

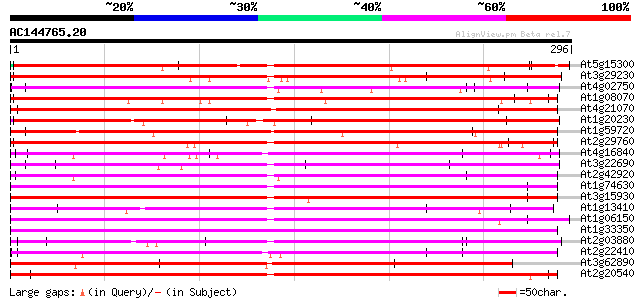
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.20 + phase: 0
(296 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g15300 unknown protein 239 2e-63
At3g29230 hypothetical protein 232 2e-61
At4g02750 hypothetical protein 228 4e-60
At1g08070 unknown protein 227 7e-60
At4g21070 putative protein (fragment) 226 9e-60
At1g20230 hypothetical protein 226 1e-59
At1g59720 hypothetical protein 225 3e-59
At2g29760 hypothetical protein 223 7e-59
At4g16840 hypothetical protein 223 1e-58
At3g22690 hypothetical protein 222 2e-58
At2g42920 hypothetical protein 222 2e-58
At1g74630 hypothetical protein 221 3e-58
At3g15930 hypothetical protein 220 8e-58
At1g13410 hypothetical protein 220 8e-58
At1g06150 unknown protein 220 8e-58
At1g33350 unknown protein 219 2e-57
At2g03880 putative selenium-binding protein 218 2e-57
At2g22410 putative protein 218 3e-57
At3g62890 putative protein 218 4e-57
At2g20540 unknown protein 217 5e-57
>At5g15300 unknown protein
Length = 514
Score = 239 bits (609), Expect = 2e-63
Identities = 125/298 (41%), Positives = 188/298 (62%), Gaps = 10/298 (3%)
Query: 3 ERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHK 62
E++ V+WN+MI+G V+ AL +F EM++AG P VT++S+L ACA G LE G +
Sbjct: 203 EKDVVTWNAMISGYVNCGYPKEALGIFKEMRDAGEHPDVVTILSLLSACAVLGDLETGKR 262
Query: 63 IY-ESLKVCEHKIESYLG----NALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGL 117
++ L+ Y+G NAL++MY KCG++ A E+F G+K + +S WN +++GL
Sbjct: 263 LHIYILETASVSSSIYVGTPIWNALIDMYAKCGSIDRAIEVFRGVKDRDLSTWNTLIVGL 322
Query: 118 AVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKI 177
A+H + E ++F EM+ + P+ VTFIGV++ACSH G VD+ R YF M Y I
Sbjct: 323 ALH-HAEGSIEMFEEMQRL---KVWPNEVTFIGVILACSHSGRVDEGRKYFSLMRDMYNI 378
Query: 178 VPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFK 237
P+ KHYGCMVD+L R G LEEA+ + + + + ++WRTLLGAC+ N EL + + +
Sbjct: 379 EPNIKHYGCMVDMLGRAGQLEEAFMFVESMKIEPNAIVWRTLLGACKIYGNVELGKYANE 438
Query: 238 QLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINMKESDRL 295
+L +++ GDYVLLSNIYA G+WD V+++R D V + G S I ++ D+L
Sbjct: 439 KLLSMRKDESGDYVLLSNIYASTGQWDGVQKVRKMFDDTRVKKPTGVSLIE-EDDDKL 495
Score = 69.3 bits (168), Expect = 2e-12
Identities = 65/284 (22%), Positives = 114/284 (39%), Gaps = 22/284 (7%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
+P+ + N ++ G + L++EM+ GV P T +L AC++ G
Sbjct: 38 IPKPDVSICNHVLRGSAQSMKPEKTVSLYTEMEKRGVSPDRYTFTFVLKACSKLEWRSNG 97
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
+ + + Y+ NAL+ + CG+L +A E+F+ W++M G A
Sbjct: 98 FAFHGKVVRHGFVLNEYVKNALILFHANCGDLGIASELFDDSAKAHKVAWSSMTSGYAKR 157
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G +E +LF EM D+V + ++ C +D AR FD ++ D
Sbjct: 158 GKIDEAMRLFDEMPYK-------DQVAWNVMITGCLKCKEMDSARELFDRFTEK-----D 205
Query: 181 SKHYGCMVDLLSRWGLLEEA---YQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFK 237
+ M+ G +EA ++ + A VV +LL AC + E +
Sbjct: 206 VVTWNAMISGYVNCGYPKEALGIFKEMRDAGEHPDVVTILSLLSACAVLGDLETGKRLHI 265
Query: 238 QLAKLKQLIDGDYV------LLSNIYAEAGRWDE-VERLRSEMD 274
+ + + YV L ++YA+ G D +E R D
Sbjct: 266 YILETASVSSSIYVGTPIWNALIDMYAKCGSIDRAIEVFRGVKD 309
Score = 42.0 bits (97), Expect = 4e-04
Identities = 49/186 (26%), Positives = 77/186 (41%), Gaps = 39/186 (20%)
Query: 90 GNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFI 149
G L A ++F+ + VS N ++ G A E+ L+TEME+ + PDR TF
Sbjct: 26 GALKYAHKLFDEIPKPDVSICNHVLRGSAQSMKPEKTVSLYTEMEKR---GVSPDRYTFT 82
Query: 150 GVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPF 209
VL ACS K W + K+V R G + Y
Sbjct: 83 FVLKACS------KLEWRSNGFAFHGKVV--------------RHGFVLNEY-------V 115
Query: 210 QNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERL 269
+N+++L+ G + +A F AK ++ + +++ YA+ G+ DE RL
Sbjct: 116 KNALILFHANCG------DLGIASELFDDSAKAHKVA---WSSMTSGYAKRGKIDEAMRL 166
Query: 270 RSEMDY 275
EM Y
Sbjct: 167 FDEMPY 172
>At3g29230 hypothetical protein
Length = 600
Score = 232 bits (591), Expect = 2e-61
Identities = 120/291 (41%), Positives = 176/291 (60%), Gaps = 3/291 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
+P +N V+W +IAG A L +M +G+K +ISIL AC E+G L +G
Sbjct: 275 LPAKNVVTWTIIIAGYAEKGLLKEADRLVDQMVASGLKFDAAAVISILAACTESGLLSLG 334
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
+I+ LK +Y+ NAL++MY KCGNL A+++FN + K + WN M+ GL VH
Sbjct: 335 MRIHSILKRSNLGSNAYVLNALLDMYAKCGNLKKAFDVFNDIPKKDLVSWNTMLHGLGVH 394
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G+ +E +LF+ M IRPD+VTFI VL +C+H GL+D+ YF M K Y +VP
Sbjct: 395 GHGKEAIELFSRMRRE---GIRPDKVTFIAVLCSCNHAGLIDEGIDYFYSMEKVYDLVPQ 451
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
+HYGC+VDLL R G L+EA +++ T P + +VV+W LLGACR + ++A+ L
Sbjct: 452 VEHYGCLVDLLGRVGRLKEAIKVVQTMPMEPNVVIWGALLGACRMHNEVDIAKEVLDNLV 511
Query: 241 KLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINMKE 291
KL G+Y LLSNIYA A W+ V +RS+M + V + +G S + +++
Sbjct: 512 KLDPCDPGNYSLLSNIYAAAEDWEGVADIRSKMKSMGVEKPSGASSVELED 562
Score = 74.3 bits (181), Expect = 7e-14
Identities = 56/222 (25%), Positives = 106/222 (47%), Gaps = 15/222 (6%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
M ER+ VSWNSM+ G V + A LF EM + ++ ++L A +
Sbjct: 180 MSERDTVSWNSMLGGLVKAGELRDARRLFDEMPQRDL----ISWNTMLDGYARCREMSKA 235
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKM--KTVSCWNAMVIGLA 118
+++E K+ E S+ + +V Y K G++ +A +F+ M + K V W ++ G A
Sbjct: 236 FELFE--KMPERNTVSW--STMVMGYSKAGDMEMARVMFDKMPLPAKNVVTWTIIIAGYA 291
Query: 119 VHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIV 178
G +E +L +M + ++ D I +L AC+ GL+ ++KR +
Sbjct: 292 EKGLLKEADRLVDQM---VASGLKFDAAAVISILAACTESGLLSLG-MRIHSILKRSNLG 347
Query: 179 PDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLL 220
++ ++D+ ++ G L++A+ + P + +V W T+L
Sbjct: 348 SNAYVLNALLDMYAKCGNLKKAFDVFNDIP-KKDLVSWNTML 388
Score = 61.2 bits (147), Expect = 6e-10
Identities = 76/292 (26%), Positives = 126/292 (43%), Gaps = 39/292 (13%)
Query: 3 ERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHK 62
E N NS+I A +FSEMQ G+ T +L AC+ L +
Sbjct: 79 EPNVHLCNSLIRAHAQNSQPYQAFFVFSEMQRFGLFADNFTYPFLLKACSGQSWLPVVKM 138
Query: 63 IYESLKVCEHKIESYLGNALVNMYCKCGNLSL--AWEIFNGMKMKTVSCWNAMVIGLAVH 120
++ ++ + Y+ NAL++ Y +CG L + A ++F M + WN+M+ GL
Sbjct: 139 MHNHIEKLGLSSDIYVPNALIDCYSRCGGLGVRDAMKLFEKMSERDTVSWNSMLGGLVKA 198
Query: 121 GYCEEVFQLFTEMEE--------SLGGSIR--------------PDR--VTFIGVLVACS 156
G + +LF EM + L G R P+R V++ +++ S
Sbjct: 199 GELRDARRLFDEMPQRDLISWNTMLDGYARCREMSKAFELFEKMPERNTVSWSTMVMGYS 258
Query: 157 HKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQII--MTA---PFQN 211
G ++ AR FD M K V + ++ + GLL+EA +++ M A F
Sbjct: 259 KAGDMEMARVMFDKMPLPAKNVVT---WTIIIAGYAEKGLLKEADRLVDQMVASGLKFDA 315
Query: 212 SVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVL--LSNIYAEAG 261
+ V+ ++L AC T+S + + K L YVL L ++YA+ G
Sbjct: 316 AAVI--SILAAC-TESGLLSLGMRIHSILKRSNLGSNAYVLNALLDMYAKCG 364
Score = 36.2 bits (82), Expect = 0.022
Identities = 31/147 (21%), Positives = 66/147 (44%), Gaps = 7/147 (4%)
Query: 77 YLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEMEES 136
++ L++ C +LA +FN ++ V N+++ A + + F +F+EM+
Sbjct: 52 HIAPKLISALSLCRQTNLAVRVFNQVQEPNVHLCNSLIRAHAQNSQPYQAFFVFSEMQRF 111
Query: 137 LGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGL 196
+ D T+ +L ACS + + + +H +++ + D ++D SR G
Sbjct: 112 ---GLFADNFTYPFLLKACSGQSWLPVVKMMHNH-IEKLGLSSDIYVPNALIDCYSRCGG 167
Query: 197 L--EEAYQIIMTAPFQNSVVLWRTLLG 221
L +A ++ + V W ++LG
Sbjct: 168 LGVRDAMKLFEKMS-ERDTVSWNSMLG 193
>At4g02750 hypothetical protein
Length = 781
Score = 228 bits (580), Expect = 4e-60
Identities = 119/290 (41%), Positives = 172/290 (59%), Gaps = 3/290 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MP+R+ VSW +MIAG AL LF +M+ G + + S L CA+ ALE+G
Sbjct: 369 MPKRDPVSWAAMIAGYSQSGHSFEALRLFVQMEREGGRLNRSSFSSALSTCADVVALELG 428
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
+++ L ++ ++GNAL+ MYCKCG++ A ++F M K + WN M+ G + H
Sbjct: 429 KQLHGRLVKGGYETGCFVGNALLLMYCKCGSIEEANDLFKEMAGKDIVSWNTMIAGYSRH 488
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G+ E + F M+ ++PD T + VL ACSH GLVDK R YF M + Y ++P+
Sbjct: 489 GFGEVALRFFESMKRE---GLKPDDATMVAVLSACSHTGLVDKGRQYFYTMTQDYGVMPN 545
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
S+HY CMVDLL R GLLE+A+ ++ PF+ +W TLLGA R NTELAE + ++
Sbjct: 546 SQHYACMVDLLGRAGLLEDAHNLMKNMPFEPDAAIWGTLLGASRVHGNTELAETAADKIF 605
Query: 241 KLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINMK 290
++ G YVLLSN+YA +GRW +V +LR M V + GYS I ++
Sbjct: 606 AMEPENSGMYVLLSNLYASSGRWGDVGKLRVRMRDKGVKKVPGYSWIEIQ 655
Score = 64.3 bits (155), Expect = 7e-11
Identities = 53/245 (21%), Positives = 107/245 (43%), Gaps = 23/245 (9%)
Query: 2 PERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGH 61
P ++ +W +M++G + R A ELF +M EV+ ++L + +E+
Sbjct: 277 PVQDVFTWTAMVSGYIQNRMVEEARELFDKMPERN----EVSWNAMLAGYVQGERMEMAK 332
Query: 62 KIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHG 121
++++ + + ++ N ++ Y +CG +S A +F+ M + W AM+ G + G
Sbjct: 333 ELFDVMPC--RNVSTW--NTMITGYAQCGKISEAKNLFDKMPKRDPVSWAAMIAGYSQSG 388
Query: 122 YCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDS 181
+ E +LF +ME G R +R +F L C+ ++ + +VK
Sbjct: 389 HSFEALRLFVQMEREGG---RLNRSSFSSALSTCADVVALELGKQLHGRLVK------GG 439
Query: 182 KHYGCMVD-----LLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISF 236
GC V + + G +EEA + ++ +V W T++ E+A F
Sbjct: 440 YETGCFVGNALLLMYCKCGSIEEANDLFKEMAGKD-IVSWNTMIAGYSRHGFGEVALRFF 498
Query: 237 KQLAK 241
+ + +
Sbjct: 499 ESMKR 503
Score = 63.9 bits (154), Expect = 1e-10
Identities = 69/278 (24%), Positives = 119/278 (41%), Gaps = 34/278 (12%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MPER+ VSWN MI G V R+ A ELF M V + ++L A+ G ++
Sbjct: 121 MPERDLVSWNVMIKGYVRNRNLGKARELFEIMPERDV----CSWNTMLSGYAQNGCVDDA 176
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
+++ ++ E S+ NAL++ Y + + A +F + + WN ++ G
Sbjct: 177 RSVFD--RMPEKNDVSW--NALLSAYVQNSKMEEACMLFKSRENWALVSWNCLLGGFVKK 232
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
E Q F M ++R D V++ ++ + G +D+AR FD V D
Sbjct: 233 KKIVEARQFFDSM------NVR-DVVSWNTIITGYAQSGKIDEARQLFDE-----SPVQD 280
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISF---- 236
+ MV + ++EEA ++ P +N V W +L E+A+ F
Sbjct: 281 VFTWTAMVSGYIQNRMVEEARELFDKMPERNE-VSWNAMLAGYVQGERMEMAKELFDVMP 339
Query: 237 -KQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEM 273
+ ++ +I G YA+ G+ E + L +M
Sbjct: 340 CRNVSTWNTMITG--------YAQCGKISEAKNLFDKM 369
Score = 45.8 bits (107), Expect = 3e-05
Identities = 56/261 (21%), Positives = 101/261 (38%), Gaps = 38/261 (14%)
Query: 9 WNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESLK 68
WN I+ + AL +F M + + V+ ++ G E+ K+++ +
Sbjct: 67 WNVAISSYMRTGRCNEALRVFKRMP----RWSSVSYNGMISGYLRNGEFELARKLFDEMP 122
Query: 69 VCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQ 128
E + S+ N ++ Y + NL A E+F M + V WN M+ G A +G ++
Sbjct: 123 --ERDLVSW--NVMIKGYVRNRNLGKARELFEIMPERDVCSWNTMLSGYAQNGCVDDARS 178
Query: 129 LFTEMEESLGGS-------------------IRPDRVTFIGVLVACSHKGLVDK-----A 164
+F M E S + R + V C G V K A
Sbjct: 179 VFDRMPEKNDVSWNALLSAYVQNSKMEEACMLFKSRENWALVSWNCLLGGFVKKKKIVEA 238
Query: 165 RWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACR 224
R +FD M R D + ++ ++ G ++EA Q+ +P Q+ V W ++
Sbjct: 239 RQFFDSMNVR-----DVVSWNTIITGYAQSGKIDEARQLFDESPVQD-VFTWTAMVSGYI 292
Query: 225 TQSNTELAEISFKQLAKLKQL 245
E A F ++ + ++
Sbjct: 293 QNRMVEEARELFDKMPERNEV 313
>At1g08070 unknown protein
Length = 741
Score = 227 bits (578), Expect = 7e-60
Identities = 117/291 (40%), Positives = 176/291 (60%), Gaps = 5/291 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
+P ++ +SWN++I G + Y AL LF EM +G P +VT++SIL ACA GA++IG
Sbjct: 327 LPYKDVISWNTLIGGYTHMNLYKEALLLFQEMLRSGETPNDVTMLSILPACAHLGAIDIG 386
Query: 61 H--KIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLA 118
+Y ++ S L +L++MY KCG++ A ++FN + K++S WNAM+ G A
Sbjct: 387 RWIHVYIDKRLKGVTNASSLRTSLIDMYAKCGDIEAAHQVFNSILHKSLSSWNAMIFGFA 446
Query: 119 VHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIV 178
+HG + F LF+ M + I+PD +TF+G+L ACSH G++D R F M + YK+
Sbjct: 447 MHGRADASFDLFSRMRKI---GIQPDDITFVGLLSACSHSGMLDLGRHIFRTMTQDYKMT 503
Query: 179 PDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQ 238
P +HYGCM+DLL GL +EA ++I + V+W +LL AC+ N EL E +
Sbjct: 504 PKLEHYGCMIDLLGHSGLFKEAEEMINMMEMEPDGVIWCSLLKACKMHGNVELGESFAEN 563
Query: 239 LAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
L K++ G YVLLSNIYA AGRW+EV + R+ ++ + + G S I +
Sbjct: 564 LIKIEPENPGSYVLLSNIYASAGRWNEVAKTRALLNDKGMKKVPGCSSIEI 614
Score = 132 bits (331), Expect = 3e-31
Identities = 86/290 (29%), Positives = 148/290 (50%), Gaps = 12/290 (4%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
+P ++ VSWN+MI+G +Y ALELF +M V+P E T+++++ ACA++G++E+G
Sbjct: 226 IPVKDVVSWNAMISGYAETGNYKEALELFKDMMKTNVRPDESTMVTVVSACAQSGSIELG 285
Query: 61 HKIYESLKVCEHKIESYLG--NALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLA 118
+++ L + +H S L NAL+++Y KCG L A +F + K V WN ++ G
Sbjct: 286 RQVH--LWIDDHGFGSNLKIVNALIDLYSKCGELETACGLFERLPYKDVISWNTLIGGYT 343
Query: 119 VHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIV 178
+E LF EM L P+ VT + +L AC+H G +D RW ++ KR K V
Sbjct: 344 HMNLYKEALLLFQEM---LRSGETPNDVTMLSILPACAHLGAIDIGRWIHVYIDKRLKGV 400
Query: 179 PD-SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFK 237
+ S ++D+ ++ G +E A+Q + + S+ W ++ + + F
Sbjct: 401 TNASSLRTSLIDMYAKCGDIEAAHQ-VFNSILHKSLSSWNAMIFGFAMHGRADASFDLFS 459
Query: 238 QLAKLK-QLIDGDYVLLSNIYAEAGRWDEVERLRSEM--DYLHVPRQAGY 284
++ K+ Q D +V L + + +G D + M DY P+ Y
Sbjct: 460 RMRKIGIQPDDITFVGLLSACSHSGMLDLGRHIFRTMTQDYKMTPKLEHY 509
Score = 95.1 bits (235), Expect = 4e-20
Identities = 81/307 (26%), Positives = 138/307 (44%), Gaps = 54/307 (17%)
Query: 3 ERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHK 62
E N + WN+M G D AL+L+ M + G+ P T +L +CA++ A + G +
Sbjct: 96 EPNLLIWNTMFRGHALSSDPVSALKLYVCMISLGLLPNSYTFPFVLKSCAKSKAFKEGQQ 155
Query: 63 IYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIF----------------------- 99
I+ + ++ Y+ +L++MY + G L A ++F
Sbjct: 156 IHGHVLKLGCDLDLYVHTSLISMYVQNGRLEDAHKVFDKSPHRDVVSYTALIKGYASRGY 215
Query: 100 --NGMKM------KTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGV 151
N K+ K V WNAM+ G A G +E +LF +M ++ ++RPD T + V
Sbjct: 216 IENAQKLFDEIPVKDVVSWNAMISGYAETGNYKEALELFKDMMKT---NVRPDESTMVTV 272
Query: 152 LVACSHKGLVDKAR----WYFDH-MVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMT 206
+ AC+ G ++ R W DH KIV ++DL S+ G LE A +
Sbjct: 273 VSACAQSGSIELGRQVHLWIDDHGFGSNLKIV------NALIDLYSKCGELETACGLFER 326
Query: 207 APFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYA-------E 259
P+++ V+ W TL+G + + A + F+++ + + D +LS + A +
Sbjct: 327 LPYKD-VISWNTLIGGYTHMNLYKEALLLFQEMLRSGE-TPNDVTMLSILPACAHLGAID 384
Query: 260 AGRWDEV 266
GRW V
Sbjct: 385 IGRWIHV 391
>At4g21070 putative protein (fragment)
Length = 1495
Score = 226 bits (577), Expect = 9e-60
Identities = 114/289 (39%), Positives = 182/289 (62%), Gaps = 2/289 (0%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MPE++ V+WNS+I G AL L++EM + G+KP T++S+L ACA+ GAL +G
Sbjct: 182 MPEKDLVAWNSVINGFAENGKPEEALALYTEMNSKGIKPDGFTIVSLLSACAKIGALTLG 241
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
+++ + + N L+++Y +CG + A +F+ M K W ++++GLAV+
Sbjct: 242 KRVHVYMIKVGLTRNLHSSNVLLDLYARCGRVEEAKTLFDEMVDKNSVSWTSLIVGLAVN 301
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G+ +E +LF ME + G + P +TF+G+L ACSH G+V + YF M + YKI P
Sbjct: 302 GFGKEAIELFKYMESTEG--LLPCEITFVGILYACSHCGMVKEGFEYFRRMREEYKIEPR 359
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
+H+GCMVDLL+R G +++AY+ I + P Q +VV+WRTLLGAC +++LAE + Q+
Sbjct: 360 IEHFGCMVDLLARAGQVKKAYEYIKSMPMQPNVVIWRTLLGACTVHGDSDLAEFARIQIL 419
Query: 241 KLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
+L+ GDYVLLSN+YA RW +V+++R +M V + G+S + +
Sbjct: 420 QLEPNHSGDYVLLSNMYASEQRWSDVQKIRKQMLRDGVKKVPGHSLVEV 468
Score = 94.4 bits (233), Expect = 7e-20
Identities = 65/255 (25%), Positives = 122/255 (47%), Gaps = 6/255 (2%)
Query: 5 NAVSWNSMIAGCVSVRDYAGALELFSEMQNAG-VKPTEVTLISILGACAETGALEIGHKI 63
N WN++I G + + A L+ EM+ +G V+P T ++ A + +G I
Sbjct: 84 NVFIWNTLIRGYAEIGNSISAFSLYREMRVSGLVEPDTHTYPFLIKAVTTMADVRLGETI 143
Query: 64 YESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYC 123
+ + Y+ N+L+++Y CG+++ A+++F+ M K + WN+++ G A +G
Sbjct: 144 HSVVIRSGFGSLIYVQNSLLHLYANCGDVASAYKVFDKMPEKDLVAWNSVINGFAENGKP 203
Query: 124 EEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKH 183
EE L+TEM I+PD T + +L AC+ G + + +M+K + +
Sbjct: 204 EEALALYTEMNSK---GIKPDGFTIVSLLSACAKIGALTLGKRVHVYMIK-VGLTRNLHS 259
Query: 184 YGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLK 243
++DL +R G +EEA + +NSV ++G E E+ FK + +
Sbjct: 260 SNVLLDLYARCGRVEEAKTLFDEMVDKNSVSWTSLIVGLAVNGFGKEAIEL-FKYMESTE 318
Query: 244 QLIDGDYVLLSNIYA 258
L+ + + +YA
Sbjct: 319 GLLPCEITFVGILYA 333
>At1g20230 hypothetical protein
Length = 760
Score = 226 bits (576), Expect = 1e-59
Identities = 122/289 (42%), Positives = 175/289 (60%), Gaps = 5/289 (1%)
Query: 3 ERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHK 62
E N VSW S+IAGC ALELF EMQ AGVKP VT+ S+L AC AL G
Sbjct: 350 ELNVVSWTSIIAGCAQNGKDIEALELFREMQVAGVKPNHVTIPSMLPACGNIAALGHGRS 409
Query: 63 IYESLKVCEHKIES-YLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHG 121
+ V H +++ ++G+AL++MY KCG ++L+ +FN M K + CWN+++ G ++HG
Sbjct: 410 TH-GFAVRVHLLDNVHVGSALIDMYAKCGRINLSQIVFNMMPTKNLVCWNSLMNGFSMHG 468
Query: 122 YCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDS 181
+EV +F E + ++PD ++F +L AC GL D+ YF M + Y I P
Sbjct: 469 KAKEVMSIF---ESLMRTRLKPDFISFTSLLSACGQVGLTDEGWKYFKMMSEEYGIKPRL 525
Query: 182 KHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAK 241
+HY CMV+LL R G L+EAY +I PF+ +W LL +CR Q+N +LAEI+ ++L
Sbjct: 526 EHYSCMVNLLGRAGKLQEAYDLIKEMPFEPDSCVWGALLNSCRLQNNVDLAEIAAEKLFH 585
Query: 242 LKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINMK 290
L+ G YVLLSNIYA G W EV+ +R++M+ L + + G S I +K
Sbjct: 586 LEPENPGTYVLLSNIYAAKGMWTEVDSIRNKMESLGLKKNPGCSWIQVK 634
Score = 67.8 bits (164), Expect = 7e-12
Identities = 41/159 (25%), Positives = 83/159 (51%), Gaps = 3/159 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
+P+ S++S+I + + ++ +FS M + G+ P L ++ CAE A ++G
Sbjct: 76 IPDPTIYSFSSLIYALTKAKLFTQSIGVFSRMFSHGLIPDSHVLPNLFKVCAELSAFKVG 135
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
+I+ V +++++ ++ +MY +CG + A ++F+ M K V +A++ A
Sbjct: 136 KQIHCVSCVSGLDMDAFVQGSMFHMYMRCGRMGDARKVFDRMSDKDVVTCSALLCAYARK 195
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKG 159
G EEV ++ +EME S I + V++ G+L + G
Sbjct: 196 GCLEEVVRILSEMESS---GIEANIVSWNGILSGFNRSG 231
Score = 67.4 bits (163), Expect = 9e-12
Identities = 59/293 (20%), Positives = 127/293 (43%), Gaps = 38/293 (12%)
Query: 3 ERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHK 62
E N VSWN +++G + A+ +F ++ + G P +VT+ S+L + ++ L +G
Sbjct: 214 EANIVSWNGILSGFNRSGYHKEAVVMFQKIHHLGFCPDQVTVSSVLPSVGDSEMLNMGRL 273
Query: 63 IYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGY 122
I+ + + + +A+++MY K G++ +FN +M NA + GL+ +G
Sbjct: 274 IHGYVIKQGLLKDKCVISAMIDMYGKSGHVYGIISLFNQFEMMEAGVCNAYITGLSRNGL 333
Query: 123 CE---EVFQLFTEMEESLG-----------------------------GSIRPDRVTFIG 150
+ E+F+LF E L ++P+ VT
Sbjct: 334 VDKALEMFELFKEQTMELNVVSWTSIIAGCAQNGKDIEALELFREMQVAGVKPNHVTIPS 393
Query: 151 VLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYG-CMVDLLSRWGLLEEAYQIIMTAPF 209
+L AC + + R V+ + + D+ H G ++D+ ++ G + + + P
Sbjct: 394 MLPACGNIAALGHGRSTHGFAVRVHLL--DNVHVGSALIDMYAKCGRINLSQIVFNMMPT 451
Query: 210 QNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYAEAGR 262
+N +V W +L+ + F+ L + + + D++ +++ + G+
Sbjct: 452 KN-LVCWNSLMNGFSMHGKAKEVMSIFESLMRTR--LKPDFISFTSLLSACGQ 501
Score = 53.9 bits (128), Expect = 1e-07
Identities = 30/118 (25%), Positives = 58/118 (48%), Gaps = 6/118 (5%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MP +N V WNS++ G + +F + +KP ++ S+L AC + G + G
Sbjct: 449 MPTKNLVCWNSLMNGFSMHGKAKEVMSIFESLMRTRLKPDFISFTSLLSACGQVGLTDEG 508
Query: 61 HKIYESLKV---CEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSC-WNAMV 114
K ++ + + ++E Y + +VN+ + G L A+++ M + SC W A++
Sbjct: 509 WKYFKMMSEEYGIKPRLEHY--SCMVNLLGRAGKLQEAYDLIKEMPFEPDSCVWGALL 564
>At1g59720 hypothetical protein
Length = 638
Score = 225 bits (573), Expect = 3e-59
Identities = 120/300 (40%), Positives = 184/300 (61%), Gaps = 13/300 (4%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MPER+ VSWNSMI V +Y AL+LF EMQ + +P T+ S+L ACA G+L +G
Sbjct: 212 MPERSLVSWNSMIDALVRFGEYDSALQLFREMQRS-FEPDGYTMQSVLSACAGLGSLSLG 270
Query: 61 HKIYESL-KVCEHKI--ESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGL 117
+ L + C+ + + + N+L+ MYCKCG+L +A ++F GM+ + ++ WNAM++G
Sbjct: 271 TWAHAFLLRKCDVDVAMDVLVKNSLIEMYCKCGSLRMAEQVFQGMQKRDLASWNAMILGF 330
Query: 118 AVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKI 177
A HG EE F M + ++RP+ VTF+G+L+AC+H+G V+K R YFD MV+ Y I
Sbjct: 331 ATHGRAEEAMNFFDRMVDKRE-NVRPNSVTFVGLLIACNHRGFVNKGRQYFDMMVRDYCI 389
Query: 178 VPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGA-CRTQSNTELAEISF 236
P +HYGC+VDL++R G + EA ++M+ P + V+WR+LL A C+ ++ EL+E
Sbjct: 390 EPALEHYGCIVDLIARAGYITEAIDMVMSMPMKPDAVIWRSLLDACCKKGASVELSEEIA 449
Query: 237 KQLAKLKQ-------LIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
+ + K+ G YVLLS +YA A RW++V +R M + ++ G S I +
Sbjct: 450 RNIIGTKEDNESSNGNCSGAYVLLSRVYASASRWNDVGIVRKLMSEHGIRKEPGCSSIEI 509
Score = 75.9 bits (185), Expect = 2e-14
Identities = 58/242 (23%), Positives = 106/242 (42%), Gaps = 13/242 (5%)
Query: 9 WNSMIAGCV-SVRDYAGALELFSEMQNAGVK-PTEVTLISILGACAETGALEIGHKIYES 66
WN++I C V A L+ +M G P + T +L ACA G +++
Sbjct: 117 WNTLIRACAHDVSRKEEAFMLYRKMLERGESSPDKHTFPFVLKACAYIFGFSEGKQVH-- 174
Query: 67 LKVCEHKI--ESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCE 124
++ +H + Y+ N L+++Y CG L LA ++F+ M +++ WN+M+ L G +
Sbjct: 175 CQIVKHGFGGDVYVNNGLIHLYGSCGCLDLARKVFDEMPERSLVSWNSMIDALVRFGEYD 234
Query: 125 EVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKR--YKIVPDSK 182
QLF EM+ S PD T VL AC+ G + W ++++ + D
Sbjct: 235 SALQLFREMQR----SFEPDGYTMQSVLSACAGLGSLSLGTWAHAFLLRKCDVDVAMDVL 290
Query: 183 HYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKL 242
++++ + G L A Q+ + + W ++ T E A F ++
Sbjct: 291 VKNSLIEMYCKCGSLRMAEQVFQGMQ-KRDLASWNAMILGFATHGRAEEAMNFFDRMVDK 349
Query: 243 KQ 244
++
Sbjct: 350 RE 351
>At2g29760 hypothetical protein
Length = 738
Score = 223 bits (569), Expect = 7e-59
Identities = 116/290 (40%), Positives = 179/290 (61%), Gaps = 4/290 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQ-NAGVKPTEVTLISILGACAETGALEI 59
MP+++ V+WN++I+ AL +F E+Q +K ++TL+S L ACA+ GALE+
Sbjct: 324 MPQKDIVAWNALISAYEQNGKPNEALIVFHELQLQKNMKLNQITLVSTLSACAQVGALEL 383
Query: 60 GHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAV 119
G I+ +K ++ ++ +AL++MY KCG+L + E+FN ++ + V W+AM+ GLA+
Sbjct: 384 GRWIHSYIKKHGIRMNFHVTSALIHMYSKCGDLEKSREVFNSVEKRDVFVWSAMIGGLAM 443
Query: 120 HGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVP 179
HG E +F +M+E+ +++P+ VTF V ACSH GLVD+A F M Y IVP
Sbjct: 444 HGCGNEAVDMFYKMQEA---NVKPNGVTFTNVFCACSHTGLVDEAESLFHQMESNYGIVP 500
Query: 180 DSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQL 239
+ KHY C+VD+L R G LE+A + I P S +W LLGAC+ +N LAE++ +L
Sbjct: 501 EEKHYACIVDVLGRSGYLEKAVKFIEAMPIPPSTSVWGALLGACKIHANLNLAEMACTRL 560
Query: 240 AKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
+L+ DG +VLLSNIYA+ G+W+ V LR M + ++ G S I +
Sbjct: 561 LELEPRNDGAHVLLSNIYAKLGKWENVSELRKHMRVTGLKKEPGCSSIEI 610
Score = 109 bits (273), Expect = 2e-24
Identities = 82/323 (25%), Positives = 153/323 (46%), Gaps = 46/323 (14%)
Query: 3 ERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHK 62
E++ VSWNSMI G V ALELF +M++ VK + VT++ +L ACA+ LE G +
Sbjct: 194 EKDVVSWNSMINGFVQKGSPDKALELFKKMESEDVKASHVTMVGVLSACAKIRNLEFGRQ 253
Query: 63 IYESLKVCEHKIESYLGNALVNMYCKCGNL--------------SLAW------------ 96
+ ++ + L NA+++MY KCG++ ++ W
Sbjct: 254 VCSYIEENRVNVNLTLANAMLDMYTKCGSIEDAKRLFDAMEEKDNVTWTTMLDGYAISED 313
Query: 97 -----EIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGV 151
E+ N M K + WNA++ +G E +F E++ L +++ +++T +
Sbjct: 314 YEAAREVLNSMPQKDIVAWNALISAYEQNGKPNEALIVFHELQ--LQKNMKLNQITLVST 371
Query: 152 LVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQN 211
L AC+ G ++ RW + +K++ I + ++ + S+ G LE++ ++ + +
Sbjct: 372 LSACAQVGALELGRWIHSY-IKKHGIRMNFHVTSALIHMYSKCGDLEKSREVFNSVE-KR 429
Query: 212 SVVLWRTLLGACRTQS-NTELAEISFK-QLAKLKQLIDGDYVLLSNIY---AEAGRWDEV 266
V +W ++G E ++ +K Q A +K + V +N++ + G DE
Sbjct: 430 DVFVWSAMIGGLAMHGCGNEAVDMFYKMQEANVKP----NGVTFTNVFCACSHTGLVDEA 485
Query: 267 ERL--RSEMDYLHVPRQAGYSQI 287
E L + E +Y VP + Y+ I
Sbjct: 486 ESLFHQMESNYGIVPEEKHYACI 508
Score = 85.5 bits (210), Expect = 3e-17
Identities = 66/301 (21%), Positives = 128/301 (41%), Gaps = 42/301 (13%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEM-QNAGVKPTEVTLISILGACAETGALEI 59
+P+ N+ +WN++I S D ++ F +M + P + T ++ A AE +L +
Sbjct: 90 IPKPNSFAWNTLIRAYASGPDPVLSIWAFLDMVSESQCYPNKYTFPFLIKAAAEVSSLSL 149
Query: 60 GHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAV 119
G ++ + ++ N+L++ Y CG+L A ++F +K K V WN+M+ G
Sbjct: 150 GQSLHGMAVKSAVGSDVFVANSLIHCYFSCGDLDSACKVFTTIKEKDVVSWNSMINGFVQ 209
Query: 120 HGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVP 179
G ++ +LF +ME ++ VT +GVL AC+ ++ R + ++ ++
Sbjct: 210 KGSPDKALELFKKMESE---DVKASHVTMVGVLSACAKIRNLEFGRQVCSY-IEENRVNV 265
Query: 180 DSKHYGCMVDLLSRWGLLEEAYQI------------------------------IMTAPF 209
+ M+D+ ++ G +E+A ++ ++ +
Sbjct: 266 NLTLANAMLDMYTKCGSIEDAKRLFDAMEEKDNVTWTTMLDGYAISEDYEAAREVLNSMP 325
Query: 210 QNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGDYVLLSNIYA-------EAGR 262
Q +V W L+ A A I F +L K + L+S + A E GR
Sbjct: 326 QKDIVAWNALISAYEQNGKPNEALIVFHELQLQKNMKLNQITLVSTLSACAQVGALELGR 385
Query: 263 W 263
W
Sbjct: 386 W 386
>At4g16840 hypothetical protein
Length = 661
Score = 223 bits (567), Expect = 1e-58
Identities = 120/292 (41%), Positives = 170/292 (58%), Gaps = 8/292 (2%)
Query: 4 RNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKI 63
+N V+WN+MI+G V L+LF M G++P L S L C+E AL++G +I
Sbjct: 247 KNLVTWNAMISGYVENSRPEDGLKLFRAMLEEGIRPNSSGLSSALLGCSELSALQLGRQI 306
Query: 64 YESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYC 123
++ + + +L++MYCKCG L AW++F MK K V WNAM+ G A HG
Sbjct: 307 HQIVSKSTLCNDVTALTSLISMYCKCGELGDAWKLFEVMKKKDVVAWNAMISGYAQHGNA 366
Query: 124 EEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKH 183
++ LF EM + IRPD +TF+ VL+AC+H GLV+ YF+ MV+ YK+ P H
Sbjct: 367 DKALCLFREM---IDNKIRPDWITFVAVLLACNHAGLVNIGMAYFESMVRDYKVEPQPDH 423
Query: 184 YGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLK 243
Y CMVDLL R G LEEA ++I + PF+ ++ TLLGACR N ELAE + ++L +L
Sbjct: 424 YTCMVDLLGRAGKLEEALKLIRSMPFRPHAAVFGTLLGACRVHKNVELAEFAAEKLLQLN 483
Query: 244 QLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHV-----PRQAGYSQINMK 290
YV L+NIYA RW++V R+R M +V + GYS I ++
Sbjct: 484 SQNAAGYVQLANIYASKNRWEDVARVRKRMKESNVVKVERVKVPGYSWIEIR 535
Score = 53.9 bits (128), Expect = 1e-07
Identities = 58/276 (21%), Positives = 102/276 (36%), Gaps = 52/276 (18%)
Query: 10 NSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESLKV 69
N +IA CV D GAL +F M+ K T ++G + + H++++ +
Sbjct: 65 NKIIARCVRSGDIDGALRVFHGMR---AKNTITWNSLLIGISKDPSRMMEAHQLFDEIP- 120
Query: 70 CEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQL 129
+ +++ N +++ Y + N A F+ M K + WN M+ G A G E+ +L
Sbjct: 121 ---EPDTFSYNIMLSCYVRNVNFEKAQSFFDRMPFKDAASWNTMITGYARRGEMEKAREL 177
Query: 130 FTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVD 189
F M E ++ W M+
Sbjct: 178 FYSMMEK-------------------------NEVSW------------------NAMIS 194
Query: 190 LLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDGD 249
G LE+A AP + VV W ++ ELAE FK + K L+ +
Sbjct: 195 GYIECGDLEKASHFFKVAPVR-GVVAWTAMITGYMKAKKVELAEAMFKDMTVNKNLVTWN 253
Query: 250 YVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYS 285
++S + D ++ R+ ++ P +G S
Sbjct: 254 -AMISGYVENSRPEDGLKLFRAMLEEGIRPNSSGLS 288
Score = 52.8 bits (125), Expect = 2e-07
Identities = 31/107 (28%), Positives = 59/107 (54%), Gaps = 3/107 (2%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
M +++ V+WN+MI+G + AL LF EM + ++P +T +++L AC G + IG
Sbjct: 345 MKKKDVVAWNAMISGYAQHGNADKALCLFREMIDNKIRPDWITFVAVLLACNHAGLVNIG 404
Query: 61 HKIYESLKVCEHKIESYLGN--ALVNMYCKCGNLSLAWEIFNGMKMK 105
+ES+ V ++K+E + +V++ + G L A ++ M +
Sbjct: 405 MAYFESM-VRDYKVEPQPDHYTCMVDLLGRAGKLEEALKLIRSMPFR 450
Score = 45.1 bits (105), Expect = 5e-05
Identities = 59/273 (21%), Positives = 106/273 (38%), Gaps = 49/273 (17%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEM--QNAGVKPTEVTLISILGACAETGALE 58
+PE + S+N M++ V ++ A F M ++A T +T A G +E
Sbjct: 119 IPEPDTFSYNIMLSCYVRNVNFEKAQSFFDRMPFKDAASWNTMIT------GYARRGEME 172
Query: 59 IGHKIYESLKVCEHKIESYLGNALVNMYCKCGNLS--------------LAWE--IFNGM 102
+++ S+ E S+ NA+++ Y +CG+L +AW I M
Sbjct: 173 KARELFYSMM--EKNEVSW--NAMISGYIECGDLEKASHFFKVAPVRGVVAWTAMITGYM 228
Query: 103 KMKTVS----------------CWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRV 146
K K V WNAM+ G + E+ +LF M E IRP+
Sbjct: 229 KAKKVELAEAMFKDMTVNKNLVTWNAMISGYVENSRPEDGLKLFRAMLEE---GIRPNSS 285
Query: 147 TFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMT 206
L+ CS + R +V + + D ++ + + G L +A+++
Sbjct: 286 GLSSALLGCSELSALQLGR-QIHQIVSKSTLCNDVTALTSLISMYCKCGELGDAWKLFEV 344
Query: 207 APFQNSVVLWRTLLGACRTQSNTELAEISFKQL 239
+ VV W ++ N + A F+++
Sbjct: 345 MK-KKDVVAWNAMISGYAQHGNADKALCLFREM 376
>At3g22690 hypothetical protein
Length = 978
Score = 222 bits (565), Expect = 2e-58
Identities = 117/291 (40%), Positives = 174/291 (59%), Gaps = 4/291 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNA-GVKPTEVTLISILGACAETGALEI 59
MPE+N VSWN++I+G V + A+E+F MQ+ GV VT++SI AC GAL++
Sbjct: 429 MPEKNIVSWNTIISGLVQGSLFEEAIEVFCSMQSQEGVNADGVTMMSIASACGHLGALDL 488
Query: 60 GHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAV 119
IY ++ +++ LG LV+M+ +CG+ A IFN + + VS W A + +A+
Sbjct: 489 AKWIYYYIEKNGIQLDVRLGTTLVDMFSRCGDPESAMSIFNSLTNRDVSAWTAAIGAMAM 548
Query: 120 HGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVP 179
G E +LF +M E ++PD V F+G L ACSH GLV + + F M+K + + P
Sbjct: 549 AGNAERAIELFDDMIEQ---GLKPDGVAFVGALTACSHGGLVQQGKEIFYSMLKLHGVSP 605
Query: 180 DSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQL 239
+ HYGCMVDLL R GLLEEA Q+I P + + V+W +LL ACR Q N E+A + +++
Sbjct: 606 EDVHYGCMVDLLGRAGLLEEAVQLIEDMPMEPNDVIWNSLLAACRVQGNVEMAAYAAEKI 665
Query: 240 AKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINMK 290
L G YVLLSN+YA AGRW+++ ++R M + + G S I ++
Sbjct: 666 QVLAPERTGSYVLLSNVYASAGRWNDMAKVRLSMKEKGLRKPPGTSSIQIR 716
Score = 90.1 bits (222), Expect = 1e-18
Identities = 53/158 (33%), Positives = 84/158 (52%), Gaps = 6/158 (3%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYA-GALELFSEM-QNAGVKPTEVTLISILGACAETGALE 58
M ERN VSW SMI G RD+A A++LF M ++ V P VT++ ++ ACA+ LE
Sbjct: 195 MSERNVVSWTSMICGYAR-RDFAKDAVDLFFRMVRDEEVTPNSVTMVCVISACAKLEDLE 253
Query: 59 IGHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLA 118
G K+Y ++ ++ + +ALV+MY KC + +A +F+ + NAM
Sbjct: 254 TGEKVYAFIRNSGIEVNDLMVSALVDMYMKCNAIDVAKRLFDEYGASNLDLCNAMASNYV 313
Query: 119 VHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACS 156
G E +F M +S +RPDR++ + + +CS
Sbjct: 314 RQGLTREALGVFNLMMDS---GVRPDRISMLSAISSCS 348
Score = 87.0 bits (214), Expect = 1e-17
Identities = 63/241 (26%), Positives = 105/241 (43%), Gaps = 39/241 (16%)
Query: 25 ALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESLKVCEHKIESY--LGNAL 82
AL +F+ M ++GV+P ++++S + +C++ + G + V + ES+ + NAL
Sbjct: 321 ALGVFNLMMDSGVRPDRISMLSAISSCSQLRNILWGKSCHGY--VLRNGFESWDNICNAL 378
Query: 83 VNMYCKC-------------------------------GNLSLAWEIFNGMKMKTVSCWN 111
++MY KC G + AWE F M K + WN
Sbjct: 379 IDMYMKCHRQDTAFRIFDRMSNKTVVTWNSIVAGYVENGEVDAAWETFETMPEKNIVSWN 438
Query: 112 AMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHM 171
++ GL EE ++F M+ G + D VT + + AC H G +D A+W + +
Sbjct: 439 TIISGLVQGSLFEEAIEVFCSMQSQEG--VNADGVTMMSIASACGHLGALDLAKWIY-YY 495
Query: 172 VKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTEL 231
+++ I D + +VD+ SR G E A I + V W +GA N E
Sbjct: 496 IEKNGIQLDVRLGTTLVDMFSRCGDPESAMSIFNSLT-NRDVSAWTAAIGAMAMAGNAER 554
Query: 232 A 232
A
Sbjct: 555 A 555
Score = 74.7 bits (182), Expect = 6e-14
Identities = 44/148 (29%), Positives = 76/148 (50%), Gaps = 2/148 (1%)
Query: 9 WNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESLK 68
+NS+I G S A+ LF M N+G+ P + T L ACA++ A G +I+ +
Sbjct: 102 YNSLIRGYASSGLCNEAILLFLRMMNSGISPDKYTFPFGLSACAKSRAKGNGIQIHGLIV 161
Query: 69 VCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQ 128
+ + ++ N+LV+ Y +CG L A ++F+ M + V W +M+ G A + ++
Sbjct: 162 KMGYAKDLFVQNSLVHFYAECGELDSARKVFDEMSERNVVSWTSMICGYARRDFAKDAVD 221
Query: 129 LFTEMEESLGGSIRPDRVTFIGVLVACS 156
LF M + P+ VT + V+ AC+
Sbjct: 222 LFFRMVRD--EEVTPNSVTMVCVISACA 247
>At2g42920 hypothetical protein
Length = 559
Score = 222 bits (565), Expect = 2e-58
Identities = 122/289 (42%), Positives = 169/289 (58%), Gaps = 3/289 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MP+RN VSWNSMI+G V + AL++F EMQ VKP T++S+L ACA GA E G
Sbjct: 218 MPQRNGVSWNSMISGFVRNGRFKDALDMFREMQEKDVKPDGFTMVSLLNACAYLGASEQG 277
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
I+E + ++ S + AL++MYCKCG + +F K +SCWN+M++GLA +
Sbjct: 278 RWIHEYIVRNRFELNSIVVTALIDMYCKCGCIEEGLNVFECAPKKQLSCWNSMILGLANN 337
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G+ E LF+E+E S + PD V+FIGVL AC+H G V +A +F M ++Y I P
Sbjct: 338 GFEERAMDLFSELERS---GLEPDSVSFIGVLTACAHSGEVHRADEFFRLMKEKYMIEPS 394
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
KHY MV++L GLLEEA +I P + V+W +LL ACR N E+A+ + K L
Sbjct: 395 IKHYTLMVNVLGGAGLLEEAEALIKNMPVEEDTVIWSSLLSACRKIGNVEMAKRAAKCLK 454
Query: 241 KLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
KL YVLLSN YA G ++E R M + ++ G S I +
Sbjct: 455 KLDPDETCGYVLLSNAYASYGLFEEAVEQRLLMKERQMEKEVGCSSIEV 503
Score = 89.0 bits (219), Expect = 3e-18
Identities = 65/268 (24%), Positives = 117/268 (43%), Gaps = 32/268 (11%)
Query: 4 RNAVSWNSMIAGCVSVRDYAGALELFSEM--QNAGVKPTEVTLISILGACAETGALEIGH 61
+N WN++I G A+ +F +M + VKP +T S+ A G G
Sbjct: 87 KNPFVWNTIIRGFSRSSFPEMAISIFIDMLCSSPSVKPQRLTYPSVFKAYGRLGQARDGR 146
Query: 62 KIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHG 121
+++ + + +S++ N +++MY CG L AW IF GM V WN+M++G A G
Sbjct: 147 QLHGMVIKEGLEDDSFIRNTMLHMYVTCGCLIEAWRIFLGMIGFDVVAWNSMIMGFAKCG 206
Query: 122 YCEEVFQLFTEMEESLGGS----------------------------IRPDRVTFIGVLV 153
++ LF EM + G S ++PD T + +L
Sbjct: 207 LIDQAQNLFDEMPQRNGVSWNSMISGFVRNGRFKDALDMFREMQEKDVKPDGFTMVSLLN 266
Query: 154 ACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSV 213
AC++ G ++ RW +++V R + +S ++D+ + G +EE + AP + +
Sbjct: 267 ACAYLGASEQGRWIHEYIV-RNRFELNSIVVTALIDMYCKCGCIEEGLNVFECAP-KKQL 324
Query: 214 VLWRTLLGACRTQSNTELAEISFKQLAK 241
W +++ E A F +L +
Sbjct: 325 SCWNSMILGLANNGFEERAMDLFSELER 352
>At1g74630 hypothetical protein
Length = 643
Score = 221 bits (564), Expect = 3e-58
Identities = 113/290 (38%), Positives = 171/290 (58%), Gaps = 4/290 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MP R+ VSW++MI G + + F E+Q AG+ P EV+L +L AC+++G+ E G
Sbjct: 229 MPHRDDVSWSTMIVGIAHNGSFNESFLYFRELQRAGMSPNEVSLTGVLSACSQSGSFEFG 288
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKT-VSCWNAMVIGLAV 119
++ ++ + + NAL++MY +CGN+ +A +F GM+ K + W +M+ GLA+
Sbjct: 289 KILHGFVEKAGYSWIVSVNNALIDMYSRCGNVPMARLVFEGMQEKRCIVSWTSMIAGLAM 348
Query: 120 HGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVP 179
HG EE +LF EM + PD ++FI +L ACSH GL+++ YF M + Y I P
Sbjct: 349 HGQGEEAVRLFNEMTAY---GVTPDGISFISLLHACSHAGLIEEGEDYFSEMKRVYHIEP 405
Query: 180 DSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQL 239
+ +HYGCMVDL R G L++AY I P + ++WRTLLGAC + N ELAE ++L
Sbjct: 406 EIEHYGCMVDLYGRSGKLQKAYDFICQMPIPPTAIVWRTLLGACSSHGNIELAEQVKQRL 465
Query: 240 AKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
+L GD VLLSN YA AG+W +V +R M + + +S + +
Sbjct: 466 NELDPNNSGDLVLLSNAYATAGKWKDVASIRKSMIVQRIKKTTAWSLVEV 515
Score = 69.3 bits (168), Expect = 2e-12
Identities = 65/275 (23%), Positives = 109/275 (39%), Gaps = 46/275 (16%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
M + N V+WN++I C D AGA E+F +M L
Sbjct: 167 MHQPNLVAWNAVITACFRGNDVAGAREIFDKM------------------------LVRN 202
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
H + N ++ Y K G L A IF+ M + W+ M++G+A +
Sbjct: 203 HTSW---------------NVMLAGYIKAGELESAKRIFSEMPHRDDVSWSTMIVGIAHN 247
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKR-YKIVP 179
G E F F E++ + + P+ V+ GVL ACS G + + + K Y +
Sbjct: 248 GSFNESFLYFRELQRA---GMSPNEVSLTGVLSACSQSGSFEFGKILHGFVEKAGYSWIV 304
Query: 180 DSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQL 239
+ ++D+ SR G + A + + +V W +++ E A F ++
Sbjct: 305 SVNN--ALIDMYSRCGNVPMARLVFEGMQEKRCIVSWTSMIAGLAMHGQGEEAVRLFNEM 362
Query: 240 AKLKQLIDG-DYVLLSNIYAEAGRWDEVERLRSEM 273
DG ++ L + + AG +E E SEM
Sbjct: 363 TAYGVTPDGISFISLLHACSHAGLIEEGEDYFSEM 397
>At3g15930 hypothetical protein
Length = 687
Score = 220 bits (560), Expect = 8e-58
Identities = 113/289 (39%), Positives = 180/289 (62%), Gaps = 3/289 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MP R+ +SW MI G + + +LE+F EMQ+AG+ P E T++S+L ACA G+LEIG
Sbjct: 328 MPVRDRISWTIMIDGYLRAGCFNESLEIFREMQSAGMIPDEFTMVSVLTACAHLGSLEIG 387
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
I + + K + +GNAL++MY KCG A ++F+ M + W AMV+GLA +
Sbjct: 388 EWIKTYIDKNKIKNDVVVGNALIDMYFKCGCSEKAQKVFHDMDQRDKFTWTAMVVGLANN 447
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G +E ++F +M++ SI+PD +T++GVL AC+H G+VD+AR +F M ++I P
Sbjct: 448 GQGQEAIKVFFQMQDM---SIQPDDITYLGVLSACNHSGMVDQARKFFAKMRSDHRIEPS 504
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
HYGCMVD+L R GL++EAY+I+ P + ++W LLGA R ++ +AE++ K++
Sbjct: 505 LVHYGCMVDMLGRAGLVKEAYEILRKMPMNPNSIVWGALLGASRLHNDEPMAELAAKKIL 564
Query: 241 KLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
+L+ Y LL NIYA RW ++ +R ++ + + + G+S I +
Sbjct: 565 ELEPDNGAVYALLCNIYAGCKRWKDLREVRRKIVDVAIKKTPGFSLIEV 613
Score = 85.5 bits (210), Expect = 3e-17
Identities = 76/277 (27%), Positives = 124/277 (44%), Gaps = 15/277 (5%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISIL-GACAETGALEI 59
+PE + V WN+MI G V + L+ M GV P T +L G + GAL
Sbjct: 94 IPEPDVVVWNNMIKGWSKVDCDGEGVRLYLNMLKEGVTPDSHTFPFLLNGLKRDGGALAC 153
Query: 60 GHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAV 119
G K++ + Y+ NALV MY CG + +A +F+ + V WN M+ G
Sbjct: 154 GKKLHCHVVKFGLGSNLYVQNALVKMYSLCGLMDMARGVFDRRCKEDVFSWNLMISGYNR 213
Query: 120 HGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACS---HKGLVDKARWYFDHMVKRYK 176
EE +L EME +L + P VT + VL ACS K L + Y V K
Sbjct: 214 MKEYEESIELLVEMERNL---VSPTSVTLLLVLSACSKVKDKDLCKRVHEY----VSECK 266
Query: 177 IVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISF 236
P + +V+ + G ++ A +I + ++ V+ W +++ + N +LA F
Sbjct: 267 TEPSLRLENALVNAYAACGEMDIAVRIFRSMKARD-VISWTSIVKGYVERGNLKLARTYF 325
Query: 237 KQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEM 273
Q+ ++ + ++ + Y AG ++E + EM
Sbjct: 326 DQMPVRDRI---SWTIMIDGYLRAGCFNESLEIFREM 359
>At1g13410 hypothetical protein
Length = 511
Score = 220 bits (560), Expect = 8e-58
Identities = 118/291 (40%), Positives = 172/291 (58%), Gaps = 9/291 (3%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAG-VKPTEVTLISILGACAETGALEI 59
MPERN SWN +I G + L F M + G V P + T+ +L ACA+ GA +
Sbjct: 184 MPERNVFSWNGLIKGYAQNGRVSEVLGSFKRMVDEGSVVPNDATMTLVLSACAKLGAFDF 243
Query: 60 G---HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIG 116
G HK E+L +K++ + NAL++MY KCG + +A E+F G+K + + WN M+ G
Sbjct: 244 GKWVHKYGETLGY--NKVDVNVKNALIDMYGKCGAIEIAMEVFKGIKRRDLISWNTMING 301
Query: 117 LAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYK 176
LA HG+ E LF EM+ S I PD+VTF+GVL AC H GLV+ YF+ M +
Sbjct: 302 LAAHGHGTEALNLFHEMKNS---GISPDKVTFVGVLCACKHMGLVEDGLAYFNSMFTDFS 358
Query: 177 IVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISF 236
I+P+ +H GC+VDLLSR G L +A + I P + V+W TLLGA + ++ E++
Sbjct: 359 IMPEIEHCGCVVDLLSRAGFLTQAVEFINKMPVKADAVIWATLLGASKVYKKVDIGEVAL 418
Query: 237 KQLAKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQI 287
++L KL+ ++V+LSNIY +AGR+D+ RL+ M ++AG S I
Sbjct: 419 EELIKLEPRNPANFVMLSNIYGDAGRFDDAARLKVAMRDTGFKKEAGVSWI 469
Score = 63.5 bits (153), Expect = 1e-10
Identities = 49/220 (22%), Positives = 90/220 (40%), Gaps = 11/220 (5%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
M E+N V W SMI G + +D A F + V +++ E G +
Sbjct: 91 MVEKNVVLWTSMINGYLLNKDLVSARRYFDLSPERDI----VLWNTMISGYIEMGNMLEA 146
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
+++ + C + N ++ Y G++ +F+ M + V WN ++ G A +
Sbjct: 147 RSLFDQMP-CRDVMS---WNTVLEGYANIGDMEACERVFDDMPERNVFSWNGLIKGYAQN 202
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G EV F M + GS+ P+ T VL AC+ G D +W + D
Sbjct: 203 GRVSEVLGSFKRMVDE--GSVVPNDATMTLVLSACAKLGAFDFGKWVHKYGETLGYNKVD 260
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLL 220
++D+ + G +E A ++ + ++ W T++
Sbjct: 261 VNVKNALIDMYGKCGAIEIAMEVFKGIK-RRDLISWNTMI 299
Score = 44.3 bits (103), Expect = 8e-05
Identities = 54/241 (22%), Positives = 95/241 (39%), Gaps = 15/241 (6%)
Query: 26 LELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESLKVCEHKIESYLGNALVNM 85
L LF M++ V P TL +L AC L G ++ + +YLG L+ +
Sbjct: 15 LLLFKRMRSFDVMPNCFTLPVVLKACVMVKDLRQGEELQCFSIKTGFRSNAYLGTKLIEI 74
Query: 86 YCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGGSIRPDR 145
Y G ++ A ++F M K V W +M+ G ++ + F E D
Sbjct: 75 YSCAGVIASANKVFCEMVEKNVVLWTSMINGYLLNKDLVSARRYFDLSPER-------DI 127
Query: 146 VTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIM 205
V + ++ G + +AR FD M R D + +++ + G +E ++
Sbjct: 128 VLWNTMISGYIEMGNMLEARSLFDQMPCR-----DVMSWNTVLEGYANIGDMEACERVFD 182
Query: 206 TAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLI--DGDYVLLSNIYAEAGRW 263
P +N V W L+ SFK++ ++ D L+ + A+ G +
Sbjct: 183 DMPERN-VFSWNGLIKGYAQNGRVSEVLGSFKRMVDEGSVVPNDATMTLVLSACAKLGAF 241
Query: 264 D 264
D
Sbjct: 242 D 242
>At1g06150 unknown protein
Length = 1288
Score = 220 bits (560), Expect = 8e-58
Identities = 112/296 (37%), Positives = 175/296 (58%), Gaps = 4/296 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MP ++ +SW +MI G + Y A+ +F +M G+ P EVT+ +++ ACA G LEIG
Sbjct: 958 MPVKDIISWTTMIKGYSQNKRYREAIAVFYKMMEEGIIPDEVTMSTVISACAHLGVLEIG 1017
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
+++ ++ Y+G+ALV+MY KCG+L A +F + K + CWN+++ GLA H
Sbjct: 1018 KEVHMYTLQNGFVLDVYIGSALVDMYSKCGSLERALLVFFNLPKKNLFCWNSIIEGLAAH 1077
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G+ +E ++F +ME S++P+ VTF+ V AC+H GLVD+ R + M+ Y IV +
Sbjct: 1078 GFAQEALKMFAKMEME---SVKPNAVTFVSVFTACTHAGLVDEGRRIYRSMIDDYSIVSN 1134
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
+HYG MV L S+ GL+ EA ++I F+ + V+W LL CR N +AEI+F +L
Sbjct: 1135 VEHYGGMVHLFSKAGLIYEALELIGNMEFEPNAVIWGALLDGCRIHKNLVIAEIAFNKLM 1194
Query: 241 KLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQA-GYSQINMKESDRL 295
L+ + G Y LL ++YAE RW +V +R M L + + G S I + + D L
Sbjct: 1195 VLEPMNSGYYFLLVSMYAEQNRWRDVAEIRGRMRELGIEKICPGTSSIRIDKRDHL 1250
Score = 67.0 bits (162), Expect = 1e-11
Identities = 65/277 (23%), Positives = 112/277 (39%), Gaps = 51/277 (18%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MPER+ ++W +M++ V D A L ++M + E T
Sbjct: 896 MPERDDIAWTTMVSAYRRVLDMDSANSLANQMS----EKNEAT----------------- 934
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
N L+N Y GNL A +FN M +K + W M+ G + +
Sbjct: 935 ------------------SNCLINGYMGLGNLEQAESLFNQMPVKDIISWTTMIKGYSQN 976
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
E +F +M E I PD VT V+ AC+H G+++ + + ++ V D
Sbjct: 977 KRYREAIAVFYKMMEE---GIIPDEVTMSTVISACAHLGVLEIGKEVHMYTLQN-GFVLD 1032
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
+VD+ S+ G LE A + P +N + W +++ A+ + K A
Sbjct: 1033 VYIGSALVDMYSKCGSLERALLVFFNLPKKN-LFCWNSIIEGLAAHG---FAQEALKMFA 1088
Query: 241 KLK-QLIDGDYVLLSNIY---AEAGRWDEVERLRSEM 273
K++ + + + V +++ AG DE R+ M
Sbjct: 1089 KMEMESVKPNAVTFVSVFTACTHAGLVDEGRRIYRSM 1125
>At1g33350 unknown protein
Length = 538
Score = 219 bits (557), Expect = 2e-57
Identities = 107/290 (36%), Positives = 170/290 (57%), Gaps = 1/290 (0%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNA-GVKPTEVTLISILGACAETGALEI 59
MPER+ SWN+++A C + A+ LF M N ++P EVT++ +L ACA+TG L++
Sbjct: 219 MPERDVPSWNAILAACTQNGLFLEAVSLFRRMINEPSIRPNEVTVVCVLSACAQTGTLQL 278
Query: 60 GHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAV 119
I+ + + ++ N+LV++Y KCGNL A +F K+++ WN+M+ A+
Sbjct: 279 AKGIHAFAYRRDLSSDVFVSNSLVDLYGKCGNLEEASSVFKMASKKSLTAWNSMINCFAL 338
Query: 120 HGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVP 179
HG EE +F EM + I+PD +TFIG+L AC+H GLV K R YFD M R+ I P
Sbjct: 339 HGRSEEAIAVFEEMMKLNINDIKPDHITFIGLLNACTHGGLVSKGRGYFDLMTNRFGIEP 398
Query: 180 DSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQL 239
+HYGC++DLL R G +EA +++ T + +W +LL AC+ + +LAE++ K L
Sbjct: 399 RIEHYGCLIDLLGRAGRFDEALEVMSTMKMKADEAIWGSLLNACKIHGHLDLAEVAVKNL 458
Query: 240 AKLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
L G +++N+Y E G W+E R R + + + + G+S+I +
Sbjct: 459 VALNPNNGGYVAMMANLYGEMGNWEEARRARKMIKHQNAYKPPGWSRIEI 508
>At2g03880 putative selenium-binding protein
Length = 630
Score = 218 bits (556), Expect = 2e-57
Identities = 117/287 (40%), Positives = 169/287 (58%), Gaps = 5/287 (1%)
Query: 5 NAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIY 64
+A+ WNS+I G ALELF M+ AG + TL S+L AC LE+G + +
Sbjct: 224 DAIVWNSIIGGFAQNSRSDVALELFKRMKRAGFIAEQATLTSVLRACTGLALLELGMQAH 283
Query: 65 ESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCE 124
+ + ++ + L NALV+MYCKCG+L A +FN MK + V W+ M+ GLA +GY +
Sbjct: 284 --VHIVKYDQDLILNNALVDMYCKCGSLEDALRVFNQMKERDVITWSTMISGLAQNGYSQ 341
Query: 125 EVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHY 184
E +LF M+ S +P+ +T +GVL ACSH GL++ +YF M K Y I P +HY
Sbjct: 342 EALKLFERMKSS---GTKPNYITIVGVLFACSHAGLLEDGWYYFRSMKKLYGIDPVREHY 398
Query: 185 GCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQ 244
GCM+DLL + G L++A +++ + V WRTLLGACR Q N LAE + K++ L
Sbjct: 399 GCMIDLLGKAGKLDDAVKLLNEMECEPDAVTWRTLLGACRVQRNMVLAEYAAKKVIALDP 458
Query: 245 LIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINMKE 291
G Y LLSNIYA + +WD VE +R+ M + ++ G S I + +
Sbjct: 459 EDAGTYTLLSNIYANSQKWDSVEEIRTRMRDRGIKKEPGCSWIEVNK 505
Score = 74.3 bits (181), Expect = 7e-14
Identities = 57/241 (23%), Positives = 110/241 (44%), Gaps = 14/241 (5%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
MP+RN +SW +MI+ + + ALEL M V+P T S+L +C + +
Sbjct: 122 MPQRNVISWTTMISAYSKCKIHQKALELLVLMLRDNVRPNVYTYSSVLRSCNGMSDVRML 181
Query: 61 HKIYESLKVCEHKIES--YLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLA 118
H + + +ES ++ +AL++++ K G A +F+ M WN+++ G A
Sbjct: 182 H-----CGIIKEGLESDVFVRSALIDVFAKLGEPEDALSVFDEMVTGDAIVWNSIIGGFA 236
Query: 119 VHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIV 178
+ + +LF M+ + ++ T VL AC+ L++ H+VK +
Sbjct: 237 QNSRSDVALELFKRMKRA---GFIAEQATLTSVLRACTGLALLELGMQAHVHIVKYDQ-- 291
Query: 179 PDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQ 238
D +VD+ + G LE+A ++ + V+ W T++ ++ A F++
Sbjct: 292 -DLILNNALVDMYCKCGSLEDALRVFNQMK-ERDVITWSTMISGLAQNGYSQEALKLFER 349
Query: 239 L 239
+
Sbjct: 350 M 350
Score = 63.9 bits (154), Expect = 1e-10
Identities = 50/222 (22%), Positives = 100/222 (44%), Gaps = 8/222 (3%)
Query: 20 RDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESLKVCEHKIESYLG 79
RD A++ +Q+ G+ T ++ C A+ G+ I L H+ +L
Sbjct: 40 RDLPRAMKAMDSLQSHGLWADSATYSELIKCCISNRAVHEGNLICRHLYFNGHRPMMFLV 99
Query: 80 NALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEMEESLGG 139
N L+NMY K L+ A ++F+ M + V W M+ + ++ +L M L
Sbjct: 100 NVLINMYVKFNLLNDAHQLFDQMPQRNVISWTTMISAYSKCKIHQKALELLVLM---LRD 156
Query: 140 SIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEE 199
++RP+ T+ VL +C+ G+ D R ++K + D ++D+ ++ G E+
Sbjct: 157 NVRPNVYTYSSVLRSCN--GMSD-VRMLHCGIIKE-GLESDVFVRSALIDVFAKLGEPED 212
Query: 200 AYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAK 241
A + ++W +++G S +++A FK++ +
Sbjct: 213 ALS-VFDEMVTGDAIVWNSIIGGFAQNSRSDVALELFKRMKR 253
Score = 58.9 bits (141), Expect = 3e-09
Identities = 32/105 (30%), Positives = 58/105 (54%), Gaps = 3/105 (2%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
M ER+ ++W++MI+G AL+LF M+++G KP +T++ +L AC+ G LE G
Sbjct: 319 MKERDVITWSTMISGLAQNGYSQEALKLFERMKSSGTKPNYITIVGVLFACSHAGLLEDG 378
Query: 61 HKIYESLKVCE--HKIESYLGNALVNMYCKCGNLSLAWEIFNGMK 103
+ S+K + + G ++++ K G L A ++ N M+
Sbjct: 379 WYYFRSMKKLYGIDPVREHYG-CMIDLLGKAGKLDDAVKLLNEME 422
>At2g22410 putative protein
Length = 681
Score = 218 bits (555), Expect = 3e-57
Identities = 114/289 (39%), Positives = 164/289 (56%), Gaps = 3/289 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
M E++ V WN+MI G V + AL LF EMQ + KP E+T+I L AC++ GAL++G
Sbjct: 349 MEEKDVVLWNAMIGGSVQAKRGQDALALFQEMQTSNTKPDEITMIHCLSACSQLGALDVG 408
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
I+ ++ + LG +LV+MY KCGN+S A +F+G++ + + A++ GLA+H
Sbjct: 409 IWIHRYIEKYSLSLNVALGTSLVDMYAKCGNISEALSVFHGIQTRNSLTYTAIIGGLALH 468
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G F EM + I PD +TFIG+L AC H G++ R YF M R+ + P
Sbjct: 469 GDASTAISYFNEM---IDAGIAPDEITFIGLLSACCHGGMIQTGRDYFSQMKSRFNLNPQ 525
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
KHY MVDLL R GLLEEA +++ + P + +W LL CR N EL E + K+L
Sbjct: 526 LKHYSIMVDLLGRAGLLEEADRLMESMPMEADAAVWGALLFGCRMHGNVELGEKAAKKLL 585
Query: 241 KLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
+L G YVLL +Y EA W++ +R R M+ V + G S I +
Sbjct: 586 ELDPSDSGIYVLLDGMYGEANMWEDAKRARRMMNERGVEKIPGCSSIEV 634
Score = 110 bits (275), Expect = 9e-25
Identities = 71/266 (26%), Positives = 128/266 (47%), Gaps = 30/266 (11%)
Query: 2 PERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGH 61
P R+ VSWN +I G + + A+ ++ M++ GVKP +VT+I ++ +C+ G L G
Sbjct: 218 PVRDLVSWNCLINGYKKIGEAEKAIYVYKLMESEGVKPDDVTMIGLVSSCSMLGDLNRGK 277
Query: 62 KIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHG 121
+ YE +K ++ L NAL++M+ KCG++ A IF+ ++ +T+ W M+ G A G
Sbjct: 278 EFYEYVKENGLRMTIPLVNALMDMFSKCGDIHEARRIFDNLEKRTIVSWTTMISGYARCG 337
Query: 122 YCEEVFQLFTEMEES--------LGGSI--------------------RPDRVTFIGVLV 153
+ +LF +MEE +GGS+ +PD +T I L
Sbjct: 338 LLDVSRKLFDDMEEKDVVLWNAMIGGSVQAKRGQDALALFQEMQTSNTKPDEITMIHCLS 397
Query: 154 ACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSV 213
ACS G +D W +++Y + + +VD+ ++ G + EA + +NS+
Sbjct: 398 ACSQLGALDVGIW-IHRYIEKYSLSLNVALGTSLVDMYAKCGNISEALSVFHGIQTRNSL 456
Query: 214 VLWRTLLGACRTQSNTELAEISFKQL 239
+ ++G + A F ++
Sbjct: 457 T-YTAIIGGLALHGDASTAISYFNEM 481
Score = 80.1 bits (196), Expect = 1e-15
Identities = 51/219 (23%), Positives = 105/219 (47%), Gaps = 8/219 (3%)
Query: 5 NAVSWNSMIAGCVSVRDYAGALELFSEMQNAGV---KPTEVTLISILGACAETGALEIGH 61
N SWN I G + + L+ +M G +P T + CA+ +GH
Sbjct: 117 NIFSWNVTIRGFSESENPKESFLLYKQMLRHGCCESRPDHFTYPVLFKVCADLRLSSLGH 176
Query: 62 KIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHG 121
I + ++ S++ NA ++M+ CG++ A ++F+ ++ + WN ++ G G
Sbjct: 177 MILGHVLKLRLELVSHVHNASIHMFASCGDMENARKVFDESPVRDLVSWNCLINGYKKIG 236
Query: 122 YCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDS 181
E+ ++ ME ++PD VT IG++ +CS G +++ + ++++ VK +
Sbjct: 237 EAEKAIYVYKLMESE---GVKPDDVTMIGLVSSCSMLGDLNRGKEFYEY-VKENGLRMTI 292
Query: 182 KHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLL 220
++D+ S+ G + EA +I + ++V W T++
Sbjct: 293 PLVNALMDMFSKCGDIHEARRIFDNLE-KRTIVSWTTMI 330
>At3g62890 putative protein
Length = 558
Score = 218 bits (554), Expect = 4e-57
Identities = 114/271 (42%), Positives = 167/271 (61%), Gaps = 8/271 (2%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQ-----NAGVKPTEVTLISILGACAETG 55
MPERN +SW+ +I G V Y AL+LF EMQ A V+P E T+ ++L AC G
Sbjct: 154 MPERNVISWSCLINGYVMCGKYKEALDLFREMQLPKPNEAFVRPNEFTMSTVLSACGRLG 213
Query: 56 ALEIGHKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGM-KMKTVSCWNAMV 114
ALE G ++ + +I+ LG AL++MY KCG+L A +FN + K V ++AM+
Sbjct: 214 ALEQGKWVHAYIDKYHVEIDIVLGTALIDMYAKCGSLERAKRVFNALGSKKDVKAYSAMI 273
Query: 115 IGLAVHGYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKR 174
LA++G +E FQLF+EM S +I P+ VTF+G+L AC H+GL+++ + YF M++
Sbjct: 274 CCLAMYGLTDECFQLFSEMTTS--DNINPNSVTFVGILGACVHRGLINEGKSYFKMMIEE 331
Query: 175 YKIVPDSKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEI 234
+ I P +HYGCMVDL R GL++EA I + P + V++W +LL R + + E
Sbjct: 332 FGITPSIQHYGCMVDLYGRSGLIKEAESFIASMPMEPDVLIWGSLLSGSRMLGDIKTCEG 391
Query: 235 SFKQLAKLKQLIDGDYVLLSNIYAEAGRWDE 265
+ K+L +L + G YVLLSN+YA+ GRW E
Sbjct: 392 ALKRLIELDPMNSGAYVLLSNVYAKTGRWME 422
Score = 60.5 bits (145), Expect = 1e-09
Identities = 34/126 (26%), Positives = 65/126 (50%), Gaps = 3/126 (2%)
Query: 80 NALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFTEME--ESL 137
N++VN Y K G + A ++F+ M + V W+ ++ G + G +E LF EM+ +
Sbjct: 132 NSVVNAYAKAGLIDDARKLFDEMPERNVISWSCLINGYVMCGKYKEALDLFREMQLPKPN 191
Query: 138 GGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLLSRWGLL 197
+RP+ T VL AC G +++ +W + + +Y + D ++D+ ++ G L
Sbjct: 192 EAFVRPNEFTMSTVLSACGRLGALEQGKWVHAY-IDKYHVEIDIVLGTALIDMYAKCGSL 250
Query: 198 EEAYQI 203
E A ++
Sbjct: 251 ERAKRV 256
>At2g20540 unknown protein
Length = 534
Score = 217 bits (553), Expect = 5e-57
Identities = 110/289 (38%), Positives = 178/289 (61%), Gaps = 3/289 (1%)
Query: 1 MPERNAVSWNSMIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIG 60
M ++ VSW +MI+G + Y A++ F EMQ AG++P E++LIS+L +CA+ G+LE+G
Sbjct: 201 MLDKTIVSWTAMISGYTGIGCYVEAMDFFREMQLAGIEPDEISLISVLPSCAQLGSLELG 260
Query: 61 HKIYESLKVCEHKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVH 120
I+ + ++ + NAL+ MY KCG +S A ++F M+ K V W+ M+ G A H
Sbjct: 261 KWIHLYAERRGFLKQTGVCNALIEMYSKCGVISQAIQLFGQMEGKDVISWSTMISGYAYH 320
Query: 121 GYCEEVFQLFTEMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPD 180
G + F EM+ + ++P+ +TF+G+L ACSH G+ + YFD M + Y+I P
Sbjct: 321 GNAHGAIETFNEMQRA---KVKPNGITFLGLLSACSHVGMWQEGLRYFDMMRQDYQIEPK 377
Query: 181 SKHYGCMVDLLSRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLA 240
+HYGC++D+L+R G LE A +I T P + +W +LL +CRT N ++A ++ L
Sbjct: 378 IEHYGCLIDVLARAGKLERAVEITKTMPMKPDSKIWGSLLSSCRTPGNLDVALVAMDHLV 437
Query: 241 KLKQLIDGDYVLLSNIYAEAGRWDEVERLRSEMDYLHVPRQAGYSQINM 289
+L+ G+YVLL+NIYA+ G+W++V RLR + ++ + G S I +
Sbjct: 438 ELEPEDMGNYVLLANIYADLGKWEDVSRLRKMIRNENMKKTPGGSLIEV 486
Score = 69.3 bits (168), Expect = 2e-12
Identities = 63/276 (22%), Positives = 123/276 (43%), Gaps = 12/276 (4%)
Query: 12 MIAGCVSVRDYAGALELFSEMQNAGVKPTEVTLISILGACAETGALEIGHKIYESLKVCE 71
M C S+ ++ + G + VT +++ + L HK+++ + E
Sbjct: 115 MFKSCASLGSCYLGKQVHGHLCKFGPRFHVVTENALIDMYMKFDDLVDAHKVFDEMY--E 172
Query: 72 HKIESYLGNALVNMYCKCGNLSLAWEIFNGMKMKTVSCWNAMVIGLAVHGYCEEVFQLFT 131
+ S+ N+L++ Y + G + A +F+ M KT+ W AM+ G G E F
Sbjct: 173 RDVISW--NSLLSGYARLGQMKKAKGLFHLMLDKTIVSWTAMISGYTGIGCYVEAMDFFR 230
Query: 132 EMEESLGGSIRPDRVTFIGVLVACSHKGLVDKARWYFDHMVKRYKIVPDSKHYGCMVDLL 191
EM+ L G I PD ++ I VL +C+ G ++ +W + +R + + ++++
Sbjct: 231 EMQ--LAG-IEPDEISLISVLPSCAQLGSLELGKWIHLY-AERRGFLKQTGVCNALIEMY 286
Query: 192 SRWGLLEEAYQIIMTAPFQNSVVLWRTLLGACRTQSNTELAEISFKQLAKLKQLIDG-DY 250
S+ G++ +A Q+ ++ V+ W T++ N A +F ++ + K +G +
Sbjct: 287 SKCGVISQAIQLFGQMEGKD-VISWSTMISGYAYHGNAHGAIETFNEMQRAKVKPNGITF 345
Query: 251 VLLSNIYAEAGRWDEVERLRSEM--DYLHVPRQAGY 284
+ L + + G W E R M DY P+ Y
Sbjct: 346 LGLLSACSHVGMWQEGLRYFDMMRQDYQIEPKIEHY 381
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.136 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 6,532,030
Number of Sequences: 26719
Number of extensions: 267056
Number of successful extensions: 4444
Number of sequences better than 10.0: 427
Number of HSP's better than 10.0 without gapping: 384
Number of HSP's successfully gapped in prelim test: 43
Number of HSP's that attempted gapping in prelim test: 606
Number of HSP's gapped (non-prelim): 1755
length of query: 296
length of database: 11,318,596
effective HSP length: 99
effective length of query: 197
effective length of database: 8,673,415
effective search space: 1708662755
effective search space used: 1708662755
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144765.20


