

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144759.6 + phase: 0
(579 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
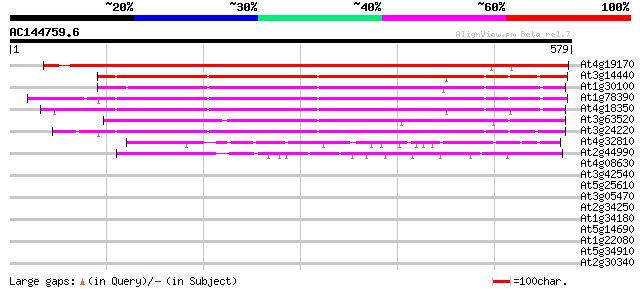
Score E
Sequences producing significant alignments: (bits) Value
At4g19170 neoxanthin cleavage enzyme-like protein 750 0.0
At3g14440 9-cis-epoxycarotenoid dioxygenase, putative 384 e-107
At1g30100 hypothetical protein 376 e-104
At1g78390 similar to 9-cis-epoxycarotenoid dioxygenase gb|AAF263... 366 e-101
At4g18350 neoxanthin cleavage enzyme - like protein 365 e-101
At3g63520 neoxanthin cleavage enzyme nc1 353 2e-97
At3g24220 9-cis-epoxycarotenoid dioxygenase, putative 327 9e-90
At4g32810 unknown protein 118 8e-27
At2g44990 hypothetical protein 94 3e-19
At4g08630 hypothetical protein 33 0.57
At3g42540 hypothetical protein 30 2.8
At5g25610 dehydration-induced protein RD22 30 3.7
At3g05470 unknown protein 30 3.7
At2g34250 putative protein transport protein SEC61 alpha subunit 30 3.7
At1g34180 unknown protein 30 4.8
At5g14690 putative protein 29 6.3
At1g22080 hypothetical protein 29 6.3
At5g34910 putative protein 29 8.2
At2g30340 unknown protein 29 8.2
>At4g19170 neoxanthin cleavage enzyme-like protein
Length = 595
Score = 750 bits (1937), Expect = 0.0
Identities = 358/551 (64%), Positives = 440/551 (78%), Gaps = 20/551 (3%)
Query: 36 SQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDDIINTFIDPPIKPS 95
++PKTL TL++ + K +++ + T++D+INTFIDPP +PS
Sbjct: 55 NKPKTLHNRTNHTLVS----------SPPKLRPEMTLATALFTTVEDVINTFIDPPSRPS 104
Query: 96 VDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDG 155
VDP+HVLS NFAPVLDELPPT C++I GTLP SLNG YIRNGPNPQFLPRGPYHLFDGDG
Sbjct: 105 VDPKHVLSDNFAPVLDELPPTDCEIIHGTLPLSLNGAYIRNGPNPQFLPRGPYHLFDGDG 164
Query: 156 MLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVAVARVL 215
MLHA+KI NGKATLCSRYV+TYKY +E + G ++PNVFSGFN + ARG++ ARVL
Sbjct: 165 MLHAIKIHNGKATLCSRYVKTYKYNVEKQTGAPVMPNVFSGFNGVTASVARGALTAARVL 224
Query: 216 TGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNM 275
TGQYNP+NGIGL NTSLA F NRLFALGESDLPY +++T +GDI+TIGRYDF+GKL M+M
Sbjct: 225 TGQYNPVNGIGLANTSLAFFSNRLFALGESDLPYAVRLTESGDIETIGRYDFDGKLAMSM 284
Query: 276 TAHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFAITKKY 335
TAHPK D TGE FA+RYG + PFLTYFRFDS G K DVP+FSM PSFLHDFAITK++
Sbjct: 285 TAHPKTDPITGETFAFRYGPVPPFLTYFRFDSAGKKQRDVPIFSMTSPSFLHDFAITKRH 344
Query: 336 ALFADIQL--EINPLDMIF-GGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMH 392
A+FA+IQL +N LD++ GGSPV +D K PR+G++P+YA +ES+MKWF+VPG NI+H
Sbjct: 345 AIFAEIQLGMRMNMLDLVLEGGSPVGTDNGKTPRLGVIPKYAGDESEMKWFEVPGFNIIH 404
Query: 393 LINAWDEEDGKTVTIVAPNILCVEHMMERLELIHEMIEKVRINVETGIVTRQPLSARNLD 452
INAWDE+DG +V ++APNI+ +EH +ER++L+H ++EKV+I++ TGIV R P+SARNLD
Sbjct: 405 AINAWDEDDGNSVVLIAPNIMSIEHTLERMDLVHALVEKVKIDLVTGIVRRHPISARNLD 464
Query: 453 LAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGC----RLYGEDCYGGEPF 508
AVIN FLG+ R++YAAIG+PMPK SGVVK+DV KG+ C R+YG CYGGEPF
Sbjct: 465 FAVINPAFLGRCSRYVYAAIGDPMPKISGVVKLDVSKGDRDDCTVARRMYGSGCYGGEPF 524
Query: 509 FVAREDGE---EEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFHG 565
FVAR+ G EEDDGY+V+YVHDE GES+FLVMDAKSPE EIVA V+LPRRVPYGFHG
Sbjct: 525 FVARDPGNPEAEEDDGYVVTYVHDEVTGESKFLVMDAKSPELEIVAAVRLPRRVPYGFHG 584
Query: 566 LFVKESDITRL 576
LFVKESD+ +L
Sbjct: 585 LFVKESDLNKL 595
>At3g14440 9-cis-epoxycarotenoid dioxygenase, putative
Length = 599
Score = 384 bits (986), Expect = e-107
Identities = 200/492 (40%), Positives = 299/492 (60%), Gaps = 14/492 (2%)
Query: 91 PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHL 150
P+ + DP ++ NFAPV +E P + + G LP S+ GVY+RNG NP P +H
Sbjct: 112 PLPKTADPSVQIAGNFAPV-NEQPVRRNLPVVGKLPDSIKGVYVRNGANPLHEPVTGHHF 170
Query: 151 FDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVA 210
FDGDGM+HAVK +G A+ R+ QT ++ E + G + P + G+A R +
Sbjct: 171 FDGDGMVHAVKFEHGSASYACRFTQTNRFVQERQLGRPVFPKAIGELHGHTGIA-RLMLF 229
Query: 211 VARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGK 270
AR G +P +G G+ N L F RL A+ E DLPY++++TPNGD++T+GR+DF+G+
Sbjct: 230 YARAAAGIVDPAHGTGVANAGLVYFNGRLLAMSEDDLPYQVQITPNGDLKTVGRFDFDGQ 289
Query: 271 LFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDF 329
L M AHPK+D ++GE FA Y ++ +P+L YFRF +G K DV + + +P+ +HDF
Sbjct: 290 LESTMIAHPKVDPESGELFALSYDVVSKPYLKYFRFSPDGTKSPDVEI-QLDQPTMMHDF 348
Query: 330 AITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLN 389
AIT+ + + D Q+ +MI GGSPV D +KV R GIL +YA++ S +KW D P
Sbjct: 349 AITENFVVVPDQQVVFKLPEMIRGGSPVVYDKNKVARFGILDKYAEDSSNIKWIDAPDCF 408
Query: 390 IMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIEKVRINVETGIVTRQPLSA 448
HL NAW+E + V ++ + + + E E + ++ ++R+N++TG TR+P+ +
Sbjct: 409 CFHLWNAWEEPETDEVVVIGSCMTPPDSIFNESDENLKSVLSEIRLNLKTGESTRRPIIS 468
Query: 449 R-----NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCY 503
NL+ ++N LG+K +F Y A+ P PK SG K+D+ G EV LYG++ Y
Sbjct: 469 NEDQQVNLEAGMVNRNMLGRKTKFAYLALAEPWPKVSGFAKVDLTTG-EVKKHLYGDNRY 527
Query: 504 GGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGF 563
GGEP F+ E G EED+GY++ +VHDEK +S +++A S E+ A VKLP RVPYGF
Sbjct: 528 GGEPLFLPGEGG-EEDEGYILCFVHDEKTWKSELQIVNAVS--LEVEATVKLPSRVPYGF 584
Query: 564 HGLFVKESDITR 575
HG F+ D+ +
Sbjct: 585 HGTFIGADDLAK 596
>At1g30100 hypothetical protein
Length = 589
Score = 376 bits (966), Expect = e-104
Identities = 205/490 (41%), Positives = 293/490 (58%), Gaps = 13/490 (2%)
Query: 91 PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHL 150
P+ +VDPRH +S N+APV ++ + V G +P ++GVY+RNG NP F P +HL
Sbjct: 102 PLPKTVDPRHQISGNYAPVPEQSVKSSLSV-DGKIPDCIDGVYLRNGANPLFEPVSGHHL 160
Query: 151 FDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVA 210
FDGDGM+HAVKITNG A+ R+ +T + E + G I P + G+A R +
Sbjct: 161 FDGDGMVHAVKITNGDASYSCRFTETERLVQEKQLGSPIFPKAIGELHGHSGIA-RLMLF 219
Query: 211 VARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGK 270
AR L G N NG G+ N L F +RL A+ E DLPY+++VT NGD++TIGR+DF+G+
Sbjct: 220 YARGLFGLLNHKNGTGVANAGLVYFHDRLLAMSEDDLPYQVRVTDNGDLETIGRFDFDGQ 279
Query: 271 LFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDF 329
L M AHPKID T E FA Y ++ +P+L YF+F G K DV + + P+ +HDF
Sbjct: 280 LSSAMIAHPKIDPVTKELFALSYDVVKKPYLKYFKFSPEGEKSPDVEI-PLASPTMMHDF 338
Query: 330 AITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLN 389
AIT+ + + D Q+ DM G SPV+ D K+ R GILPR A + S+M W + P
Sbjct: 339 AITENFVVIPDQQVVFKLSDMFLGKSPVKYDGEKISRFGILPRNAKDASEMVWVESPETF 398
Query: 390 IMHLINAWDEEDGKTVTIVAPNILCVEHMMERL-ELIHEMIEKVRINVETGIVTRQPL-- 446
HL NAW+ + V ++ + + + E ++ ++ ++R+N++TG TR+ +
Sbjct: 399 CFHLWNAWESPETDEVVVIGSCMTPADSIFNECDEQLNSVLSEIRLNLKTGKSTRRTIIP 458
Query: 447 --SARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCYG 504
NL+ ++N LG+K R+ Y AI P PK SG K+D+ G EV YG YG
Sbjct: 459 GSVQMNLEAGMVNRNLLGRKTRYAYLAIAEPWPKVSGFAKVDLSTG-EVKNHFYGGKKYG 517
Query: 505 GEPFFVARE-DGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGF 563
GEPFF+ R + + EDDGY++S+VHDE+ ES +++A + E E A VKLP RVPYGF
Sbjct: 518 GEPFFLPRGLESDGEDDGYIMSFVHDEESWESELHIVNAVTLELE--ATVKLPSRVPYGF 575
Query: 564 HGLFVKESDI 573
HG FV +D+
Sbjct: 576 HGTFVNSADM 585
>At1g78390 similar to 9-cis-epoxycarotenoid dioxygenase
gb|AAF26356.1
Length = 657
Score = 366 bits (939), Expect = e-101
Identities = 211/567 (37%), Positives = 317/567 (55%), Gaps = 19/567 (3%)
Query: 19 IPQFNTASVRTKEKPQTS-QPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALIL 77
+P + ++SV K +S Q T + K L +T K N N L +A ++
Sbjct: 96 VPCYVSSSVNKKSSVSSSLQSPTFKPPSWKKLCNDVTNLIPKTTNQNPKLNPVQRTAAMV 155
Query: 78 NTLDDIINTFIDP-----PIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGV 132
LD + N I P + DP ++ NF PV E P + GT+P + GV
Sbjct: 156 --LDAVENAMISHERRRHPHPKTADPAVQIAGNFFPV-PEKPVVHNLPVTGTVPECIQGV 212
Query: 133 YIRNGPNPQFLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPN 192
Y+RNG NP P +HLFDGDGM+HAV+ NG + R+ +T + E E G + P
Sbjct: 213 YVRNGANPLHKPVSGHHLFDGDGMVHAVRFDNGSVSYACRFTETNRLVQERECGRPVFPK 272
Query: 193 VFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIK 252
+ +G+A + + R L G +P G+G+ N L F L A+ E DLPY +K
Sbjct: 273 AIGELHGHLGIA-KLMLFNTRGLFGLVDPTGGLGVANAGLVYFNGHLLAMSEDDLPYHVK 331
Query: 253 VTPNGDIQTIGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVK 311
VT GD++T GRYDF+G+L M AHPKID +T E FA Y ++ +P+L YFRF S+G K
Sbjct: 332 VTQTGDLETSGRYDFDGQLKSTMIAHPKIDPETRELFALSYDVVSKPYLKYFRFTSDGEK 391
Query: 312 HNDVPVFSMKRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILP 371
DV + + +P+ +HDFAIT+ + + D Q+ +MI GGSPV D K R GIL
Sbjct: 392 SPDVEI-PLDQPTMIHDFAITENFVVIPDQQVVFRLPEMIRGGSPVVYDEKKKSRFGILN 450
Query: 372 RYADNESKMKWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIE 430
+ A + S ++W +VP HL N+W+E + V ++ + + + E E + ++
Sbjct: 451 KNAKDASSIQWIEVPDCFCFHLWNSWEEPETDEVVVIGSCMTPPDSIFNEHDETLQSVLS 510
Query: 431 KVRINVETGIVTRQPLSAR--NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVL 488
++R+N++TG TR+P+ + NL+ ++N LG+K R+ Y A+ P PK SG K+D+
Sbjct: 511 EIRLNLKTGESTRRPVISEQVNLEAGMVNRNLLGRKTRYAYLALTEPWPKVSGFAKVDLS 570
Query: 489 KGEEVGCRLYGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFE 548
G E+ +YGE YGGEP F+ DG EED GY++ +VHDE+K +S +++A + + E
Sbjct: 571 TG-EIRKYIYGEGKYGGEPLFLPSGDG-EEDGGYIMVFVHDEEKVKSELQLINAVNMKLE 628
Query: 549 IVAEVKLPRRVPYGFHGLFVKESDITR 575
A V LP RVPYGFHG F+ + D+++
Sbjct: 629 --ATVTLPSRVPYGFHGTFISKEDLSK 653
>At4g18350 neoxanthin cleavage enzyme - like protein
Length = 583
Score = 365 bits (937), Expect = e-101
Identities = 209/560 (37%), Positives = 317/560 (56%), Gaps = 24/560 (4%)
Query: 32 KPQTSQPKTLRTT---------PTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDD 82
+P QPK ++ T + L TT T N + +A+ ++ +
Sbjct: 26 RPIKRQPKVIKCTVQIDVTELTKKRQLFTPRTTATPPQHNPLRLNIFQKAAAIAIDAAER 85
Query: 83 -IINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQ 141
+I+ D P+ + DPR ++ N++PV E + ++GT+P ++GVYIRNG NP
Sbjct: 86 ALISHEQDSPLPKTADPRVQIAGNYSPV-PESSVRRNLTVEGTIPDCIDGVYIRNGANPM 144
Query: 142 FLPRGPYHLFDGDGMLHAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLI 201
F P +HLFDGDGM+HAVKITNG A+ R+ +T + E G + P +
Sbjct: 145 FEPTAGHHLFDGDGMVHAVKITNGSASYACRFTKTERLVQEKRLGRPVFPKAIGELHGHS 204
Query: 202 GLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQT 261
G+A R + AR L G N NG+G+ N L F NRL A+ E DLPY++K+T GD+QT
Sbjct: 205 GIA-RLMLFYARGLCGLINNQNGVGVANAGLVYFNNRLLAMSEDDLPYQLKITQTGDLQT 263
Query: 262 IGRYDFNGKLFMNMTAHPKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSM 320
+GRYDF+G+L M AHPK+D T E A Y ++ +P+L YFRF +GVK ++ + +
Sbjct: 264 VGRYDFDGQLKSAMIAHPKLDPVTKELHALSYDVVKKPYLKYFRFSPDGVKSPELEI-PL 322
Query: 321 KRPSFLHDFAITKKYALFADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKM 380
+ P+ +HDFAIT+ + + D Q+ +MI G SPV D KV R+GI+P+ A S++
Sbjct: 323 ETPTMIHDFAITENFVVIPDQQVVFKLGEMISGKSPVVFDGEKVSRLGIMPKDATEASQI 382
Query: 381 KWFDVPGLNIMHLINAWDEEDGKTVTIVAPNILCVEHMM-ERLELIHEMIEKVRINVETG 439
W + P HL NAW+ + + + ++ + + + ER E + ++ ++RIN+ T
Sbjct: 383 IWVNSPETFCFHLWNAWESPETEEIVVIGSCMSPADSIFNERDESLRSVLSEIRINLRTR 442
Query: 440 IVTRQPLSAR---NLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCR 496
TR+ L NL++ ++N LG+K RF + AI P PK SG K+D+ G E+
Sbjct: 443 KTTRRSLLVNEDVNLEIGMVNRNRLGRKTRFAFLAIAYPWPKVSGFAKVDLCTG-EMKKY 501
Query: 497 LYGEDCYGGEPFFVAREDG---EEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEV 553
+YG + YGGEPFF+ G E EDDGY+ +VHDE+ S +++A + + E A +
Sbjct: 502 IYGGEKYGGEPFFLPGNSGNGEENEDDGYIFCHVHDEETKTSELQIINAVNLKLE--ATI 559
Query: 554 KLPRRVPYGFHGLFVKESDI 573
KLP RVPYGFHG FV +++
Sbjct: 560 KLPSRVPYGFHGTFVDSNEL 579
>At3g63520 neoxanthin cleavage enzyme nc1
Length = 538
Score = 353 bits (905), Expect = 2e-97
Identities = 200/495 (40%), Positives = 284/495 (56%), Gaps = 25/495 (5%)
Query: 98 PRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGML 157
P H LS NFAP+ DE PP + + G LP LNG ++R GPNP+F YH FDGDGM+
Sbjct: 44 PLHYLSGNFAPIRDETPPVKDLPVHGFLPECLNGEFVRVGPNPKFDAVAGYHWFDGDGMI 103
Query: 158 HAVKITNGKATLCSRYVQTYKYKIENEAGYQILPNV--FSGFNSLIGLAARGSVAVARVL 215
H V+I +GKAT SRYV+T + K E G + GF L+ + + ++L
Sbjct: 104 HGVRIKDGKATYVSRYVKTSRLKQEEFFGAAKFMKIGDLKGFFGLLMVNVQQLRTKLKIL 163
Query: 216 TGQYNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNM 275
Y G G NT+L +L AL E+D PY IKV +GD+QT+G D++ +L +
Sbjct: 164 DNTY----GNGTANTALVYHHGKLLALQEADKPYVIKVLEDGDLQTLGIIDYDKRLTHSF 219
Query: 276 TAHPKIDSDTGECFAYRYGLIRPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFAITKKY 335
TAHPK+D TGE F + Y P+LTY +G+ H+ VP+ ++ P +HDFAIT+ Y
Sbjct: 220 TAHPKVDPVTGEMFTFGYSHTPPYLTYRVISKDGIMHDPVPI-TISEPIMMHDFAITETY 278
Query: 336 ALFADIQLEINPLDMIFGGSPVRS-DPSKVPRIGILPRYADNESKMKWFDVPGLNIMHLI 394
A+F D+ + P +M+ + S DP+K R G+LPRYA +E ++WF++P I H
Sbjct: 279 AIFMDLPMHFRPKEMVKEKKMIYSFDPTKKARFGVLPRYAKDELMIRWFELPNCFIFHNA 338
Query: 395 NAWDEEDGK---TVTIVAPNILCVE-HMMERLELIHEMIEKVRINVETGIVTRQPLSARN 450
NAW+EED T + P++ V + E+LE + ++R N++TG +++ LSA
Sbjct: 339 NAWEEEDEVVLITCRLENPDLDMVSGKVKEKLENFGNELYEMRFNMKTGSASQKKLSASA 398
Query: 451 LDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRL------------Y 498
+D IN + GKK R++Y I + + K +G++K D+ E G R+
Sbjct: 399 VDFPRINECYTGKKQRYVYGTILDSIAKVTGIIKFDLHAEAETGKRMLEVGGNIKGIYDL 458
Query: 499 GEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRR 558
GE YG E +V RE EEDDGYL+ +VHDE G+S V+DAK+ E VA V+LP R
Sbjct: 459 GEGRYGSEAIYVPRETA-EEDDGYLIFFVHDENTGKSCVTVIDAKTMSAEPVAVVELPHR 517
Query: 559 VPYGFHGLFVKESDI 573
VPYGFH LFV E +
Sbjct: 518 VPYGFHALFVTEEQL 532
>At3g24220 9-cis-epoxycarotenoid dioxygenase, putative
Length = 577
Score = 327 bits (839), Expect = 9e-90
Identities = 200/541 (36%), Positives = 296/541 (53%), Gaps = 21/541 (3%)
Query: 45 PTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDDIINTFIDP-----PIKPSVDPR 99
PT L + KLK T L + L LD I ++ + P P+ DP
Sbjct: 41 PTLPDLTSPVPSPVKLKPTYPNLNL--LQKLAATMLDKIESSIVIPMEQNRPLPKPTDPA 98
Query: 100 HVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGMLHA 159
LS NFAPV +E P + G +P L GVYIRNG NP F P +HLFDGDGM+HA
Sbjct: 99 VQLSGNFAPV-NECPVQNGLEVVGQIPSCLKGVYIRNGANPMFPPLAGHHLFDGDGMIHA 157
Query: 160 VKIT-NGKATLCSRYVQTYKYKIENEAGYQILPNVFSGFNSLIGLAARGSVAVARVLTGQ 218
V I + + + RY +T + E G + P + GLA R ++ AR G
Sbjct: 158 VSIGFDNQVSYSCRYTKTNRLVQETALGRSVFPKPIGELHGHSGLA-RLALFTARAGIGL 216
Query: 219 YNPINGIGLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNMTAH 278
+ G+G+ N + F RL A+ E DLPY++K+ GD++TIGR+ F+ ++ ++ AH
Sbjct: 217 VDGTRGMGVANAGVVFFNGRLLAMSEDDLPYQVKIDGQGDLETIGRFGFDDQIDSSVIAH 276
Query: 279 PKIDSDTGECFAYRYGLI-RPFLTYFRFDSNGVKHNDVPVFSMKRPSFLHDFAITKKYAL 337
PK+D+ TG+ Y ++ +P L Y +F++ G K DV + ++ P+ +HDFAIT+ + +
Sbjct: 277 PKVDATTGDLHTLSYNVLKKPHLRYLKFNTCGKKTRDVEI-TLPEPTMIHDFAITENFVV 335
Query: 338 FADIQLEINPLDMIFGGSPVRSDPSKVPRIGILPRYADNESKMKWFDVPGLNIMHLINAW 397
D Q+ +MI GGSPV K+ R G+L + S + W DVP HL NAW
Sbjct: 336 IPDQQMVFKLSEMIRGGSPVIYVKEKMARFGVLSKQDLTGSDINWVDVPDCFCFHLWNAW 395
Query: 398 DE--EDGKTVTIVAPNILCVEHMM--ERLELIHEMIEKVRINVETGIVTRQPL-SARNLD 452
+E E+G V +V + + + E E + ++R+N+ T R+ + + NL+
Sbjct: 396 EERTEEGDPVIVVIGSCMSPPDTIFSESGEPTRVELSEIRLNMRTKESNRKVIVTGVNLE 455
Query: 453 LAVINNEFLGKKHRFIYAAIGNPMPKFSGVVKIDVLKGEEVGCRLYGEDCYGGEPFFVAR 512
IN ++G+K +F+Y AI +P PK SG+ K+D+ G V YG +GGEP FV
Sbjct: 456 AGHINRSYVGRKSQFVYIAIADPWPKCSGIAKVDIQNG-TVSEFNYGPSRFGGEPCFVPE 514
Query: 513 EDGEEEDDGYLVSYVHDEKKGESRFLVMDAKSPEFEIVAEVKLPRRVPYGFHGLFVKESD 572
+G EED GY++ +V DE+K ES F+V+DA + + VA V+LP RVPYGFHG FV E+
Sbjct: 515 GEG-EEDKGYVMGFVRDEEKDESEFVVVDA--TDMKQVAAVRLPERVPYGFHGTFVSENQ 571
Query: 573 I 573
+
Sbjct: 572 L 572
>At4g32810 unknown protein
Length = 570
Score = 118 bits (296), Expect = 8e-27
Identities = 125/506 (24%), Positives = 220/506 (42%), Gaps = 93/506 (18%)
Query: 121 IKGTLPPSLNGVYIRNGPNPQFLPRGPY-HLFDGDGMLHAVKITNGKATLCSRYVQTYKY 179
++G +P LNG Y+RNGP + + HLFDG L ++ G+ R +++ Y
Sbjct: 97 VQGKIPTWLNGTYLRNGPGLWNIGDHDFRHLFDGYSTLVKLQFDGGRIFAAHRLLESDAY 156
Query: 180 KI-------------ENEAGYQILPNVFSGFNSLIGLAARGSVAVARVLTGQYNPINGIG 226
K E I N FSG ++ R+ +G+ N
Sbjct: 157 KAAKKHNRLCYREFSETPKSVIINKNPFSGIGEIV-----------RLFSGESLTDNA-- 203
Query: 227 LINTSLALFGN-RLFALGESDLPYEIKVTPNGDIQTIGRYDFNGKLFMNM--TAHPKIDS 283
NT + G+ R+ L E+ + + ++TIG+++++ L +M +AHP +
Sbjct: 204 --NTGVIKLGDGRVMCLTETQKGSIL--VDHETLETIGKFEYDDVLSDHMIQSAHPIVTE 259
Query: 284 DTGECFAYRYGLIRPFLTYFRFDSNGVKHNDVPVFSMKR----PSFLHDFAITKKYALFA 339
E + L++P R ++ K V + P ++H FA+T+ Y +
Sbjct: 260 T--EMWTLIPDLVKPGYRVVRMEAGSNKREVVGRVRCRSGSWGPGWVHSFAVTENYVVIP 317
Query: 340 DIQLEINPLDMIFGGSPVRSDPSKVPRIGILPR---YADNESKMKW-----FDVPGLNIM 391
++ L + +++ R++P+ + + P+ + SK+ +VP
Sbjct: 318 EMPLRYSVKNLL------RAEPTPLYKFEWCPQDGAFIHVMSKLTGEVVASVEVPAYVTF 371
Query: 392 HLINAWDEE---DGKTVTIVAPNILCVEHM-------MERLELI-----HEMIEKVRIN- 435
H INA++E+ DGK I+A C EH M RL+ + H+++ RI
Sbjct: 372 HFINAYEEDKNGDGKATVIIAD---CCEHNADTRILDMLRLDTLRSSHGHDVLPDARIGR 428
Query: 436 ------------VETGIVTRQPLSARNLDLAVINNEFLGKKHRFIYAAIGNPMPKFSGVV 483
+ET + + R +D+ IN +LG+K+R++YA F +
Sbjct: 429 FRIPLDGSKYGKLETAVEAEK--HGRAMDMCSINPLYLGQKYRYVYACGAQRPCNFPNAL 486
Query: 484 -KIDVLKGEEVGCRLYGEDCYGGEPFFVAREDGEEEDDGYLVSYVHDEKKGESRFLVMDA 542
K+D+++ + +G EPFFV R EDDG ++S V E+ G S +++D
Sbjct: 487 SKVDIVEKKVKNWHEHG--MIPSEPFFVPRPGATHEDDGVVISIV-SEENGGSFAILLDG 543
Query: 543 KSPEFEIVAEVKLPRRVPYGFHGLFV 568
S FE +A K P +PYG HG ++
Sbjct: 544 SS--FEEIARAKFPYGLPYGLHGCWI 567
>At2g44990 hypothetical protein
Length = 618
Score = 93.6 bits (231), Expect = 3e-19
Identities = 149/566 (26%), Positives = 231/566 (40%), Gaps = 124/566 (21%)
Query: 111 DELPPTQCKVIKGTLPPSL-NGVYIRNGPNPQFLPRGP-YHLFDGDGMLHAVKITNGK-- 166
+ + P K I+G++P + +G Y GP G H DG G L A I K
Sbjct: 71 ETIEPVVIKPIEGSIPVNFPSGTYYLAGPGLFTDDHGSTVHPLDGHGYLRAFHIDGNKRK 130
Query: 167 ATLCSRYVQTYKYKIENEAGYQILPNVFSG-FNSLIGLAARGSVAVARVLTGQYNPINGI 225
AT ++YV+T K E++ G F+ L G G+ V +
Sbjct: 131 ATFTAKYVKTEAKKEEHDPVTDTWRFTHRGPFSVLKGGKRFGNTKVMK------------ 178
Query: 226 GLINTSLALFGNRLFALGESDLPYEIKVTPNGDIQTIGRY------------------DF 267
+ NTS+ + RL L E PYEI+ +G + T+GR+ D
Sbjct: 179 NVANTSVLKWAGRLLCLWEGGEPYEIE---SGSLDTVGRFNVENNGCESCDDDDSSDRDL 235
Query: 268 NGKLFMNMTA--------------------HPKIDSD-----TGECFAYRYGLIRPFLTY 302
+G + A H K+D T C A L R T+
Sbjct: 236 SGHDIWDTAADLLKPILQGVFKMPPKRFLSHYKVDGRRKRLLTVTCNAEDMLLPRSNFTF 295
Query: 303 FRFDSNGVKHNDVPVFSMKRPSFLHDFAITKK-YALFADIQLEINPLDMIF---GGSPVR 358
+DS K F + +HD+A T Y LFA+ ++++NP+ I G SP+
Sbjct: 296 CEYDSE-FKLIQTKEFKIDDHMMIHDWAFTDTHYILFAN-RVKLNPIGSIAAMCGMSPMV 353
Query: 359 SDPSKVPR-----IGILPRYADNESKM-KWFDVP-----GLNIMHLINAWD-EEDGKTVT 406
S S P I ILPR++D S+ + + VP L ++H NA++ ED +
Sbjct: 354 SALSLNPSNESSPIYILPRFSDKYSRGGRDWRVPVEVSSQLWLIHSGNAYETREDNGDLK 413
Query: 407 IVAPNILC-----------------------VEHMMERLELIHEMIEKVRINVE-TGIVT 442
I C V ++ + + + KV + ++ TG
Sbjct: 414 IQIQASACSYRWFDFQKMFGYDWQSNKLDPSVMNLNRGDDKLLPHLVKVSMTLDSTGNCN 473
Query: 443 R---QPLSARNL--DLAVINNEFLGKKHRFIYAAIGN------PMPKFSGVVKIDVLKGE 491
+PL+ N D VIN+ + GKK++++Y+A + P F VVK D L
Sbjct: 474 SCDVEPLNGWNKPSDFPVINSSWSGKKNKYMYSAASSGTRSELPHFPFDMVVKFD-LDSN 532
Query: 492 EVGCRLYGEDCYGGEPFFVAR---EDGEEEDDGYLVSYVHDEKKGESRFLVMDAK--SPE 546
V G + GEP FV + E+GEEEDDGY+V + +++DAK
Sbjct: 533 LVRTWSTGARRFVGEPMFVPKNSVEEGEEEDDGYIVVVEYAVSVERCYLVILDAKKIGES 592
Query: 547 FEIVAEVKLPRRV--PYGFHGLFVKE 570
+V+ +++PR + P GFHGL+ +
Sbjct: 593 DAVVSRLEVPRNLTFPMGFHGLWASD 618
>At4g08630 hypothetical protein
Length = 779
Score = 32.7 bits (73), Expect = 0.57
Identities = 26/100 (26%), Positives = 46/100 (46%), Gaps = 7/100 (7%)
Query: 26 SVRTKEKPQTSQPKTLRTT---PTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDD 82
S+RTKE+PQT P + + T + ++T T + TN+ Q+ S + I N
Sbjct: 307 SIRTKEQPQTGLPTSGSRSSLYEDSTESSAMSTLTHPSQTTNQVEQSPSARSAISNKSSQ 366
Query: 83 IINTFIDPP-IKPSVDPRHVLSQNFAPVLDELPPTQCKVI 121
++ PP + S + R + + AP++ P K +
Sbjct: 367 SLSAMDQPPSARSSFNGRPIRT---APLMPSSVPISLKPV 403
>At3g42540 hypothetical protein
Length = 208
Score = 30.4 bits (67), Expect = 2.8
Identities = 14/39 (35%), Positives = 20/39 (50%)
Query: 118 CKVIKGTLPPSLNGVYIRNGPNPQFLPRGPYHLFDGDGM 156
CK+ +L PS +Y+ +GPNP H GDG+
Sbjct: 99 CKIGNKSLKPSGRALYVVSGPNPYEYGVNKAHNVGGDGL 137
>At5g25610 dehydration-induced protein RD22
Length = 392
Score = 30.0 bits (66), Expect = 3.7
Identities = 12/31 (38%), Positives = 20/31 (63%)
Query: 486 DVLKGEEVGCRLYGEDCYGGEPFFVAREDGE 516
D+++G+E+ R ED YGG+ F+ R + E
Sbjct: 181 DLVRGKEMNVRFNAEDGYGGKTAFLPRGEAE 211
>At3g05470 unknown protein
Length = 884
Score = 30.0 bits (66), Expect = 3.7
Identities = 26/111 (23%), Positives = 44/111 (39%), Gaps = 1/111 (0%)
Query: 14 PPTTYIPQFNTASVRTKEKPQTSQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSIS 73
PP +PQF+ + T P+T+ +TL + + L A + T+ + N +
Sbjct: 373 PPPPPLPQFSNKRIHTLSSPETANLQTLSSQLCEKLCASSSKTSFPINVPNSQPRPPPPP 432
Query: 74 ALILNTLDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGT 124
IN PP+ R L ++ AP L +L P ++ T
Sbjct: 433 PPPQQLQVAGINKTPPPPLSLDFSERRPLGKDGAP-LPKLKPLHWDKVRAT 482
>At2g34250 putative protein transport protein SEC61 alpha subunit
Length = 475
Score = 30.0 bits (66), Expect = 3.7
Identities = 17/51 (33%), Positives = 27/51 (52%), Gaps = 2/51 (3%)
Query: 193 VFSGFNSLIGLAARGSVAVARVLTGQYNPINGIGLINTSLALFGNRLFALG 243
+ +G L+G+ AVA VL+G Y P+ +G+ N L + +LF G
Sbjct: 114 LLNGAQKLLGILIAIGEAVAYVLSGMYGPVGQLGVGNAILIIL--QLFFAG 162
>At1g34180 unknown protein
Length = 564
Score = 29.6 bits (65), Expect = 4.8
Identities = 27/100 (27%), Positives = 41/100 (41%), Gaps = 7/100 (7%)
Query: 80 LDDIINTFIDPPIKPSVDPRHVLSQNFAPVLDELPPTQCKVIKGTLPPSLNGVYIRNGPN 139
L+DI + P P V+ R F + +VI+ TL + +G YI N
Sbjct: 226 LEDIEKLLNEIPDAPGVNQRQ-----FDEFVGVPQGNSAEVIQSTLLNNSSGEYIDPRTN 280
Query: 140 PQFLPRGPYHLFDG--DGMLHAVKITNGKATLCSRYVQTY 177
FLP G + D L++ + T+G A L + Y
Sbjct: 281 GMFLPNGQLYNRDSSFQSHLNSFEATSGMAPLLDNEKEEY 320
>At5g14690 putative protein
Length = 217
Score = 29.3 bits (64), Expect = 6.3
Identities = 21/59 (35%), Positives = 28/59 (46%), Gaps = 11/59 (18%)
Query: 489 KGEEVGCRLYGEDCYGGEPF----FVAREDG----EEEDDGYLVSYV---HDEKKGESR 536
+GE+ GC G CYGG + FV DG EEE++ S HDE + +R
Sbjct: 132 EGEKCGCGGGGGGCYGGGDYDDHRFVVERDGSLTKEEEEEERTTSCKGEDHDESRISAR 190
>At1g22080 hypothetical protein
Length = 475
Score = 29.3 bits (64), Expect = 6.3
Identities = 12/28 (42%), Positives = 17/28 (59%)
Query: 92 IKPSVDPRHVLSQNFAPVLDELPPTQCK 119
++PS PRH+LS P++DE P K
Sbjct: 288 LRPSSTPRHLLSDFNCPLMDEAPRANLK 315
>At5g34910 putative protein
Length = 203
Score = 28.9 bits (63), Expect = 8.2
Identities = 18/68 (26%), Positives = 31/68 (45%), Gaps = 1/68 (1%)
Query: 4 KPIIIIACKQPPTTYIPQFNTASVRTKEKPQTSQPKTLRTTPTKTLLALLTTTTQKLKNT 63
K ++ + K P ++ + P T+ P TL TPT T A++ T L +T
Sbjct: 134 KLVLFMKEKDPELAAYLSTDSTEPEPETHPTTTTPSTL-PTPTTTAPAIIVAGTTPLVST 192
Query: 64 NKTLQTSS 71
+ T+ +S
Sbjct: 193 SATISNTS 200
>At2g30340 unknown protein
Length = 268
Score = 28.9 bits (63), Expect = 8.2
Identities = 27/81 (33%), Positives = 34/81 (41%), Gaps = 6/81 (7%)
Query: 36 SQPKTLRTTPTKTLLALLTTTTQKLKNTNKTLQTSSISALILNTLDDIINTFIDPPIKPS 95
S+ T+RT + TT T N N T TSS+S + L I PP P+
Sbjct: 143 SELTTVRTEILRHKYQEATTITSLQNNFNSTTTTSSVSC-DQHALASAILLPPPPPPPPT 201
Query: 96 VDPRHVLSQNFAPVLDELPPT 116
P +LS AP PPT
Sbjct: 202 PRPPRLLSSQPAP-----PPT 217
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.140 0.415
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 14,154,780
Number of Sequences: 26719
Number of extensions: 682409
Number of successful extensions: 1509
Number of sequences better than 10.0: 19
Number of HSP's better than 10.0 without gapping: 13
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1434
Number of HSP's gapped (non-prelim): 21
length of query: 579
length of database: 11,318,596
effective HSP length: 105
effective length of query: 474
effective length of database: 8,513,101
effective search space: 4035209874
effective search space used: 4035209874
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144759.6


