

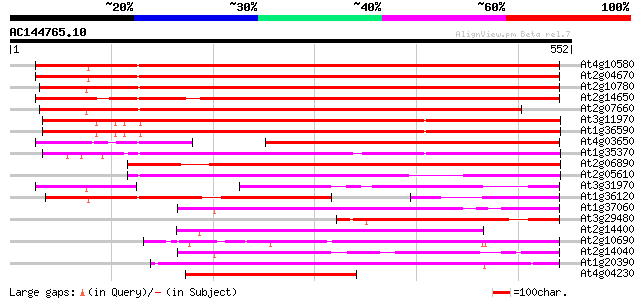
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144765.10 - phase: 0
(552 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g10580 putative reverse-transcriptase -like protein 596 e-171
At2g04670 putative retroelement pol polyprotein 593 e-170
At2g10780 pseudogene 591 e-169
At2g14650 putative retroelement pol polyprotein 555 e-158
At2g07660 putative retroelement pol polyprotein 526 e-149
At3g11970 hypothetical protein 420 e-118
At1g36590 hypothetical protein 420 e-118
At4g03650 putative reverse transcriptase 396 e-110
At1g35370 hypothetical protein 387 e-108
At2g06890 putative retroelement integrase 376 e-104
At2g05610 putative retroelement pol polyprotein 346 2e-95
At3g31970 hypothetical protein 241 6e-64
At1g36120 putative reverse transcriptase gb|AAD22339.1 241 1e-63
At1g37060 Athila retroelment ORF 1, putative 236 2e-62
At3g29480 hypothetical protein 213 2e-55
At2g14400 putative retroelement pol polyprotein 206 3e-53
At2g10690 putative retroelement pol polyprotein 206 4e-53
At2g14040 putative retroelement pol polyprotein 199 3e-51
At1g20390 hypothetical protein 199 3e-51
At4g04230 putative transposon protein 196 3e-50
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 596 bits (1537), Expect = e-171
Identities = 290/526 (55%), Positives = 377/526 (71%), Gaps = 11/526 (2%)
Query: 26 LSMFGRDFEVDLVCLPLLGMDVIFGMNWLEYNRVCINCFNKTVHFSSAEEE--------- 76
+ + G DL+ P+ DVI GM+WL+Y RV ++ V F E
Sbjct: 358 IQIAGESMPADLIISPVELYDVILGMDWLDYYRVHLDWHRGRVFFERPEGRLVYQGVRPI 417
Query: 77 SGAQFLTTKQLKQLERDGILMFSLMASLSLE-NQVVIDRLPVVNEFHEVFPDEIPDVPPE 135
SG+ ++ Q +++ G + + S+ QV + + VV EF +VF + +PP
Sbjct: 418 SGSLVISAVQAEKMIEKGCEAYLVTISMPESVGQVAVSDIRVVQEFQDVF-QSLQGLPPS 476
Query: 136 REVEFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGAPVLLVKK 195
+ F+I+L PG +S APY M+ +E+AELKKQL+DLL K F+RPS SPWGAPVL VKK
Sbjct: 477 QSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKK 536
Query: 196 KDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDM 255
KDGS RLCIDYR+LN+VT+KNRYPLPRID+L+DQL GA FSKIDL SGYHQI + + D+
Sbjct: 537 KDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADV 596
Query: 256 QKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEE 315
+KTAFRTRYGH+E+ VMPFG+TNAP VFM MN +F FLD+FV++FIDDIL+YSK EE
Sbjct: 597 RKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEE 656
Query: 316 HAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPK 375
HL+ V++ L+E+KL+AKLSKC FW E+ FLGH+ S G++VDP K++A+ W P
Sbjct: 657 QEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWPRPT 716
Query: 376 SVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAP 435
+ TEIRSFLG AGYYRRF++GF+ +A P+T+LT K FVW +CE F LK+ LT+ P
Sbjct: 717 NATEIRSFLGWAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSQECEEGFVSLKEMLTSTP 776
Query: 436 ILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFV 495
+L LP+ +P++VY DAS+ GLG VLMQ GKV+AYASRQL HE N PTHDLE+AAV+F
Sbjct: 777 VLALPEHGQPYMVYTDASRVGLGCVLMQHGKVIAYASRQLMKHEGNYPTHDLEMAAVIFA 836
Query: 496 LKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
LKIWR YLYG + +VF+DHKSLKY+F Q ELN+RQRRW+EL+ DYD
Sbjct: 837 LKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYD 882
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 593 bits (1529), Expect = e-170
Identities = 287/526 (54%), Positives = 373/526 (70%), Gaps = 11/526 (2%)
Query: 26 LSMFGRDFEVDLVCLPLLGMDVIFGMNWLEYNRVCINCFNKTVHFSSAEEE--------- 76
+ + G DL+ P+ DVI GM+WL++ RV ++C V F E
Sbjct: 384 IQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPT 443
Query: 77 SGAQFLTTKQLKQLERDGILMFSLMASLSLE-NQVVIDRLPVVNEFHEVFPDEIPDVPPE 135
SG+ ++ Q K++ G + + S+ QV + + V+ EF +VF + +PP
Sbjct: 444 SGSLVISAVQAKKMIEKGCEAYLVTISMPESLGQVAVSDIRVIQEFEDVF-QSLQGLPPS 502
Query: 136 REVEFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGAPVLLVKK 195
R F+I+L PG +S APY M+ +E+ ELKKQLEDLL K F+RPS SPWGAPVL VKK
Sbjct: 503 RSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKK 562
Query: 196 KDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDM 255
KDGS RLCIDYR LN VT+KN+YPLPRID+L+DQL GA FSKIDL SGYHQI + + D+
Sbjct: 563 KDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADV 622
Query: 256 QKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEE 315
+KTAFRTRYGH+E+ VMPF +TNAP FM MN +F FLD+FV++FIDDIL+YSK EE
Sbjct: 623 RKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSKSPEE 682
Query: 316 HAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPK 375
H HL+ V++ L+E+KL+AKLSKC FW E+ FLGH+ S G++VDP K++A+ W P
Sbjct: 683 HEVHLRRVMEKLREQKLFAKLSKCSFWQREIGFLGHIVSAEGVSVDPEKIEAIRDWPRPT 742
Query: 376 SVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAP 435
+ TEIRSFL L GYYRRF++GF+ +A P+T+LT K FVW +CE F LK+ LT+ P
Sbjct: 743 NATEIRSFLRLTGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTP 802
Query: 436 ILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFV 495
+L LP+ +P++VY DAS+ GLG VLMQ GKV+AYASRQLR HE N PTHDLE+A V+F
Sbjct: 803 VLALPEHGQPYMVYTDASRVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAVVIFA 862
Query: 496 LKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
LKIWR YLYG + +VF+DHKSLKY+F+Q ELN+RQ RW+EL+ DYD
Sbjct: 863 LKIWRSYLYGGKVQVFTDHKSLKYIFNQPELNLRQMRWMELVADYD 908
>At2g10780 pseudogene
Length = 1611
Score = 591 bits (1524), Expect = e-169
Identities = 290/522 (55%), Positives = 378/522 (71%), Gaps = 11/522 (2%)
Query: 30 GRDFEVDLVCLPLLGMDVIFGMNWLEYNRVCINCFNKTVHFSSAE---------EESGAQ 80
G + DL+ +PL DVI GM+WL + I+C + F E SG+
Sbjct: 530 GVNMPADLIIVPLKKHDVILGMDWLGKYKAHIDCHRGRIQFERDEGMLKFQGIRTTSGSL 589
Query: 81 FLTTKQLKQLERDGILMF-SLMASLSLENQVVIDRLPVVNEFHEVFPDEIPDVPPEREVE 139
++ Q +++ G + + + + + + +P+VNEF +VF + VPP+R
Sbjct: 590 VISAIQAERMLGKGCEAYLATITTKEVGASAELKDIPIVNEFSDVFA-AVSGVPPDRSDP 648
Query: 140 FSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGAPVLLVKKKDGS 199
F+I+L PG +S APY M+ +E+A+LKKQLE+LLDK F+RPS SPWGAPVL VKKKDGS
Sbjct: 649 FTIELEPGTTPISKAPYRMAPAEMAKLKKQLEELLDKGFIRPSSSPWGAPVLFVKKKDGS 708
Query: 200 MRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTA 259
RLCIDYR LNKVT+KN+YPLPRID+LMDQL GA+ FSKIDL SGYHQI ++ D++KTA
Sbjct: 709 FRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTA 768
Query: 260 FRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKH 319
FRTRY H+E+ VMPFG+TNAP FM+ MN +F FLD+FV++FI+DIL+YSK E H +H
Sbjct: 769 FRTRYDHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEH 828
Query: 320 LKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTE 379
L+ VL+ L+E +L+AKLSKC FW V FLGHV S G++VDP K+ ++ +W P++ TE
Sbjct: 829 LRAVLERLREHELFAKLSKCSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATE 888
Query: 380 IRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILIL 439
IRSFLGLAGYYRRF+ F+ +A PLT+LT K +F W +CE SF ELK LT AP+L+L
Sbjct: 889 IRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLTNAPVLVL 948
Query: 440 PKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFVLKIW 499
P+ EP+ VY DAS GLG VLMQ G V+AYASRQLR HEKN PTHDLE+AAVVF LKIW
Sbjct: 949 PEEGEPYTVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFFLKIW 1008
Query: 500 RHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
R YLYG++ ++++DHKSLKY+F Q ELN+RQRRW+EL+ DY+
Sbjct: 1009 RSYLYGAKVQIYTDHKSLKYIFTQPELNLRQRRWMELVADYN 1050
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 555 bits (1431), Expect = e-158
Identities = 279/516 (54%), Positives = 355/516 (68%), Gaps = 24/516 (4%)
Query: 26 LSMFGRDFEVDLVCLPLLGMDVIFGMNWLEYNRVCINCFNKTVHFSSAEEESGAQFLTTK 85
+ + G DL+ P+ DVI GM+WL++ RV ++C V F E Q +
Sbjct: 366 IQIAGESMPADLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGSLVYQGVRPT 425
Query: 86 QLKQLERDGILMFSLMASLSLENQVVIDRLPVVNEFHEVFPDEIPDVPPEREVEFSIDLV 145
SL+ S +++ EF +VF + +PP R F+I+L
Sbjct: 426 S-----------GSLVISAVQAEKMIEKGCEAYLEFEDVF-QSLQGLPPSRSDPFTIELE 473
Query: 146 PGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGAPVLLVKKKDGSMRLCID 205
PG +S APY M+ +E+AELKKQLEDLL GAPVL VKKKDGS RLCID
Sbjct: 474 PGTAPLSKAPYRMAPAEMAELKKQLEDLL------------GAPVLFVKKKDGSFRLCID 521
Query: 206 YRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYG 265
YR LN VT+KN+YPLPRID+L+DQL GA FSKIDL SGYH I + + D++KTAFRTRYG
Sbjct: 522 YRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAFRTRYG 581
Query: 266 HYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQ 325
H+E+ VMPFG+TNAP FM MN +F LD+FV++FIDDIL+YSK EEH HL+ V++
Sbjct: 582 HFEFVVMPFGLTNAPAAFMRLMNSVFQEVLDEFVIIFIDDILVYSKSLEEHEVHLRRVME 641
Query: 326 ILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLG 385
L+E+KL+AKLSKC FW E+ FLGH+ S G++VDP K++A+ W TP + TEIRSFLG
Sbjct: 642 KLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDWHTPTNATEIRSFLG 701
Query: 386 LAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILILPKPEEP 445
LAGYYRRF++GF+ +A P+T+LT K FVW +CE F LK+ LT+ P+L LP+ EP
Sbjct: 702 LAGYYRRFVKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEMLTSTPVLALPEHGEP 761
Query: 446 FVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFVLKIWRHYLYG 505
++VY DAS GLG VLMQ GKV+AYASRQLR HE N PTHDLE+AAV+F LKIWR YLYG
Sbjct: 762 YMVYTDASGVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMAAVIFALKIWRSYLYG 821
Query: 506 SRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
+VF+DHKSLKY+F Q ELN+RQR+W+EL+ DYD
Sbjct: 822 GNVQVFTDHKSLKYIFTQPELNLRQRQWMELVADYD 857
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 526 bits (1354), Expect = e-149
Identities = 265/484 (54%), Positives = 342/484 (69%), Gaps = 11/484 (2%)
Query: 30 GRDFEVDLVCLPLLGMDVIFGMNWLEYNRVCINCFNKTVHFSSAE---------EESGAQ 80
G + DL+ +PL DVI GM+WL + I+C + + F E SG+
Sbjct: 25 GVNMPADLIIVPLKKHDVILGMDWLGKYKGHIDCHRERIQFERDEGMLKFQGIRTTSGSL 84
Query: 81 FLTTKQLKQLERDGILMF-SLMASLSLENQVVIDRLPVVNEFHEVFPDEIPDVPPEREVE 139
++ Q +++ G + + + + + + + +VNEF +VF + VPP+R
Sbjct: 85 VISAIQAERMLGKGCEAYLATITTKEVGASAELKDILIVNEFSDVFA-AVSGVPPDRSDP 143
Query: 140 FSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGAPVLLVKKKDGS 199
F+I+L PG +S APY M+ +E+AELKKQLE+LL K F+RPS SPWGAPVL VKKKDGS
Sbjct: 144 FTIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGS 203
Query: 200 MRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTA 259
RLCIDYR LNKVT+KN+YPLPRID+LMDQL GA+ FSKIDL SGYHQI ++ D++KTA
Sbjct: 204 FRLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTA 263
Query: 260 FRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKH 319
FRTRYGH+E+ VMPFG+TNAP FM+ MN +F FLD+FV++FIDDIL++SK E H +H
Sbjct: 264 FRTRYGHFEFVVMPFGLTNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEH 323
Query: 320 LKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTE 379
L+ VL+ L+E +L+AKLSK FW V FLGHV S G++VDP K+ ++ +W P++ TE
Sbjct: 324 LRAVLERLREHELFAKLSKFSFWQRSVGFLGHVISDQGVSVDPEKIRSIKEWPRPRNATE 383
Query: 380 IRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILIL 439
IRSFLGLAGYYRRF+ F+ +A PLT+LT K +F W +CE SF ELK L AP+L+L
Sbjct: 384 IRSFLGLAGYYRRFVMSFASMAQPLTRLTGKDTAFNWSDECEKSFLELKAMLINAPVLVL 443
Query: 440 PKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFVLKIW 499
P+ EP+ VY DAS GLG VLMQ G V+AYASRQLR HEKN PTHDLE+AAVVF LKIW
Sbjct: 444 PEEGEPYTVYTDASIVGLGCVLMQKGSVIAYASRQLRKHEKNYPTHDLEMAAVVFALKIW 503
Query: 500 RHYL 503
R YL
Sbjct: 504 RSYL 507
>At3g11970 hypothetical protein
Length = 1499
Score = 420 bits (1080), Expect = e-118
Identities = 229/534 (42%), Positives = 332/534 (61%), Gaps = 25/534 (4%)
Query: 33 FEVDLVCLPLLGMDVIFGMNWLE-YNRVCINCFNKTVHFSSAEEESGAQFLTT------- 84
F+ D++ +PL G+D++ G+ WLE R+ + F ++ LT+
Sbjct: 460 FQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVREVK 519
Query: 85 -KQLKQLERDGILMFSLMA---SLSLENQVV--------IDRLPVVNEFHEVFPD---EI 129
++L++L+ D + + L S S E ++ + VV E +PD E
Sbjct: 520 AQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVLNEYPDIFIEP 579
Query: 130 PDVPPEREVE-FSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGA 188
+PP RE I L+ G+ V+ PY S + E+ K +EDLL V+ S SP+ +
Sbjct: 580 TALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYAS 639
Query: 189 PVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQI 248
PV+LVKKKDG+ RLC+DYR+LN +T+K+ +P+P I+DLMD+L GA +FSKIDLR+GYHQ+
Sbjct: 640 PVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQV 699
Query: 249 KVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILI 308
++ +D+QKTAF+T GH+EY VMPFG+TNAP F MN IF FL KFV+VF DDIL+
Sbjct: 700 RMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILV 759
Query: 309 YSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAV 368
YS EEH +HLK V ++++ KL+AKLSKC F + +V +LGH S GI DP+K+ AV
Sbjct: 760 YSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAV 819
Query: 369 SQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELK 428
+W P ++ ++R FLGLAGYYRRF+ F +A PL LT K +F W A + +F +LK
Sbjct: 820 KEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALT-KTDAFEWTAVAQQAFEDLK 878
Query: 429 QRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLE 488
L AP+L LP ++ FVV DA G+G VLMQ+G +AY SRQL+ + + ++ E
Sbjct: 879 AALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQLHLSIYEKE 938
Query: 489 LAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDF 542
L AV+F ++ WRHYL S F + +D +SLKYL +Q+ Q++WL L ++D+
Sbjct: 939 LLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDY 992
>At1g36590 hypothetical protein
Length = 1499
Score = 420 bits (1080), Expect = e-118
Identities = 229/534 (42%), Positives = 332/534 (61%), Gaps = 25/534 (4%)
Query: 33 FEVDLVCLPLLGMDVIFGMNWLE-YNRVCINCFNKTVHFSSAEEESGAQFLTT------- 84
F+ D++ +PL G+D++ G+ WLE R+ + F ++ LT+
Sbjct: 460 FQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVREVK 519
Query: 85 -KQLKQLERDGILMFSLMA---SLSLENQVV--------IDRLPVVNEFHEVFPD---EI 129
++L++L+ D + + L S S E ++ + VV E +PD E
Sbjct: 520 AQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVLNEYPDIFIEP 579
Query: 130 PDVPPEREVE-FSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGA 188
+PP RE I L+ G+ V+ PY S + E+ K +EDLL V+ S SP+ +
Sbjct: 580 TALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASSSPYAS 639
Query: 189 PVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQI 248
PV+LVKKKDG+ RLC+DYR+LN +T+K+ +P+P I+DLMD+L GA +FSKIDLR+GYHQ+
Sbjct: 640 PVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRAGYHQV 699
Query: 249 KVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILI 308
++ +D+QKTAF+T GH+EY VMPFG+TNAP F MN IF FL KFV+VF DDIL+
Sbjct: 700 RMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVFFDDILV 759
Query: 309 YSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAV 368
YS EEH +HLK V ++++ KL+AKLSKC F + +V +LGH S GI DP+K+ AV
Sbjct: 760 YSSSLEEHRQHLKQVFEVMRANKLFAKLSKCAFAVPKVEYLGHFISAQGIETDPAKIKAV 819
Query: 369 SQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELK 428
+W P ++ ++R FLGLAGYYRRF+ F +A PL LT K +F W A + +F +LK
Sbjct: 820 KEWPQPTTLKQLRGFLGLAGYYRRFVRSFGVIAGPLHALT-KTDAFEWTAVAQQAFEDLK 878
Query: 429 QRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLE 488
L AP+L LP ++ FVV DA G+G VLMQ+G +AY SRQL+ + + ++ E
Sbjct: 879 AALCQAPVLSLPLFDKQFVVETDACGQGIGAVLMQEGHPLAYISRQLKGKQLHLSIYEKE 938
Query: 489 LAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDF 542
L AV+F ++ WRHYL S F + +D +SLKYL +Q+ Q++WL L ++D+
Sbjct: 939 LLAVIFAVRKWRHYLLQSHFIIKTDQRSLKYLLEQRLNTPIQQQWLPKLLEFDY 992
>At4g03650 putative reverse transcriptase
Length = 839
Score = 396 bits (1018), Expect = e-110
Identities = 181/290 (62%), Positives = 228/290 (78%)
Query: 252 DEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSK 311
+ D++KTAFRTRYGH+E+ VMPFG+TNAP FM MN +F FLD+FV++FIDDIL+YSK
Sbjct: 518 EADVRKTAFRTRYGHFEFVVMPFGLTNAPAAFMRLMNSVFQEFLDEFVIIFIDDILVYSK 577
Query: 312 IEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQW 371
EEH HL+ V++ L+E+KL+AKLSKC FW E+ FLGH+ S G++VDP K++A+ W
Sbjct: 578 SPEEHEVHLRRVMEKLREQKLFAKLSKCSFWQREMGFLGHIVSAEGVSVDPEKIEAIRDW 637
Query: 372 ETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRL 431
P + TEIRSFLGLAGYYRRFI+GF+ +A P+T+LT K FVW +CE F LK+ L
Sbjct: 638 PRPTNATEIRSFLGLAGYYRRFIKGFASMAQPMTKLTGKDVPFVWSPECEEGFVSLKEML 697
Query: 432 TTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAA 491
T+ P+L LP+ EP++VY DAS GLG LMQ GKV+AYASRQLR HE N PTHDLE+AA
Sbjct: 698 TSTPVLALPEHGEPYMVYTDASGVGLGCALMQRGKVIAYASRQLRKHEGNYPTHDLEMAA 757
Query: 492 VVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
V+F LKIWR YLYG + +VF+DHKSLKY+F Q ELN+RQRRW+EL+ DYD
Sbjct: 758 VIFALKIWRSYLYGGKVQVFTDHKSLKYIFTQPELNLRQRRWMELVADYD 807
Score = 80.9 bits (198), Expect = 2e-15
Identities = 54/155 (34%), Positives = 80/155 (50%), Gaps = 9/155 (5%)
Query: 26 LSMFGRDFEVDLVCLPLLGMDVIFGMNWLEYNRVCINCFNKTVHFSSAEEESGAQFLTTK 85
+ + G DL+ + DVI GM+WL++ RV ++C V F E +G + +
Sbjct: 377 IQIAGESLPADLIISHVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGSAGREN-DRE 435
Query: 86 QLKQLERDGILMFSLMASLSLENQVVIDRLPVVNEFHEVFPDEIPDVPPEREVEFSIDLV 145
L+ L D I + QV + + VV EF VF + +PP R F+I+L
Sbjct: 436 GLRGLPGDDIYA-------GVCGQVAVSDIRVVQEFQYVF-QSLQGLPPSRSDPFTIELE 487
Query: 146 PGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVR 180
P +S APY M+ +++AELKKQLEDLL + VR
Sbjct: 488 PKTAPLSKAPYRMAPAKMAELKKQLEDLLAEADVR 522
>At1g35370 hypothetical protein
Length = 1447
Score = 387 bits (995), Expect = e-108
Identities = 218/538 (40%), Positives = 320/538 (58%), Gaps = 41/538 (7%)
Query: 33 FEVDLVCLPLLGMDVIFGMNWLE--------YNRVCINCFNKTVH------FSSAEEESG 78
F+ D++ +PL G+D++ G+ WLE + ++ + F K + + +
Sbjct: 439 FQSDILLIPLQGVDMVLGVQWLETLGRISWEFKKLEMQFFYKNQRVWLHGIITGSVRDIK 498
Query: 79 AQFLTTKQLKQL-------------ERDGILMFSLMASLSLENQVVIDRLPVVNEFHEVF 125
A L Q Q+ E I S + S +E VV + +V EF +VF
Sbjct: 499 AHKLQKTQADQIQLAMVCVREVVSDEEQEIGSISALTSDVVEESVVQN---IVEEFPDVF 555
Query: 126 PDEIPDVPPEREV-EFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVS 184
E D+PP RE + I L+ GA V+ PY + E+ K ++D++ ++ S S
Sbjct: 556 A-EPTDLPPFREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSS 614
Query: 185 PWGAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSG 244
P+ +PV+LVKKKDG+ RLC+DY +LN +T+K+R+ +P I+DLMD+L G+ VFSKIDLR+G
Sbjct: 615 PFASPVVLVKKKDGTWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAG 674
Query: 245 YHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFID 304
YHQ+++ +D+QKTAF+T GH+EY VM FG+TNAP F MN +F FL KFV+VF D
Sbjct: 675 YHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFD 734
Query: 305 DILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSK 364
DILIYS EEH +HL++V ++++ KL+AK SK LGH S I DP+K
Sbjct: 735 DILIYSSSIEEHKEHLRLVFEVMRLHKLFAKGSK--------EHLGHFISAREIETDPAK 786
Query: 365 VDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSF 424
+ AV +W TP +V ++R FLG AGYYRRF+ F +A PL LT K F W + +S+F
Sbjct: 787 IQAVKEWPTPTTVKQVRGFLGFAGYYRRFVRNFGVIAGPLHALT-KTDGFCWSLEAQSAF 845
Query: 425 NELKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPT 484
+ LK L AP+L LP ++ F+V DA G+ VLMQ G +AY SRQL+ + +
Sbjct: 846 DTLKAVLCNAPVLALPVFDKQFMVETDACGQGIRAVLMQKGHPLAYISRQLKGKQLHLSI 905
Query: 485 HDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYDF 542
++ EL A +F ++ WRHYL S F + +D +SLKYL +Q+ Q++WL L ++D+
Sbjct: 906 YEKELLAFIFAVRKWRHYLLPSHFIIKTDQRSLKYLLEQRLNTPVQQQWLPKLLEFDY 963
>At2g06890 putative retroelement integrase
Length = 1215
Score = 376 bits (966), Expect = e-104
Identities = 187/427 (43%), Positives = 272/427 (62%), Gaps = 27/427 (6%)
Query: 117 VVNEFHEVFPDEIPD-VPPEREVEFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLD 175
++ ++ +VFP++ P +PP R +E ID VPGA L + Y + E EL++Q
Sbjct: 411 LLQDYKDVFPEDNPKGLPPIRGIEHQIDFVPGASLPNRPAYRTNPVETKELQRQ------ 464
Query: 176 KKFVRPSVSPWGAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKV 235
KDGS R+C D R +N VT+K +P+PR+DD++D+L G+ +
Sbjct: 465 --------------------KDGSWRMCFDCRAINNVTVKYCHPIPRLDDMLDELHGSSI 504
Query: 236 FSKIDLRSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFL 295
FSKIDL+SGYHQI++ + D KTAF+T++G YE+ VMPFG+T+AP FM MN + AF+
Sbjct: 505 FSKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPSTFMRLMNHVLRAFI 564
Query: 296 DKFVVVFIDDILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSG 355
FV+V+ DDIL+YS+ EH +HL VL +L++ +LYA L KC F + FLG V S
Sbjct: 565 GIFVIVYFDDILVYSESLREHIEHLDSVLNVLRKEELYANLKKCTFCTDNLVFLGFVVSA 624
Query: 356 NGIAVDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFV 415
+G+ VD KV A+ W +PK+V E+RSF GLAG+YRRF + FS + PLT++ K F
Sbjct: 625 DGVKVDEEKVKAIRDWPSPKTVGEVRSFHGLAGFYRRFFKDFSTIVAPLTEVMKKDVGFK 684
Query: 416 WDAQCESSFNELKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQL 475
W+ E +F LK +LT AP+LIL + + F + CDAS G+G VLMQD K++A+ S +L
Sbjct: 685 WEKAQEEAFQSLKDKLTNAPVLILSEFLKTFEIECDASGIGIGAVLMQDQKLIAFFSEKL 744
Query: 476 RVHEKNSPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLE 535
N PT+D EL A+V L+ W+HYL+ F + +DH+SLK+L Q++LN R RW+E
Sbjct: 745 GGATLNYPTYDKELYALVRALQRWQHYLWPKVFVIHTDHESLKHLKGQQKLNKRHARWVE 804
Query: 536 LLKDYDF 542
++ + +
Sbjct: 805 FIETFAY 811
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 346 bits (887), Expect = 2e-95
Identities = 182/427 (42%), Positives = 257/427 (59%), Gaps = 54/427 (12%)
Query: 117 VVNEFHEVFPDEIPDVPPEREV-EFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLD 175
VV EF ++F E D+PP R + I+L+ + V+ PY + E+ K ++D+L
Sbjct: 10 VVTEFPDIFV-EPTDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLA 68
Query: 176 KKFVRPSVSPWGAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKV 235
++ S SP+ +PV+LVKKKDG+ RLC+DYR+LN +T+K+R+P+P I+DLMD+L G+ V
Sbjct: 69 SGTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNV 128
Query: 236 FSKIDLRSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFL 295
+SKIDLR+GYHQ+++ D+ KTAF+T GHYEY VMPFG+TNAP F MN F FL
Sbjct: 129 YSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFFKPFL 188
Query: 296 DKFVVVFIDDILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSG 355
KFV+VF DDILIYS EEH KHL+ V ++++ L+AK+SKC F + V +LGH SG
Sbjct: 189 RKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMRVHHLFAKMSKCAFAVPRVEYLGHFISG 248
Query: 356 NGIAVDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFV 415
GIA DP+K+ AV W P ++ ++ FLGL GYYRRF
Sbjct: 249 EGIATDPAKIKAVQDWPVPVNLKQLCGFLGLTGYYRRF---------------------- 286
Query: 416 WDAQCESSFNELKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQL 475
FVV DA G+G VLMQ+G +A+ SRQL
Sbjct: 287 ------------------------------FVVEMDACGHGIGAVLMQEGHPLAFISRQL 316
Query: 476 RVHEKNSPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLE 535
+ + + ++ EL AV+FV++ WRHYL S F + +D +SLKYL +Q+ Q++WL
Sbjct: 317 KGKQLHLSIYEKELLAVIFVVRKWRHYLISSHFIIKTDQRSLKYLLEQRLNTPIQQQWLP 376
Query: 536 LLKDYDF 542
L ++D+
Sbjct: 377 KLLEFDY 383
>At3g31970 hypothetical protein
Length = 1329
Score = 241 bits (616), Expect = 6e-64
Identities = 132/315 (41%), Positives = 180/315 (56%), Gaps = 67/315 (21%)
Query: 227 MDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEY 286
M + VG S I + + I + + +++KTAFRTRYGH+E+ VMPFG+TN P FM
Sbjct: 552 MPESVGQVAVSDIRVVHEFEDIPIAEANVRKTAFRTRYGHFEFVVMPFGLTNIPTAFMRL 611
Query: 287 MNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEV 346
MN +F FLD+FV++FIDDIL+YSK EEH ++ ++ S E +EV
Sbjct: 612 MNSVFQEFLDEFVIIFIDDILVYSKSPEEH--------------EVQSEESDGEAAGAEV 657
Query: 347 SFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQ 406
G++VD K++A+ W P + TEIRSFLGLAGYY+RF++ F+ +A P+T+
Sbjct: 658 ----------GVSVDLEKIEAIRDWPRPTNATEIRSFLGLAGYYKRFVKVFASMAQPMTK 707
Query: 407 LTYKGKSFVWDAQCESSFNELKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQDGK 466
LT K FVW +C+ F LK+ LT+ P+L LP+ EP++VY DAS+ GL VLMQ GK
Sbjct: 708 LTGKDVPFVWSPECDEGFMSLKEMLTSTPVLALPEHGEPYMVYTDASRVGLDCVLMQRGK 767
Query: 467 VVAYASRQLRVHEKNSPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKEL 526
VF+DHKSLKY+F Q EL
Sbjct: 768 -------------------------------------------VFTDHKSLKYIFTQPEL 784
Query: 527 NMRQRRWLELLKDYD 541
N+RQRRW+EL+ DYD
Sbjct: 785 NLRQRRWMELVADYD 799
Score = 42.4 bits (98), Expect = 7e-04
Identities = 28/109 (25%), Positives = 51/109 (46%), Gaps = 10/109 (9%)
Query: 26 LSMFGRDFEVDLVCLPLLGMDVIFGMNWLEYNRVCINCFNKTVHFSSAE---------EE 76
+ + G VDL+ P+ DVI GM+WL++ RV ++C V F E
Sbjct: 465 IQIAGESMPVDLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQRVRPT 524
Query: 77 SGAQFLTTKQLKQLERDGILMFSLMASL-SLENQVVIDRLPVVNEFHEV 124
S + ++ Q +++ G + + S+ QV + + VV+EF ++
Sbjct: 525 SRSLVISAVQAEKMIEKGCEAYLVTISMPESVGQVAVSDIRVVHEFEDI 573
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 241 bits (614), Expect = 1e-63
Identities = 131/291 (45%), Positives = 177/291 (60%), Gaps = 29/291 (9%)
Query: 36 DLVCLPLLGMDVIFGMNWLEYNRVCINCFNKTVHFSSAEEE---------SGAQFLTTKQ 86
DL+ P++ D I GM+WL++ RV ++C V F E S + ++ Q
Sbjct: 412 DLIINPVVLYDAILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPTSRSLVISEVQ 471
Query: 87 LKQLERDGILMFSLMASLSLE-NQVVIDRLPVVNEFHEVFPDEIPDVPPEREVEFSIDLV 145
++++ G + + S+S QV + + VV EF +VF + +PP R F+I+L
Sbjct: 472 VEKMIEKGCEAYLVTISMSESVGQVAVSDIWVVQEFEDVF-QSLQGLPPSRSDPFTIELE 530
Query: 146 PGAKLVSMAPYHMSASELAELKKQLEDLLDKKFVRPSVSPWGAPVLLVKKKDGSMRLCID 205
G +S PY M +E+AELKKQLEDLL K F+RPS S WGAP
Sbjct: 531 LGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRWGAP---------------- 574
Query: 206 YRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYG 265
LN+VT+KN+YPLPRID+L+DQL GA FSKIDL GYHQ + + D++KTAFRTRYG
Sbjct: 575 --GLNRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPIAEADVRKTAFRTRYG 632
Query: 266 HYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEH 316
H+E+ VMPFG+TNAP M MN +F FLD+FV++FIDDIL+Y K EEH
Sbjct: 633 HFEFVVMPFGLTNAPTALMRLMNSVFQEFLDEFVIIFIDDILVYFKSPEEH 683
Score = 132 bits (331), Expect = 7e-31
Identities = 67/147 (45%), Positives = 84/147 (56%), Gaps = 39/147 (26%)
Query: 395 EGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILILPKPEEPFVVYCDASK 454
+GF+ +A P+T+LT K FVW +CE F
Sbjct: 691 KGFASMAQPMTKLTEKDIPFVWSPECEEGFR----------------------------- 721
Query: 455 FGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDH 514
GKV+ YASRQLR HE N PTHDLE+AA++F LKIW YLYG + +VF+DH
Sbjct: 722 ----------GKVIVYASRQLRKHEGNYPTHDLEMAAIIFALKIWGSYLYGGKVQVFTDH 771
Query: 515 KSLKYLFDQKELNMRQRRWLELLKDYD 541
KSLK +F Q ELN+RQRRW+EL+ DYD
Sbjct: 772 KSLKDIFTQPELNLRQRRWMELVADYD 798
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 236 bits (603), Expect = 2e-62
Identities = 142/395 (35%), Positives = 208/395 (51%), Gaps = 43/395 (10%)
Query: 166 LKKQLEDLLDKKFVRP-SVSPWGAPVLLVKKKDGSM------------------RLCIDY 206
+KK++ LLD + P S S W +PV V KK G R+CI+Y
Sbjct: 915 VKKEILKLLDAGVIYPISDSTWVSPVHCVPKKGGMTVVKNSKDELIPTRTITGHRMCIEY 974
Query: 207 RQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYGH 266
R+LN + K +PLP ID ++++L + +D SG+ QI + D KT F YG
Sbjct: 975 RKLNVASRKEHFPLPFIDHMLERLANHPYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGT 1034
Query: 267 YEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQI 326
+ YK MPFG+ NAP F M IF +++ V VF+DD +Y +L VL+
Sbjct: 1035 FAYKRMPFGLCNAPATFQRCMTSIFSDLIEEMVEVFMDDFSVYGSSFSSCLLNLCRVLKR 1094
Query: 327 LKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLGL 386
+E L KC F + E LG S GI VD +K+D + Q + PK+V +IRSFLG
Sbjct: 1095 CEETNLVLNWEKCHFMVREGIVLGRKISEEGIEVDKAKIDVMMQLQPPKTVKDIRSFLGH 1154
Query: 387 AGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILILPKPEEPF 446
AG+YR FI+ FSKLA PLT+L K F +D +C ++F +K+ L TAPI+ P + PF
Sbjct: 1155 AGFYRIFIKDFSKLARPLTRLLCKETEFAFDDECLTAFKLIKEALITAPIVQAPNWDFPF 1214
Query: 447 VVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFVLKIWRHYLYGS 506
++ D V YA T + EL AVVF + +R YL GS
Sbjct: 1215 ------------EIITMDDAQVRYA------------TTEKELLAVVFAFEKFRSYLVGS 1250
Query: 507 RFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
+ +++DH +L++++ +K+ R RW+ LL+++D
Sbjct: 1251 KVTIYTDHAALRHIYAKKDTKPRLLRWILLLQEFD 1285
>At3g29480 hypothetical protein
Length = 718
Score = 213 bits (543), Expect = 2e-55
Identities = 112/223 (50%), Positives = 146/223 (65%), Gaps = 23/223 (10%)
Query: 322 IVLQILKERKLYAKLSKCEFWLSEVSFL--GHVTSGN-GIAVDPSKVDAVSQWETPKSVT 378
+V+ ++ K+ K CE +L +S G T G++VD K++A+ W P + T
Sbjct: 379 LVISAVQAEKMIKK--DCEAYLVTISMKSDGEATGAEAGVSVDLEKIEAIKDWPRPTNAT 436
Query: 379 EIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILI 438
EIRSFLGLAGYYRRF++GF+ +A PLT+LT K FVW +CE F LK+ LT+ P+L
Sbjct: 437 EIRSFLGLAGYYRRFVKGFASMAPPLTKLTGKNVPFVWSPECEEGFVSLKEILTSTPVLA 496
Query: 439 LPKPEEPFVVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFVLKI 498
LP+ E ++VY DAS+ GLG VLMQ GKV+AYASRQLR HE N PTHDLE++A
Sbjct: 497 LPEHGESYMVYTDASRVGLGCVLMQRGKVIAYASRQLRKHEGNYPTHDLEMSA------- 549
Query: 499 WRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
VF+DHKSLKY+ Q ELN+RQRRW+EL+ DYD
Sbjct: 550 -----------VFTDHKSLKYIITQPELNLRQRRWMELVADYD 581
Score = 32.0 bits (71), Expect = 0.92
Identities = 13/25 (52%), Positives = 17/25 (68%)
Query: 46 DVIFGMNWLEYNRVCINCFNKTVHF 70
DVI GM+WL+Y RV ++C V F
Sbjct: 336 DVILGMDWLDYYRVHLDCHRGRVSF 360
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 206 bits (524), Expect = 3e-53
Identities = 109/307 (35%), Positives = 173/307 (55%), Gaps = 5/307 (1%)
Query: 165 ELKKQLEDLLDKKFVRPSVSP-----WGAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYP 219
E K + D +DK S+ W A ++VKKK+G R+CID+ LNK K+ +P
Sbjct: 475 ERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACPKDSFP 534
Query: 220 LPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNA 279
LP ID L++ G ++ S +D SGY+QI + ED +KT F T G Y YKVMPFG+ NA
Sbjct: 535 LPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNA 594
Query: 280 PGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKC 339
+ +N++F + K + V+IDD+LI S +E+H KHL+ IL + ++ +KC
Sbjct: 595 GATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECFAILNQYQMKLNPAKC 654
Query: 340 EFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSK 399
F + FLG++ + GI +P++++A +PK+ E++ G RFI +
Sbjct: 655 TFGVPSGEFLGYIVTKRGIEANPNQINAFLNMPSPKNFKEVQRLTGRIAALNRFISRSTD 714
Query: 400 LALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILILPKPEEPFVVYCDASKFGLGG 459
+LP Q+ K F+WD +CE +F +LK LT+ P+L P+ +E +Y S + G
Sbjct: 715 KSLPFYQILKGNKEFLWDEKCEEAFGQLKAYLTSPPVLFKPELDEKLYLYVSVSNHAVSG 774
Query: 460 VLMQDGK 466
VL+++ +
Sbjct: 775 VLIREDR 781
>At2g10690 putative retroelement pol polyprotein
Length = 622
Score = 206 bits (523), Expect = 4e-53
Identities = 145/421 (34%), Positives = 219/421 (51%), Gaps = 28/421 (6%)
Query: 132 VPPEREVEFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLD---KKFVRPSVSPWGA 188
+P E EF +VP ++VS + S +AE K ++LD + +S A
Sbjct: 46 IPMEEPEEFEELIVPSEEVVSGC----NMSLIAE-KANSTEMLDHGGENISSDDLSELKA 100
Query: 189 PVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQI 248
P L +K +R + + YP+ D L D+ V ++ +++ G +
Sbjct: 101 PKLDLKPLPKGLRYAF-------LGPNDTYPVIINDGLSDEQVN-QLLNELRKYRGQLVV 152
Query: 249 KVKDEDM----QKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFID 304
K + +KT F G + YK M FG+ NAPG F M IF F+++ + VF+D
Sbjct: 153 SFKLQFTLMIKKKTTFTCPNGTFAYKRMSFGLCNAPGTFQRSMTSIFSDFIEEIMEVFMD 212
Query: 305 DILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSK 364
D +S +L VL+ +E L KC F + E LGH SG GI VD +K
Sbjct: 213 DFSGFSSC----LLNLCRVLERCEETNLVLNWEKCHFMVHEGIVLGHKISGKGIEVDKAK 268
Query: 365 VDAVSQWETPKSVTEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSF 424
+D + Q + PK+V +IRSFLG A +YRRFI+ FSK+A PLT+L K F +D C +F
Sbjct: 269 IDVMIQLQPPKTVKDIRSFLGHAEFYRRFIKDFSKIARPLTRLLCKETEFNFDEDCLKAF 328
Query: 425 NELKQRLTTAPILILPKPEEPFVVYCDASKFGLGGVLMQ--DGK--VVAYASRQLRVHEK 480
+ +K+ L +API P + PF + CDA + +G VL Q D K V+ YASR + +
Sbjct: 329 HLIKETLVSAPIFQAPNWDHPFEIMCDAFDYAVGAVLDQKIDDKLHVIYYASRTMDEAQT 388
Query: 481 NSPTHDLELAAVVFVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDY 540
T + EL AVVF + R YL G + +V+ DH +L++++ +KE R RW+ LL+++
Sbjct: 389 RYATIEKELLAVVFAFEKIRSYLVGFKVKVYIDHAALRHIYAKKETKSRLLRWILLLQEF 448
Query: 541 D 541
D
Sbjct: 449 D 449
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 199 bits (507), Expect = 3e-51
Identities = 132/395 (33%), Positives = 195/395 (48%), Gaps = 71/395 (17%)
Query: 166 LKKQLEDLLDKKFVRP-SVSPWGAPVLLVKKKDGSM------------------RLCIDY 206
+KK++ LLD + P S S W +PV +V KK G + R+CIDY
Sbjct: 390 VKKEIMKLLDAGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCIDY 449
Query: 207 RQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYGH 266
R+LN T K+ +PL ID ++++L + +D G+ QI + +D +KT F YG
Sbjct: 450 RKLNSATRKDNFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYGT 509
Query: 267 YEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQI 326
+ Y+ MPFG+ NAP F M IF ++ F+ VFIDD L+ + E++H
Sbjct: 510 FAYRRMPFGLCNAPATFQHCMKYIFSDMIEDFMEVFIDDFLVLQRCEDKH---------- 559
Query: 327 LKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSVTEIRSFLGL 386
L K F + + LGH S G+ VD +K+ EI FLG
Sbjct: 560 -----LVLNWEKSHFMVRDGIVLGHKISEKGVEVDRAKI-------------EIMRFLGH 601
Query: 387 AGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPILILPKPEEPF 446
AG+YRRFI+ FSK+A P TQL K + +D C +FN +K+ L +A I+ P+ E PF
Sbjct: 602 AGFYRRFIKDFSKIARPCTQLLCKEQKSEFDDTCLQAFNIVKESLVSALIVQPPEWELPF 661
Query: 447 VVYCDASKFGLGGVLMQDGKVVAYASRQLRVHEKNSPTHDLELAAVVFVLKIWRHYLYGS 506
V CDAS + +G VL Q D +L A+ + + L G+
Sbjct: 662 EVICDASDYVVGAVLGQ--------------------RKDKKLHAIYYASRT----LDGA 697
Query: 507 RFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
+ V + H +L+YL +K+ R RW+ LL+D+D
Sbjct: 698 QVIVHTVHAALRYLLSKKDAKPRLLRWILLLQDFD 732
>At1g20390 hypothetical protein
Length = 1791
Score = 199 bits (506), Expect = 3e-51
Identities = 122/408 (29%), Positives = 199/408 (47%), Gaps = 8/408 (1%)
Query: 139 EFSIDLVPGAKLVSMAPYHMSASELAELKKQLEDLLDK-KFVRPSVSPWGAPVLLVKKKD 197
E ++D P K V + + +++E LL + + W A ++VKKK+
Sbjct: 768 ELNVD--PTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKN 825
Query: 198 GSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGAKVFSKIDLRSGYHQIKVKDEDMQK 257
G R+C+DY LNK K+ YPLP ID L++ G + S +D SGY+QI + +D +K
Sbjct: 826 GKWRVCVDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEK 885
Query: 258 TAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHAFLDKFVVVFIDDILIYSKIEEEHA 317
T+F T G Y YKVM FG+ NA + ++N++ + + V V+IDD+L+ S E+H
Sbjct: 886 TSFVTDRGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHV 945
Query: 318 KHLKIVLQILKERKLYAKLSKCEFWLSEVSFLGHVTSGNGIAVDPSKVDAVSQWETPKSV 377
+HL +L + +KC F ++ FLG+V + GI +P ++ A+ + +P++
Sbjct: 946 EHLSKCFDVLNTYGMKLNPTKCTFGVTSGEFLGYVVTKRGIEANPKQIRAILELPSPRNA 1005
Query: 378 TEIRSFLGLAGYYRRFIEGFSKLALPLTQLTYKGKSFVWDAQCESSFNELKQRLTTAPIL 437
E++ G RFI + LP L + F WD E +F +LK L+T PIL
Sbjct: 1006 REVQRLTGRIAALNRFISRSTDKCLPFYNLLKRRAQFDWDKDSEEAFEKLKDYLSTPPIL 1065
Query: 438 ILPKPEEPFVVYCDASKFGLGGVLMQDG----KVVAYASRQLRVHEKNSPTHDLELAAVV 493
+ P+ E +Y S + VL+++ + + Y S+ L E P + AVV
Sbjct: 1066 VKPEVGETLYLYIAVSDHAVSSVLVREDRGEQRPIFYTSKSLVEAETRYPVIEKAALAVV 1125
Query: 494 FVLKIWRHYLYGSRFEVFSDHKSLKYLFDQKELNMRQRRWLELLKDYD 541
+ R Y V +D + L+ + R +W L +YD
Sbjct: 1126 TSARKLRPYFQSHTIAVLTD-QPLRVALHSPSQSGRMTKWAVELSEYD 1172
>At4g04230 putative transposon protein
Length = 315
Score = 196 bits (498), Expect = 3e-50
Identities = 88/168 (52%), Positives = 127/168 (75%)
Query: 174 LDKKFVRPSVSPWGAPVLLVKKKDGSMRLCIDYRQLNKVTIKNRYPLPRIDDLMDQLVGA 233
+ K ++R S+SP PVLLV KKDG+ R+C+D R +N +TIK R+P+PR+ D++D+L GA
Sbjct: 125 IHKGYIRESLSPCAVPVLLVPKKDGTWRMCLDCRAINNITIKYRHPIPRLYDMLDELSGA 184
Query: 234 KVFSKIDLRSGYHQIKVKDEDMQKTAFRTRYGHYEYKVMPFGVTNAPGVFMEYMNRIFHA 293
+FSK+DLRSGYHQ+++++ D KTAF+T+ G YE VMPFG+TNAP FM MN++ +
Sbjct: 185 IIFSKVDLRSGYHQVRMREGDEWKTAFKTKQGLYECLVMPFGLTNAPSTFMRLMNQVLRS 244
Query: 294 FLDKFVVVFIDDILIYSKIEEEHAKHLKIVLQILKERKLYAKLSKCEF 341
F+ KFVVV+ DDILIY+K +H + L+++L+ L++ LYA L KC F
Sbjct: 245 FIGKFVVVYFDDILIYNKSYSDHIQPLELILKTLRKEGLYANLKKCTF 292
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.140 0.418
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,450,343
Number of Sequences: 26719
Number of extensions: 534111
Number of successful extensions: 1338
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 50
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1216
Number of HSP's gapped (non-prelim): 83
length of query: 552
length of database: 11,318,596
effective HSP length: 104
effective length of query: 448
effective length of database: 8,539,820
effective search space: 3825839360
effective search space used: 3825839360
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144765.10


