

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.18 - phase: 2 /pseudo
(374 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
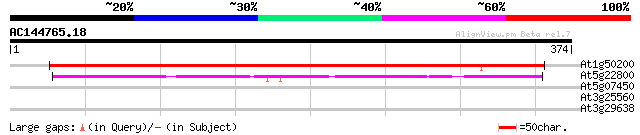
At1g50200 alanine--tRNA ligase, putative 492 e-139
At5g22800 alanyl-tRNA synthetase 163 2e-40
At5g07450 unknown protein 30 2.1
At3g25560 unknown protein 28 6.2
At3g29638 putative protein 28 8.1
>At1g50200 alanine--tRNA ligase, putative
Length = 1027
Score = 492 bits (1267), Expect = e-139
Identities = 251/354 (70%), Positives = 282/354 (78%), Gaps = 24/354 (6%)
Query: 27 NNDDPTCIEIWNLVLXQFNREADSSLKSLPAKHVDTGMGFERLTSILQRKLSNYDTDVFM 86
NNDDPTC+EIWNLV QFNRE+D SLK LPAKHVDTGMGFERLTS+LQ K+SNYDTDVFM
Sbjct: 258 NNDDPTCLEIWNLVFIQFNRESDGSLKPLPAKHVDTGMGFERLTSVLQNKMSNYDTDVFM 317
Query: 87 PIFDAIQLATGARPYSGKVGPEDADKIDMAYRVVADHIRTLSFAIADGSRPGNEGREYVL 146
PIFD IQ ATGARPYSGKVGPED D++DMAYRVVADHIRTLSFAIADGSRPGNEGREYVL
Sbjct: 318 PIFDDIQKATGARPYSGKVGPEDVDRVDMAYRVVADHIRTLSFAIADGSRPGNEGREYVL 377
Query: 147 RRILRRAVRYGREVLKAEEGFFNGLVNVVANVLGDVFPELKQQEVHIRNVIQEEEESFGR 206
RRILRRAVRYG+E+LKAEEGFFNGLV+ V V+GDVF ELK+ E I ++I+EEE SF +
Sbjct: 378 RRILRRAVRYGKEILKAEEGFFNGLVSSVIRVMGDVFTELKEHEKKITDIIKEEEASFCK 437
Query: 207 TLVKGIAKFKTAVGHVQGNILSGEATFELWDTYGFPLDLTQLMAEEKKLKVDVEGFNSAM 266
TL KGI KF+ A VQGN LSG+ F LWDTYGFPLDLTQLMAEE+ L VDV+GFN AM
Sbjct: 438 TLAKGIEKFRKAGQAVQGNTLSGDDAFILWDTYGFPLDLTQLMAEERGLLVDVDGFNKAM 497
Query: 267 EAARERSRTAQNKQAGGAIVMDADATSALQKRSIVPTDDSFKFASFR------------- 313
E ARERSR+AQNKQAGGAIVMDADATS L K + TDDSFK+ F+
Sbjct: 498 EEARERSRSAQNKQAGGAIVMDADATSTLHKAGVSATDDSFKYIWFQVCPFIPENLHVAP 557
Query: 314 -----------DHETVVKAIYTGSEFVDSVNTDGDVGVILESTSFYAEQGGQVF 356
DHE+ +KAIYTGS F++S +VG++L STSFYAEQGGQ+F
Sbjct: 558 EMFLCWRRNEEDHESELKAIYTGSTFLESSAASDNVGLVLGSTSFYAEQGGQIF 611
>At5g22800 alanyl-tRNA synthetase
Length = 954
Score = 163 bits (412), Expect = 2e-40
Identities = 111/347 (31%), Positives = 171/347 (48%), Gaps = 39/347 (11%)
Query: 29 DDPTCIEIWNLVLXQFNREADSSLKSLPAKHVDTGMGFERLTSILQRKLSNYDTDVFMPI 88
DD IE +NLV Q+N+ D L+ L K++DTG+G ER+ ILQ+ +NY+TD+ PI
Sbjct: 240 DDTRFIEFYNLVFMQYNKTEDGLLEPLKQKNIDTGLGLERIAQILQKVPNNYETDLIYPI 299
Query: 89 FDAIQLATGARPYSGKVGPEDADKIDMAYRVVADHIRTLSFAIADGSRPGNEGREYVLRR 148
I S DK + +V+ADH+R + + I+DG P N GR YV+RR
Sbjct: 300 IAKISELANISYDSAN------DKAKTSLKVIADHMRAVVYLISDGVSPSNIGRGYVVRR 353
Query: 149 ILRRAVRYGREVLKAEEGFFNG------LVNVVANVL--------------GDVFPELKQ 188
++RRAVR G+ + G NG L V V+ + E++Q
Sbjct: 354 LIRRAVRKGKSL--GINGDMNGNLKGAFLPAVAEKVIELSTYIDSDVKLKASRIIEEIRQ 411
Query: 189 QEVHIRNVIQEEEESFGRTLVKGIAKFKTAVGHVQGNILSGEATFELWDTYGFPLDLTQL 248
+E+H + ++ E+ + L ++ A L G+ F L+DT+GFP+++T
Sbjct: 412 EELHFKKTLERGEKLLDQKLNDALS---IADKTKDTPYLDGKDAFLLYDTFGFPVEITAE 468
Query: 249 MAEEKKLKVDVEGFNSAMEAARERSRTAQNKQAGGAIVMDADATSALQKRSIVPTDDSFK 308
+AEE+ + +D+ GF ME R +S+ A N + DAD T + D F
Sbjct: 469 VAEERGVSIDMNGFEVEMENQRRQSQAAHN-VVKLTVEDDADMTKNI-------ADTEFL 520
Query: 309 FASFRDHETVVKAIYTGSEFVDSVNTDGDVGVILESTSFYAEQGGQV 355
VVK++ + V V+ +V V+L+ T FYAE GGQ+
Sbjct: 521 GYDSLSARAVVKSLLVNGKPVIRVSEGSEVEVLLDRTPFYAESGGQI 567
>At5g07450 unknown protein
Length = 216
Score = 30.0 bits (66), Expect = 2.1
Identities = 19/80 (23%), Positives = 37/80 (45%), Gaps = 15/80 (18%)
Query: 295 LQKRSIVPTDDSFKFASFRDHETVVKAIYTGSEFVDSVNTDGDVGVILESTSFYAEQGGQ 354
+QK+ ++P D S H ++ ++ ++F+D + + +FYA+ GG
Sbjct: 93 IQKQPLLPIDSS------NVHRLIITSVLVSAKFMDD---------LCYNNAFYAKVGGI 137
Query: 355 VFSIVNILYMIFCMSILFYL 374
+N+L + F I F L
Sbjct: 138 TTEEMNLLELDFLFGIGFQL 157
>At3g25560 unknown protein
Length = 635
Score = 28.5 bits (62), Expect = 6.2
Identities = 23/81 (28%), Positives = 39/81 (47%), Gaps = 7/81 (8%)
Query: 176 ANVLGDVFPELKQQEVHIRNVIQEEEESFGRTLVKGIAKFKTAVGHVQGNILSGEATFEL 235
AN+L D + E + + ++ + EES T V+G VGH+ LS + E
Sbjct: 439 ANILLDDYFEAVVGDFGLAKLL-DHEESHVTTAVRG------TVGHIAPEYLSTGQSSEK 491
Query: 236 WDTYGFPLDLTQLMAEEKKLK 256
D +GF + L +L+ + L+
Sbjct: 492 TDVFGFGILLLELITGLRALE 512
>At3g29638 putative protein
Length = 412
Score = 28.1 bits (61), Expect = 8.1
Identities = 28/97 (28%), Positives = 44/97 (44%), Gaps = 14/97 (14%)
Query: 183 FPELKQQEVHIRNVIQEEEESFGRTLVKGIAKFKTAVGHVQGNILSGEATFELWDTY--- 239
F ++ E H +V+ EEESF I F T V I+S +A FE ++ Y
Sbjct: 325 FRNYREVEKHGESVMLSEEESF------KIDYFYTIVDQA---IVSLQARFEQFEEYGKH 375
Query: 240 -GFPLDLTQLMAE-EKKLKVDVEGFNSAMEAARERSR 274
GF +DL +L E E LK ++++ + +
Sbjct: 376 FGFLIDLRKLKRETEDGLKASCINLETSLKHGKNSDK 412
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.138 0.408
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,130,696
Number of Sequences: 26719
Number of extensions: 335963
Number of successful extensions: 789
Number of sequences better than 10.0: 5
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 783
Number of HSP's gapped (non-prelim): 5
length of query: 374
length of database: 11,318,596
effective HSP length: 101
effective length of query: 273
effective length of database: 8,619,977
effective search space: 2353253721
effective search space used: 2353253721
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144765.18


