 14 NISNCVTVKLTDRNYILWKSQFESFLSGQGLLGFVNGAYAAPTGTVSGPQDAGVTEAIPN 73
Query: 67 PAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSE 126
P ++ W +DQ + M+ ++ + ++ +TS ++W + S+I L+
Sbjct: 74 PDYQAWFRSDQVV-------MSEDILSVVVGSKTSHEVWMNLAKHFNRISSSRIFELQRR 126
Query: 127 FHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSDHT 186
HS+ K ME+YL +K + D+L G+P++ I ++GL EY P++ L
Sbjct: 127 LHSLSKEGKTMEEYLRYLKTICDQLASVGSPVAEKMKIFAMVHGLTREYEPLITSLEGTL 186
Query: 187 TL----SWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDH----RDNRFNSNNNW 238
S+ D+ +L F+ R+ Q +T+++ + N + R+NR N +
Sbjct: 187 DAFPGPSYEDVVYRLKNFDDRL-QGYTVTDVSPHLAFNTFRSSNRGRGGRNNRGKGNFST 245
Query: 239 RGSNFRGW--RGGRGRGRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSH 296
RG F+ S K CQ+CGK H A+ C+HRFD +Y S +A +
Sbjct: 246 RGRGFQQQFSSSSSSVSASEKPMCQICGKRGHYALQCWHRFDDSYQHSEAAA-----AAF 300
Query: 297 NAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSS 356
+A + S +D W DS A+ H+T+ +++ Q + + G ++++ +G+ L I S+
Sbjct: 301 SALHITDVS-DDSGWVPDSAATAHITNNSSRLQQMQPYLGNDTVMASDGNFLPITHIGSA 359
Query: 357 KLKS----LNLDDVLYVPNITKNLLSVSKLAADNNIFVEFD 393
L S L L DVL PNI K+LLSVSKL D FD
Sbjct: 360 NLPSTSGNLPLKDVLVCPNIAKSLLSVSKLTKDYPCSFTFD 400
>At1g59265 polyprotein, putative
Length = 1466
Score = 150 bits (379), Expect = 2e-36
Identities = 107/410 (26%), Positives = 195/410 (47%), Gaps = 28/410 (6%)
Query: 7 NNKNDLPSSVS--VKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKS 64
NN + L ++S KL NY +W V + G +L G++ G+ P I +D++
Sbjct: 11 NNTSILNVNMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATI-GTDAAPR 69
Query: 65 NNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLK 124
NP + W+ D+ + +L +++ + + T+ Q+W+ + + + + L+
Sbjct: 70 VNPDYTRWKRQDKLIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLR 129
Query: 125 SEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKL-S 183
++ KG ++DY+ + D+L L G P+ + + + + L L EY P++ ++ +
Sbjct: 130 TQLKQWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIAA 189
Query: 184 DHTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATA------------NVANKFDHRDNR 231
T + ++ +LL ES+I +++ T + + A A N N+ + DNR
Sbjct: 190 KDTPPTLTEIHERLLNHESKILAVSSATVIPITANAVSHRNTTTTNNNNNGNRNNRYDNR 249
Query: 232 FNSNNN--WRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCF---HRFDKNYSRSNY 286
N+NN+ W+ S+ + + + CQ+CG H+A C H S+
Sbjct: 250 NNNNNSKPWQQSS-TNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQPP 308
Query: 287 SADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGD 346
S + Q N + S S +W DSGA++H+T N + G + ++V +G
Sbjct: 309 SPFTPWQPRANLALGSPYS--SNNWLLDSGATHHITSDFNNLSLHQPYTGGDDVMVADGS 366
Query: 347 KLEIVAT----CSSKLKSLNLDDVLYVPNITKNLLSVSKLAADNNIFVEF 392
+ I T S+K + LNL ++LYVPNI KNL+SV +L N + VEF
Sbjct: 367 TIPISHTGSTSLSTKSRPLNLHNILYVPNIHKNLISVYRLCNANGVSVEF 416
>At1g58889 polyprotein, putative
Length = 1466
Score = 150 bits (379), Expect = 2e-36
Identities = 107/410 (26%), Positives = 195/410 (47%), Gaps = 28/410 (6%)
Query: 7 NNKNDLPSSVS--VKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKS 64
NN + L ++S KL NY +W V + G +L G++ G+ P I +D++
Sbjct: 11 NNTSILNVNMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTTMPPATI-GTDAAPR 69
Query: 65 NNPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLK 124
NP + W+ D+ + +L +++ + + T+ Q+W+ + + + + L+
Sbjct: 70 VNPDYTRWKRQDKLIYSAVLGAISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLR 129
Query: 125 SEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKL-S 183
++ KG ++DY+ + D+L L G P+ + + + + L L EY P++ ++ +
Sbjct: 130 TQLKQWTKGTKTIDDYMQGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIAA 189
Query: 184 DHTTLSWVDLQAQLLTFESRIEQLNNLTNLNLNATA------------NVANKFDHRDNR 231
T + ++ +LL ES+I +++ T + + A A N N+ + DNR
Sbjct: 190 KDTPPTLTEIHERLLNHESKILAVSSATVIPITANAVSHRNTTTTNNNNNGNRNNRYDNR 249
Query: 232 FNSNNN--WRGSNFRGWRGGRGRGRSSKAPCQVCGKTNHTAINCF---HRFDKNYSRSNY 286
N+NN+ W+ S+ + + + CQ+CG H+A C H S+
Sbjct: 250 NNNNNSKPWQQSS-TNFHPNNNQSKPYLGKCQICGVQGHSAKRCSQLQHFLSSVNSQQPP 308
Query: 287 SADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGD 346
S + Q N + S S +W DSGA++H+T N + G + ++V +G
Sbjct: 309 SPFTPWQPRANLALGSPYS--SNNWLLDSGATHHITSDFNNLSLHQPYTGGDDVMVADGS 366
Query: 347 KLEIVAT----CSSKLKSLNLDDVLYVPNITKNLLSVSKLAADNNIFVEF 392
+ I T S+K + LNL ++LYVPNI KNL+SV +L N + VEF
Sbjct: 367 TIPISHTGSTSLSTKSRPLNLHNILYVPNIHKNLISVYRLCNANGVSVEF 416
>At4g16870 retrotransposon like protein
Length = 1474
Score = 145 bits (367), Expect = 4e-35
Identities = 98/396 (24%), Positives = 192/396 (47%), Gaps = 17/396 (4%)
Query: 11 DLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFE 70
++ +S KL NNY +W + ++ G +L G++ G+ + P +T+++ S NP +
Sbjct: 25 NINTSNVTKLTSNNYLMWSLQIHALLDGYELAGHLDGSIETPAPTLTTNNVV-SANPQYT 83
Query: 71 EWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSI 130
W+ D+ + ++ +++ + + + Q+W + + I L+++ +
Sbjct: 84 LWKRQDRLIFSALIGAISPPVQPLVSRATKASQIWKTLTNTYAKSSYDHIKQLRTQIKQL 143
Query: 131 RKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKL-SDHTTLS 189
+KG +++Y++ L D+L + G P+ + + + + L GL +Y +V ++ T S
Sbjct: 144 KKGTKTIDEYVLSHTTLLDQLAILGKPMEHEEQVERILEGLPEDYKTVVDQIEGKDNTPS 203
Query: 190 WVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWR--GSNFRGWR 247
++ +L+ E+++ L++ +L +ANVA + H +NR N+ N R G+ +
Sbjct: 204 ITEIHERLINHEAKLLSTAALSSSSLPMSANVAQQRHHNNNRNNNQNKNRTQGNTYTNNW 263
Query: 248 GGRGRGRSSKAP-------CQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFI 300
+S + P CQ+C H+A C + S+ SA + A +
Sbjct: 264 QPSANNKSGQRPFKPYLGKCQICNVQGHSARRC-PQLQAMQPSSSSSASTFTPWQPRANL 322
Query: 301 ASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCS----S 356
A +W DSGA++H+T N L + + + +++ +G L+I T S S
Sbjct: 323 AMGAPYTANNWLLDSGATHHITSDLNALA-LHQPYNGDDVMIADGTSLKITKTGSTFLPS 381
Query: 357 KLKSLNLDDVLYVPNITKNLLSVSKLAADNNIFVEF 392
+ L L+ VLYVP+I KNL+SV +L N + VEF
Sbjct: 382 NARDLTLNKVLYVPDIQKNLVSVYRLCNTNQVSVEF 417
>At4g10690 retrotransposon like protein
Length = 1515
Score = 145 bits (366), Expect = 5e-35
Identities = 106/388 (27%), Positives = 173/388 (44%), Gaps = 48/388 (12%)
Query: 42 DGYMLGTKKCPEE--FITSSD-SSKSNNPAFEEWQANDQRLLGWMLNSMATEMATQLLHC 98
+G++ G P +T D S+ N F +W DQ + W+ S++ E ++
Sbjct: 39 NGFVTGATPRPASTIIVTKDDIQSEEANQEFLKWTRIDQLVKAWIFGSLSEEALKVVIGL 98
Query: 99 ETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPI 158
+++++W + ++ L+ + K M+ YL ++KN+ D+L G P+
Sbjct: 99 NSAQEVWLGLARRFNRFSTTRKYDLQKRLGTCSKAGKTMDAYLSEVKNICDQLDSIGFPV 158
Query: 159 SNSDLIIQTLNGLDSEYNPIVVKLSDHTTL----SWVDLQAQLLTFESRIEQLNNLTNLN 214
+ + I LNGL EY I + + + D+ +L TF+ ++ N
Sbjct: 159 TEQEKIFGVLNGLGKEYESIATVIEHSLDVYPGPCFDDVVYKLTTFDDKLSTYT----AN 214
Query: 215 LNATANVANKFDHRDNRFNSNNNWRGSNFRGWRG-------------------GRGRGRS 255
T ++A D + NNN RG + +RG G G
Sbjct: 215 SEVTPHLAFYTD-KSYSSRGNNNSRGGRYGNFRGRGSYSSRGRGFHQQFGSGSNNGSGNG 273
Query: 256 SKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAF----IASQNSVEDYDW 311
SK CQ+C K H+A C+ RF++NY + NAF ++ QN ++W
Sbjct: 274 SKPTCQICRKYGHSAFKCYTRFEENYLPEDLP---------NAFAAMRVSDQNQASSHEW 324
Query: 312 YFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLK----SLNLDDVL 367
DS A+ H+T+ T+ Q+ + G +S++VGNGD L I + L +L L+DVL
Sbjct: 325 LPDSAATAHITNTTDGLQNSQTYSGDDSVIVGNGDFLPITHIGTIPLNISQGTLPLEDVL 384
Query: 368 YVPNITKNLLSVSKLAADNNIFVEFDKN 395
P ITK+LLSVSKL D FD +
Sbjct: 385 VCPGITKSLLSVSKLTDDYPCSFTFDSD 412
>At2g06950 putative retroelement pol polyprotein
Length = 405
Score = 144 bits (364), Expect = 8e-35
Identities = 96/377 (25%), Positives = 170/377 (44%), Gaps = 32/377 (8%)
Query: 11 DLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSD----SSKSNN 66
++ + V+V L NY LWKS + G L G++ G+ P + SD +S S N
Sbjct: 20 NISNCVTVTLTAKNYILWKSQFESFLDGQGLLGFVTGSIPAPSQTSVVSDIDGSTSASPN 79
Query: 67 PAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSE 126
P + W D+ + W+L S ++ + +++C TS ++W + + S++ L+
Sbjct: 80 PEYYTWFKTDRVVKSWLLGSFLEDILSVVVNCNTSHEVWISVANHFNRVSSSRLFELQRR 139
Query: 127 FHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSDHT 186
++ K + M++YL +K + D+L G+P++ I LNGL EY PI + +
Sbjct: 140 LQNVSKRDKSMDEYLKDLKTICDQLASVGSPVTEKMKIFAALNGLGREYEPIKTTIENSM 199
Query: 187 TL----SWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFD-HRDNRFNSNNNWRGS 241
S D+ +L ++ R++ T ++ + N+ D + FN+ N +G
Sbjct: 200 DALPGPSLEDVIPKLTGYDDRLQGYLEETAVSPHVAFNITTSDDSNASGYFNAYNRGKGK 259
Query: 242 NFRGWRGGRGRGR------------------SSKAPCQVCGKTNHTAINCFHRFDKNYSR 283
+ RG RGR + CQ+CGK H A+ C+HRF+
Sbjct: 260 SNRGRNSFSTRGRGFHQQISSTNSSSGSQSGGTSVVCQICGKMGHPALKCWHRFN----- 314
Query: 284 SNYSADSDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVG 343
++Y + + I +W DS A+ HVT+ Q +HG ++++V
Sbjct: 315 NSYQYEELPRALAAMRITDITDQHGNEWLPDSAATAHVTNSPRSLQQSQPYHGSDAVMVA 374
Query: 344 NGDKLEIVATCSSKLKS 360
+G+ L I T S+ L S
Sbjct: 375 DGNFLPITHTGSTNLAS 391
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 135 bits (340), Expect = 5e-32
Identities = 103/407 (25%), Positives = 183/407 (44%), Gaps = 43/407 (10%)
Query: 6 NNNKNDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSN 65
N N ++ S KL NY +W V + G +L G++ G+ P I +D+
Sbjct: 12 NTNILNVNMSNVTKLTSTNYLMWSRQVHALFDGYELAGFLDGSTPMPPATI-GTDAVPRV 70
Query: 66 NPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKS 125
NP + W+ D+ + +L +++ + + T+ Q+W+ + IY
Sbjct: 71 NPDYTRWRRQDKLIYSAILGAISMSVQPAVSRATTAAQIWETLRK----------IYANP 120
Query: 126 EFHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLS-D 184
+ + + ++ D+L L G P+ + + + + L L +Y P++ +++
Sbjct: 121 SYGHVTQ---------LRFITRFDQLALLGKPMDHDEQVERVLENLPDDYKPVIDQIAAK 171
Query: 185 HTTLSWVDLQAQLLTFESRIEQLNN----------LTNLNLNATANVANKFDHRDNRFNS 234
T S ++ +L+ ES++ LN+ +T+ N N N N+ D+R+ +N+
Sbjct: 172 DTPPSLTEIHERLINRESKLLALNSAEVVPITANVVTHRNTNTNRNQNNRGDNRN--YNN 229
Query: 235 NNNWRGSNFRGWRGGRGRGRSSK---APCQVCGKTNHTAINC--FHRFDKNYSRSNYSAD 289
NNN S G R R K CQ+C H+A C H+F ++ S
Sbjct: 230 NNNRSNSWQPSSSGSRSDNRQPKPYLGRCQICSVQGHSAKRCPQLHQFQSTTNQQQ-STS 288
Query: 290 SDKQGSHNAFIASQNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLE 349
A +A + +W DSGA++H+T N + G + +++ +G +
Sbjct: 289 PFTPWQPRANLAVNSPYNANNWLLDSGATHHITSDFNNLSFHQPYTGGDDVMIADGSTIP 348
Query: 350 IVATCSSKL----KSLNLDDVLYVPNITKNLLSVSKLAADNNIFVEF 392
I T S+ L +SL+L+ VLYVPNI KNL+SV +L N + VEF
Sbjct: 349 ITHTGSASLPTSSRSLDLNKVLYVPNIHKNLISVYRLCNTNRVSVEF 395
>At1g44510 polyprotein, putative
Length = 1459
Score = 129 bits (325), Expect = 3e-30
Identities = 98/394 (24%), Positives = 177/394 (44%), Gaps = 23/394 (5%)
Query: 19 KLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQANDQR 78
KL NY +W + ++ G L GY+ + P E T+ +S S NP+F W+ D+
Sbjct: 32 KLTSTNYLMWSIQIHALLDGYDLAGYLDNSVVIPPE-TTTINSVVSANPSFTLWKRQDKL 90
Query: 79 LLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRKGEMKME 138
+ ++ +++ + + + S Q+W + + I L+ + + KG ++
Sbjct: 91 IFSALIGAISPAVQSLVSRATNSSQIWSTLNNTYAKPSYGHIKQLRQQIQRLTKGTKTID 150
Query: 139 DYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKL-SDHTTLSWVDLQAQL 197
+Y+ D+L + G P+ + + + L GL EY +V ++ T + ++ +L
Sbjct: 151 EYVQSHTTRLDQLAILGKPMEHEEQVEHILKGLPEEYKTVVDQIEGKDNTPTITEIHERL 210
Query: 198 LTFESRI--EQLNNLTNLNLNATA-------NVANKFDHRD----NRFNSNNNWRGSNFR 244
+ ES++ +++ ++ ++A A N N+ H++ N N+N N
Sbjct: 211 INHESKLLSDEVPPSSSFPMSANAVQQRNFNNNCNQNQHKNRYQGNTHNNNTNTNSQPST 270
Query: 245 GWRGGRGRGRSSKAPCQVCGKTNHTAINC--FHRFDKNYSRSNYSADSDKQGSHNAFIAS 302
+ G+ + CQ+C H+A C S S +S + Q N I S
Sbjct: 271 YNKSGQRTFKPYLGKCQICSVQGHSARRCPQLQAMQLPASSSAHSPFTPWQPRANLAIGS 330
Query: 303 QNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKSLN 362
+ W DSGA++H+T N ++G +++ +G L I T S+ L S N
Sbjct: 331 PYAAN--PWLLDSGATHHITSDLNALSLHQPYNGGEYVMIADGTGLTIKQTGSTFLPSQN 388
Query: 363 LD----DVLYVPNITKNLLSVSKLAADNNIFVEF 392
D VLYVP+I KNL+SV +L N + VEF
Sbjct: 389 RDLALHKVLYVPDIRKNLISVYRLCNTNQVSVEF 422
>At4g16010 reverse transcriptase like protein
Length = 1942
Score = 127 bits (319), Expect = 1e-29
Identities = 93/361 (25%), Positives = 161/361 (43%), Gaps = 40/361 (11%)
Query: 11 DLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSD----SSKSNN 66
++ + V+V L + NY LWKS + G L G++ G+ P I D ++ N
Sbjct: 20 NISNCVTVTLTQQNYILWKSQFESFLSGQGLLGFVTGSISAPSPTIPVPDINGVTTDRPN 79
Query: 67 PAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSE 126
P F+ W D+ + W+L S A ++ + +++ T+ +L Q+L
Sbjct: 80 PEFDVWFKTDKVVKSWLLGSFAEDILSVVVNYVTAHELQRRLQTL--------------- 124
Query: 127 FHSIRKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSDHT 186
K + M+ YL +++ + ++L G+P+ I LNGL EY PI +
Sbjct: 125 ----EKKDKPMQVYLKELQTIYEQLASVGSPVPEKMKIFAALNGLGREYEPIKTSIEGSI 180
Query: 187 ----TLSWVDLQAQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRGSN 242
T ++ +L ++ R++ ++++ + N N N RG
Sbjct: 181 DIPPTPKLDEIMPRLNGYDDRLQAYAANSDVSPHLAFNTVQA-----NSVFYTNRGRGQG 235
Query: 243 FRGWRGGRGRGRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIAS 302
R + G RG+ R + CQ+CGK H A+NC+HRFD ++Y D Q I
Sbjct: 236 NRRFGGSRGQERPT---CQICGKHGHHALNCWHRFD-----NSYQLDVLPQALPATQITD 287
Query: 303 QNSVEDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKSLN 362
+W DS A+ H+T+ Q + G +S++VGNG+ L I T S+ + S +
Sbjct: 288 ITDHSGSEWVTDSAATAHITNSPRHLQQTKSYAGSDSVMVGNGNFLPITHTGSTSIGSTS 347
Query: 363 L 363
+
Sbjct: 348 V 348
Score = 39.7 bits (91), Expect = 0.004
Identities = 18/54 (33%), Positives = 31/54 (57%)
Query: 310 DWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKSLNL 363
+W DS A+ H+T+ Q + G +S++VGNG+ L I T S+ + S ++
Sbjct: 1195 EWVTDSAATAHITNSPRHLQQTKSYAGSDSVMVGNGNFLPITHTGSTSIGSTSV 1248
>At1g60020 hypothetical protein
Length = 1194
Score = 127 bits (319), Expect = 1e-29
Identities = 104/384 (27%), Positives = 174/384 (45%), Gaps = 50/384 (13%)
Query: 19 KLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQANDQR 78
KL NY +W V ++ G L GY+ G+ P E IT++ S +N PA++ W+ D+
Sbjct: 30 KLTPTNYIMWNRQVHALLDGYDLAGYIDGSVTAPSEMITTAGVSAAN-PAYKFWKRQDKL 88
Query: 79 LLGWMLNSMATEMATQLLHCETSKQLWDEAQSLAGAHTRSQIIYLKSEFHSIRKGEMKME 138
+ +L ++ T + L T+ ++W++ +S+ + I ++ KG +
Sbjct: 89 IYSAILGTITTTIQPLLSRSNTAAEIWEKLKSIYATPSWGHIQQMRQHIKQWSKGTKTIT 148
Query: 139 DYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSDHTTL-----SWVDL 193
+Y D+L L G P+ +++ I L GL +Y +V D T + + +L
Sbjct: 149 EYFQGHTTRFDELALLGKPLEHAEQIEFLLGGLSEDYKSVV----DQTEIRDKPPTLTEL 204
Query: 194 QAQLLTFESRIEQLNNLTNLNLNATANVAN-KFDHRDNRFNSNNNWRGSNFRGWRGGRGR 252
+LL E+++ T +L ATA+ AN K + +N++N+NN R +
Sbjct: 205 LEKLLNREAKL-MCAAATTPSLPATAHAANYKGNSNNNQYNNNN----------RNNKSH 253
Query: 253 GRSSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSVEDYDWY 312
G S + + S + Q NA IAS + +W
Sbjct: 254 GSSYNSQQSIPA----------------------SPFTPWQPRANAAIASPYNAN--NWL 289
Query: 313 FDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKL----KSLNLDDVLY 368
DSGA++H+T N + G + + +G L I T S+ + +SL L DVLY
Sbjct: 290 LDSGATHHITSDLNNLSLHQPYTGGEDVTIADGSGLSISHTGSALISTPSRSLALTDVLY 349
Query: 369 VPNITKNLLSVSKLAADNNIFVEF 392
VPNI KNL+SV ++ N + VEF
Sbjct: 350 VPNIHKNLISVYRMCNANKVSVEF 373
>At3g60170 putative protein
Length = 1339
Score = 111 bits (277), Expect = 1e-24
Identities = 100/376 (26%), Positives = 177/376 (46%), Gaps = 38/376 (10%)
Query: 25 YPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAFEEWQANDQRLLGWML 84
Y W + +R +L + L + P + ++ S++ A EE + D ++ ++
Sbjct: 19 YDFWSMTMENFLRSREL--WRLVEEGIPAIVVGTTPVSEAQRSAVEEAKLKDLKVKNFLF 76
Query: 85 NSMATEMATQLLHCETSKQLWDEAQSLAGAHT---RSQIIYLKSEFH--SIRKGEMKMED 139
++ E+ +L TSK +W+ + T R+Q+ L+ EF ++++GE K++
Sbjct: 77 QAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAMKEGE-KIDT 135
Query: 140 YLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKL---SDHTTLSWVDLQAQ 196
+L + + +K+K G + S ++ + L L ++N +V + +D +TLS +L
Sbjct: 136 FLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLSTLSIDELHGS 195
Query: 197 LLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNFRGWRG-GRGRGRS 255
LL E R LN + A K H + S RG FRG RG GRGRGRS
Sbjct: 196 LLVHEQR---------LNGHVQEEQALKVTHEERP--SQGRGRGV-FRGSRGRGRGRGRS 243
Query: 256 SK----APCQVCGKTNHTAINCFHRFDKNYSRSNYSA-DSDKQGSHNAFIASQNSVEDYD 310
C C H C ++KN +NY+ + +++ A++ + D
Sbjct: 244 GTNRAIVECYKCHNLGHFQYEC-PEWEKN---ANYAELEEEEELLLMAYVEQNQANRDEV 299
Query: 311 WYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLK----SLNLDDV 366
W+ DSG SNH+T F +L E + ++ +GN ++ +V S K+K + + +V
Sbjct: 300 WFLDSGCSNHMTGSKEWFSELEEGFNR-TVKLGNDTRMSVVGKGSVKVKVNGVTQVIPEV 358
Query: 367 LYVPNITKNLLSVSKL 382
YVP + NLLS+ +L
Sbjct: 359 YYVPELRNNLLSLGQL 374
>At3g61330 copia-type polyprotein
Length = 1352
Score = 90.1 bits (222), Expect = 2e-18
Identities = 91/413 (22%), Positives = 179/413 (43%), Gaps = 46/413 (11%)
Query: 10 NDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAF 69
N++P V V L ++NY W SL + + G D + + K E S S++
Sbjct: 4 NNVPFQVPV-LTKSNYDNW-SLRMKAILGAH-DVWEIVEKGFIEPENEGS-LSQTQKDGL 59
Query: 70 EEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQ-SLAGAHT--RSQIIYLKSE 126
+ + D++ L + + + +++ ++K+ W++ + S GA + ++ L+ E
Sbjct: 60 RDSRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGE 119
Query: 127 FHSI--RKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSD 184
F ++ ++GE+ + DY ++ + + LK G + + ++ + L LD ++ IV + +
Sbjct: 120 FEALQMKEGEL-VSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEE 178
Query: 185 HTTLSWVDLQ---AQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRG- 240
L + ++ L +E + ++ ++ LN R RG
Sbjct: 179 TKDLEAMTIEQLLGSLQAYEEKKKKKEDIAEQVLNMQITKEENGQSYQRRGGGQVRGRGR 238
Query: 241 ---SNFRGWR------------GGRGRGRS--------SKAPCQVCGKTNHTAINCFHRF 277
N RGWR RGRG+ S C CGK H A C
Sbjct: 239 GGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPS 298
Query: 278 DKNYSRSNYSADSDKQGSHNAFIAS---QNSVEDYDWYFDSGASNHVTHQTNKFQDLTEH 334
+K + + + Q +AS E++ WY DSGASNH+ + + F +L E
Sbjct: 299 NKKFEEKAHYVEEKIQEEDMLLMASYKKDEQKENHKWYLDSGASNHMCGRKSMFAELDE- 357
Query: 335 HGKNSLVVGNGDKLEIVATCSSKLKSLN-----LDDVLYVPNITKNLLSVSKL 382
+ ++ +G+ K+E+ + ++ N + +V Y+P++ N+LS+ +L
Sbjct: 358 SVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQL 410
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 88.6 bits (218), Expect = 7e-18
Identities = 91/413 (22%), Positives = 178/413 (43%), Gaps = 46/413 (11%)
Query: 10 NDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAF 69
N++P V V L ++NY W SL + + G D + + K E S S++
Sbjct: 4 NNVPFQVPV-LTKSNYDNW-SLRMKAILGAH-DVWEIVEKGFIEPENEGS-LSQTQKDGL 59
Query: 70 EEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQ-SLAGAHT--RSQIIYLKSE 126
+ + D++ L + + + +++ ++K+ W++ + S GA + ++ L+ E
Sbjct: 60 RDSRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGE 119
Query: 127 FHSI--RKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSD 184
F ++ ++GE+ + DY ++ + + LK G + + ++ + L LD ++ IV + +
Sbjct: 120 FEALQMKEGEL-VSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEE 178
Query: 185 HTTLSWVDLQ---AQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRG- 240
L + ++ L +E + ++ ++ LN R RG
Sbjct: 179 TKDLEAMTIEQLLGSLQAYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGR 238
Query: 241 ---SNFRGWR------------GGRGRGRS--------SKAPCQVCGKTNHTAINCFHRF 277
N RGWR RGRG+ S C CGK H A C
Sbjct: 239 GGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPS 298
Query: 278 DKNYSRSNYSADSDKQGSHNAFIAS---QNSVEDYDWYFDSGASNHVTHQTNKFQDLTEH 334
+K + + Q +AS E++ WY DSGASNH+ + + F +L E
Sbjct: 299 NKKFKEKANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDE- 357
Query: 335 HGKNSLVVGNGDKLEIVATCSSKLKSLN-----LDDVLYVPNITKNLLSVSKL 382
+ ++ +G+ K+E+ + ++ N + +V Y+P++ N+LS+ +L
Sbjct: 358 SVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQL 410
>At1g58140 hypothetical protein
Length = 1320
Score = 88.6 bits (218), Expect = 7e-18
Identities = 91/413 (22%), Positives = 178/413 (43%), Gaps = 46/413 (11%)
Query: 10 NDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAF 69
N++P V V L ++NY W SL + + G D + + K E S S++
Sbjct: 4 NNVPFQVPV-LTKSNYDNW-SLRMKAILGAH-DVWEIVEKGFIEPENEGS-LSQTQKDGL 59
Query: 70 EEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQ-SLAGAHT--RSQIIYLKSE 126
+ + D++ L + + + +++ ++K+ W++ + S GA + ++ L+ E
Sbjct: 60 RDSRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGE 119
Query: 127 FHSI--RKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSD 184
F ++ ++GE+ + DY ++ + + LK G + + ++ + L LD ++ IV + +
Sbjct: 120 FEALQMKEGEL-VSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEE 178
Query: 185 HTTLSWVDLQ---AQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRG- 240
L + ++ L +E + ++ ++ LN R RG
Sbjct: 179 TKDLEAMTIEQLLGSLQAYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGR 238
Query: 241 ---SNFRGWR------------GGRGRGRS--------SKAPCQVCGKTNHTAINCFHRF 277
N RGWR RGRG+ S C CGK H A C
Sbjct: 239 GGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPS 298
Query: 278 DKNYSRSNYSADSDKQGSHNAFIAS---QNSVEDYDWYFDSGASNHVTHQTNKFQDLTEH 334
+K + + Q +AS E++ WY DSGASNH+ + + F +L E
Sbjct: 299 NKKFEEKANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDE- 357
Query: 335 HGKNSLVVGNGDKLEIVATCSSKLKSLN-----LDDVLYVPNITKNLLSVSKL 382
+ ++ +G+ K+E+ + ++ N + +V Y+P++ N+LS+ +L
Sbjct: 358 SVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQL 410
>At1g48710 hypothetical protein
Length = 1352
Score = 88.2 bits (217), Expect = 9e-18
Identities = 91/413 (22%), Positives = 178/413 (43%), Gaps = 46/413 (11%)
Query: 10 NDLPSSVSVKLDRNNYPLWKSLVLPVVRGCKLDGYMLGTKKCPEEFITSSDSSKSNNPAF 69
N++P V V L ++NY W SL + + G D + + K E S S++
Sbjct: 4 NNVPFQVPV-LTKSNYDNW-SLRMKAILGAH-DVWEIVEKGFIEPENEGS-LSQTQKDGL 59
Query: 70 EEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWDEAQ-SLAGAHT--RSQIIYLKSE 126
+ + D++ L + + + +++ ++K+ W++ + S GA + ++ L+ E
Sbjct: 60 RDSRKRDKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGE 119
Query: 127 FHSI--RKGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLDSEYNPIVVKLSD 184
F ++ ++GE+ + DY ++ + + LK G + + ++ + L LD ++ IV + +
Sbjct: 120 FEALQMKEGEL-VSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEE 178
Query: 185 HTTLSWVDLQ---AQLLTFESRIEQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRG- 240
L + ++ L +E + ++ ++ LN R RG
Sbjct: 179 TKDLEAMTIEQLLGSLQAYEEKKKKKEDIIEQVLNMQITKEENGQSYQRRGGGQVRGRGR 238
Query: 241 ---SNFRGWR------------GGRGRGRS--------SKAPCQVCGKTNHTAINCFHRF 277
N RGWR RGRG+ S C CGK H A C
Sbjct: 239 GGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPS 298
Query: 278 DKNYSRSNYSADSDKQGSHNAFIAS---QNSVEDYDWYFDSGASNHVTHQTNKFQDLTEH 334
+K + + Q +AS E++ WY DSGASNH+ + + F +L E
Sbjct: 299 NKKFEEKANYVEEKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDE- 357
Query: 335 HGKNSLVVGNGDKLEIVATCSSKLKSLN-----LDDVLYVPNITKNLLSVSKL 382
+ ++ +G+ K+E+ + ++ N + +V Y+P++ N+LS+ +L
Sbjct: 358 SVRGNVALGDESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQL 410
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 76.6 bits (187), Expect = 3e-14
Identities = 84/352 (23%), Positives = 151/352 (42%), Gaps = 48/352 (13%)
Query: 66 NPAFEEWQANDQRLLGWMLNSMATEMATQLLHCETSKQLWD--EAQSLAGAHTR-SQIIY 122
+P ++ + ND + S+ Q+ +TSK +W+ + ++L + +++
Sbjct: 15 DPGSDDMEKNDMAR-ALLFQSVPESTILQVGKHKTSKAMWEAIKTRNLGAERVKEAKLQT 73
Query: 123 LKSEFHSIR-KGEMKMEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLD-SEYNPIVV 180
L +EF + K +++++ ++ ++ K + G I S ++ + L L +Y I+
Sbjct: 74 LMAEFDRLNMKDNETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIHIIA 133
Query: 181 KLS---DHTTLSWVDLQAQLLTFESRI-EQLNNLTNLNLNATANVANKFDHRDNRFNSNN 236
L D T + D+ ++ T+E R+ ++ ++ AN + +D R R
Sbjct: 134 ALEQILDLNTTGFEDIVGRMKTYEDRVCDEDDSPEEQGKLMYANSESSYDTRGGR----- 188
Query: 237 NWRGSNFRGWRGGRGRGR-------SSKAPCQVCGKTNHTAINCFHRF---DKNYSRSNY 286
RG RG GRGRG SK C C KT H A C R K +
Sbjct: 189 -GRG---RGRSSGRGRGGYGYQQRDKSKVICYRCDKTGHYASECLDRLLKLIKAQEQQQN 244
Query: 287 SADSDKQGS---HNAFIASQNSVE--------DYDWYFDSGASNHVTHQTNKFQDLTE-- 333
+ D D+ S H ++ SV+ D WY D+GASNH+T F L E
Sbjct: 245 NEDDDEIESLMMHEVVYLNERSVKPKEFEACSDNSWYLDNGASNHMTGNLQWFSKLNEMI 304
Query: 334 ----HHGKNSLVVGNGDKLEIVATCSSKLKSLNLDDVLYVPNITKNLLSVSK 381
G +S + G ++ T K+L DV ++P++ N++S+ +
Sbjct: 305 TGKVRFGDDSRIDIKGKGSIVLITKGGIRKTLT--DVYFIPDLKSNIISLGQ 354
>At1g36110 hypothetical protein
Length = 745
Score = 74.7 bits (182), Expect = 1e-13
Identities = 79/320 (24%), Positives = 134/320 (41%), Gaps = 46/320 (14%)
Query: 81 GWMLNSMATEMATQLLHCETSKQLWDEAQSL---AGAHTRSQIIYLKSEFHSIRKGEM-K 136
G + S+ + Q+ T+K +WD ++ A +++ L +EF ++ E K
Sbjct: 62 GLIFQSIPESLTLQVGTLATAKLVWDSIKTRYVGADRVKEARLQTLMAEFEKMKMKESEK 121
Query: 137 MEDYLIKMKNLADKLKLAGNPISNSDLIIQTLNGLD-SEYNPIVVKLS---DHTTLSWVD 192
++ + ++ LA + G+ I S L+ + LN L +Y I+ L D S+ D
Sbjct: 122 IDVFAGRLAELATRSDALGSNIETSKLVKKFLNALPLRKYIHIIASLEQVLDLNNTSFED 181
Query: 193 LQAQLLTFESRI----EQLNNLTNLNLNATANVANKFDHRDNRFNSNNNWRGSNFRGWRG 248
+ ++ +E R+ EQ ++ L T D +D+ + S +G F G
Sbjct: 182 IVGRIKVYEERVWDGEEQEDDQGKLMYANT-------DTQDSWYASRGRGQGGGFNGRGR 234
Query: 249 GRGRGR--SSKAPCQVCGKTNHTAINCFHRFDKNYSRSNYSADSDKQGSHNAFIASQNSV 306
GRGRG +SK C C K H A NC DS K + N
Sbjct: 235 GRGRGSRDTSKVTCYRCDKLGHYASNC--------------PDSVKPSVFETELDGDNV- 279
Query: 307 EDYDWYFDSGASNHVTHQTNKFQDLTEHHGKNSLVVGNGDKLEIVATCSSKLKSLN---- 362
WY D+GASNH+T F + E + G+ ++I S S N
Sbjct: 280 ----WYLDNGASNHMTGNRAYFSKIDESI-TGKVRFGDDSCIDIKGKGSILFMSKNGEKK 334
Query: 363 -LDDVLYVPNITKNLLSVSK 381
L +V ++P++ N++S+ +
Sbjct: 335 VLAEVYFIPDVKSNIISLGQ 354
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.130 0.391
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 10,224,192
Number of Sequences: 26719
Number of extensions: 413218
Number of successful extensions: 1702
Number of sequences better than 10.0: 116
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 1418
Number of HSP's gapped (non-prelim): 168
length of query: 482
length of database: 11,318,596
effective HSP length: 103
effective length of query: 379
effective length of database: 8,566,539
effective search space: 3246718281
effective search space used: 3246718281
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144765.7


