

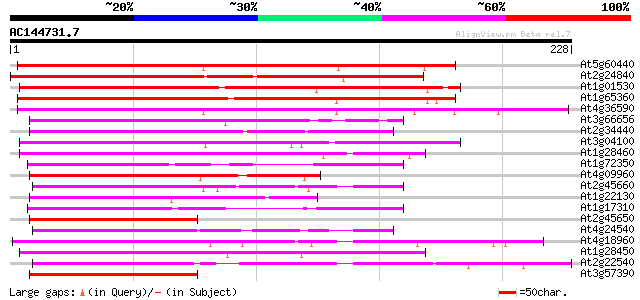
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144731.7 + phase: 0
(228 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g60440 putative MADS box protein 177 3e-45
At2g24840 putative MADS-box protein 140 6e-34
At1g01530 hypothetical protein 137 5e-33
At1g65360 135 2e-32
At4g36590 putative MADS-box protein 128 2e-30
At3g66656 SRF-type transcription factor 100 5e-22
At2g34440 MADS-box protein (AGL29) 100 1e-21
At3g04100 putative SRF-type transcription factor 96 1e-20
At1g28460 hypothetical protein 89 2e-18
At1g72350 putative SRF-type transcription factor 87 7e-18
At4g09960 MADS-box protein AGL11 84 8e-17
At2g45660 MADS-box protein (AGL20) 82 2e-16
At1g22130 hypothetical protein 82 2e-16
At1g17310 transcription factor, putative 81 4e-16
At2g45650 MADS-box protein (AGL6) 80 9e-16
At4g24540 MADS-box protein AGL24 80 1e-15
At4g18960 floral homeotic protein agamous 79 3e-15
At1g28450 hypothetical protein 77 8e-15
At2g22540 putative MADS-box protein 76 2e-14
At3g57390 MADS transcription factor-like protein 75 3e-14
>At5g60440 putative MADS box protein
Length = 299
Score = 177 bits (450), Expect = 3e-45
Identities = 98/185 (52%), Positives = 128/185 (68%), Gaps = 7/185 (3%)
Query: 4 VRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFS 63
V+K +GRQKIEM K+ NESNLQVTFSK +GLFKKASELCTLCGA+VA+VVFSP KVFS
Sbjct: 2 VKKSKGRQKIEMVKMKNESNLQVTFSKRRSGLFKKASELCTLCGAEVAIVVFSPGRKVFS 61
Query: 64 FGHPNLDTVIDRFL--SLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGD 121
FGHPN+D+VIDRF+ + +P MQ E RN+ V++LN LTQ+ + L+ EKK D
Sbjct: 62 FGHPNVDSVIDRFINNNPLPPHQHNNMQLRETRRNSIVQDLNNHLTQVLSQLETEKKKYD 121
Query: 122 ELSNLHKETEA-KFWWACVVDGMNRDQLEIFKKALEELKKLVIQHAA----TRTLPFFVG 176
EL + ++T+A WW V+ + QLE FK LE LKK+V A+ F+VG
Sbjct: 122 ELKKIREKTKALGNWWEDPVEELALSQLEGFKGNLENLKKVVTVEASRFFQANVPNFYVG 181
Query: 177 NTSSS 181
++S++
Sbjct: 182 SSSNN 186
>At2g24840 putative MADS-box protein
Length = 210
Score = 140 bits (353), Expect = 6e-34
Identities = 79/171 (46%), Positives = 111/171 (64%), Gaps = 5/171 (2%)
Query: 1 MSSVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGK 60
MS ++ GRQKI M KI ES+ QVTFSK GLFKKASELCTLCGA++ ++VFSP+ K
Sbjct: 1 MSKKKESIGRQKIPMVKIKKESHRQVTFSKRRAGLFKKASELCTLCGAEIGIIVFSPAKK 60
Query: 61 VFSFGHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIG 120
FSFGHP++++V+DR++S + Q ++ A+ ELN QLT I + ++ EKK G
Sbjct: 61 PFSFGHPSVESVLDRYVSR-NNMSLAQSQQLQGSPAASC-ELNMQLTHILSEVEEEKKKG 118
Query: 121 DELSNLHKETEAKF---WWACVVDGMNRDQLEIFKKALEELKKLVIQHAAT 168
+ + KE+ + WW V+ MN QL+ K ALEEL+K V+ + A+
Sbjct: 119 QAMEEMRKESVRRSMINWWEKPVEEMNMVQLQEMKYALEELRKTVVTNMAS 169
>At1g01530 hypothetical protein
Length = 247
Score = 137 bits (345), Expect = 5e-33
Identities = 79/183 (43%), Positives = 111/183 (60%), Gaps = 8/183 (4%)
Query: 5 RKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF 64
RK GR+KIE+ K++NESNLQVTFSK +GLFKK SELCTLC A++A++VFSPSGK +SF
Sbjct: 3 RKNLGRRKIELVKMTNESNLQVTFSKRRSGLFKKGSELCTLCDAEIAIIVFSPSGKAYSF 62
Query: 65 GHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDEL- 123
GHPN++ ++D L + N+ F E+ ++ LN LT++ + E++ +
Sbjct: 63 GHPNVNKLLDHSLGRVIRHNN--TNFAESRTKLRIQMLNESLTEVMAEKEKEQETKQSIV 120
Query: 124 SNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQHAAT---RTLPFFVGNTSS 180
N + +A+ WW +N Q K LE LKK V + A R L F+VG SS
Sbjct: 121 QNERENKDAEKWWRNSPTELNLAQSTSMKCDLEALKKEVDEKVAQLHHRNLNFYVG--SS 178
Query: 181 SNI 183
SN+
Sbjct: 179 SNV 181
>At1g65360
Length = 226
Score = 135 bits (340), Expect = 2e-32
Identities = 75/183 (40%), Positives = 116/183 (62%), Gaps = 7/183 (3%)
Query: 4 VRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFS 63
V+K GR+K+E+ K++ ESNLQVTFSK GLFKKASE CTLC A +A++VFSP+GKVFS
Sbjct: 2 VKKTLGRRKVEIVKMTKESNLQVTFSKRKAGLFKKASEFCTLCDAKIAMIVFSPAGKVFS 61
Query: 64 FGHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDEL 123
FGHPN+D ++D F + N+ + E++ +V+ LN T++ ++ E+K
Sbjct: 62 FGHPNVDVLLDHFRGCVVGHNNTNLD--ESYTKLHVQMLNKSYTEVKAEVEKEQKNKQSR 119
Query: 124 SNLHKETE-AKFWWACVVDGMNRDQLEIFKKALEELKKLVIQHAAT---RTLP-FFVGNT 178
+ +E E A+ WW+ +N Q + L++LKK+V + A +T P F+VG++
Sbjct: 120 AQNERENENAEEWWSKSPLELNLSQSTCMIRVLKDLKKIVDEKAIQLIHQTNPNFYVGSS 179
Query: 179 SSS 181
S++
Sbjct: 180 SNA 182
>At4g36590 putative MADS-box protein
Length = 248
Score = 128 bits (322), Expect = 2e-30
Identities = 86/239 (35%), Positives = 128/239 (52%), Gaps = 15/239 (6%)
Query: 4 VRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFS 63
VR +GRQKIEMKK+ NESNLQVTFSK GLFKKASELCTL GA++ L+VFSP GKVFS
Sbjct: 2 VRSTKGRQKIEMKKMENESNLQVTFSKRRFGLFKKASELCTLSGAEILLIVFSPGGKVFS 61
Query: 64 FGHPNLDTVIDRFL-----SLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKK 118
FGHP++ +I RF S I + +Q +E + N++ LN LT++ + EK+
Sbjct: 62 FGHPSVQELIHRFSNPNHNSAIVHHQNNNLQLVETRPDRNIQYLNNILTEVLANQEKEKQ 121
Query: 119 IGDELSNLHKETE-AKFWWACVVDGMNRDQLEIFKKALEELKKLVI----QHAATRTLPF 173
L L + E W+ V ++ ++ AL+++KK ++ Q++
Sbjct: 122 KRMVLDLLKESREQVGNWYEKDVKDLDMNETNQLISALQDVKKKLVREMSQYSQVNVSQN 181
Query: 174 FVGNTS----SSNIYLHHQPNPQQAEMF-PPQIFQNAMLQLQTHFFDGSMIPHHVFNNM 227
+ G +S N+ + + A + P +F N + + DG ++P N M
Sbjct: 182 YFGQSSGVIGGGNVGIDLFDQRRNAFNYNPNMVFPNHTPPMFGYNNDGVLVPISNMNYM 240
>At3g66656 SRF-type transcription factor
Length = 178
Score = 100 bits (250), Expect = 5e-22
Identities = 62/156 (39%), Positives = 89/156 (56%), Gaps = 15/156 (9%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR+KI+M+K+ + + QVTFSK GLFKKASEL TLC A+V +VVFSP K +SFG PN
Sbjct: 2 GRRKIKMEKVQDTNTKQVTFSKRRLGLFKKASELATLCNAEVGIVVFSPGNKPYSFGKPN 61
Query: 69 LDTVIDRFLSLIPTQNDG----TMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELS 124
D + +RF + + +G T + +R +++ +L I +AEKK G++
Sbjct: 62 FDVIAERFKNEFEEEEEGDSCETSGYSRGNRARQEKKICKRLNSITEEAEAEKKHGED-- 119
Query: 125 NLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKK 160
LHK E + D N+ E+ LEELK+
Sbjct: 120 -LHKWLE-----SAEQDKFNKPIEEL---TLEELKE 146
>At2g34440 MADS-box protein (AGL29)
Length = 172
Score = 99.8 bits (247), Expect = 1e-21
Identities = 57/148 (38%), Positives = 89/148 (59%), Gaps = 2/148 (1%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR+KI+M+ + + + QVTFSK GLFKKASEL TLC A++ +VVFSP GK FS+G PN
Sbjct: 2 GRRKIKMEMVQDMNTRQVTFSKRRTGLFKKASELATLCNAELGIVVFSPGGKPFSYGKPN 61
Query: 69 LDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLHK 128
LD+V +RF+ + G + +R ++ L+ +L +N ++AEK+ G E S
Sbjct: 62 LDSVAERFMREYDDSDSGDEEKSGNYR-PKLKRLSERLDLLNQEVEAEKERG-EKSQEKL 119
Query: 129 ETEAKFWWACVVDGMNRDQLEIFKKALE 156
E+ + ++ + D+L +K L+
Sbjct: 120 ESAGDERFKESIETLTLDELNEYKDRLQ 147
>At3g04100 putative SRF-type transcription factor
Length = 207
Score = 96.3 bits (238), Expect = 1e-20
Identities = 60/185 (32%), Positives = 98/185 (52%), Gaps = 8/185 (4%)
Query: 5 RKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF 64
RK +G+QKIEMKK+ N + +TFSK G+FKK +EL +C +VA ++FS K ++F
Sbjct: 10 RKTKGKQKIEMKKVENYGDRMITFSKRKTGIFKKMNELVAMCDVEVAFLIFSQPKKPYTF 69
Query: 65 GHPNLDTVIDRFLS---LIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTL--DAEK-K 118
HP++ V DR + P + D T +EA++ + +L ++ + L D EK K
Sbjct: 70 AHPSMKKVADRLKNPSRQEPLERDDTRPLVEAYKKRRLHDLVKKMEALEEELAMDLEKLK 129
Query: 119 IGDELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQHAATRTLPFFVGNT 178
+ E N K K WW +G++ +L+ +A+ EL+ + + A L G +
Sbjct: 130 LLKESRNEKKLD--KMWWNFPSEGLSAKELQQRYQAMLELRDNLCDNMAHLRLGKDCGGS 187
Query: 179 SSSNI 183
SS +
Sbjct: 188 SSVRV 192
>At1g28460 hypothetical protein
Length = 182
Score = 88.6 bits (218), Expect = 2e-18
Identities = 54/170 (31%), Positives = 92/170 (53%), Gaps = 7/170 (4%)
Query: 5 RKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF 64
+K +G+QKI +KKI + VTFSK NG++ K SEL LCG +VA + +S SGK ++F
Sbjct: 4 KKTKGKQKINIKKIEKDEGRSVTFSKRLNGIYTKISELSILCGVEVAFIGYSCSGKPYTF 63
Query: 65 GHPNLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELS 124
G P+ V +RFL+ + + + + AH+ A ++EL + ++ L ++ + +
Sbjct: 64 GSPSFQAVAERFLNGDASSSSSSSLVMNAHKQAKIQELCKKYNRLVEELKVDEVKVKKAA 123
Query: 125 NL--HKETEAKFWWACVVDGMNRDQLEIFKKALEELKKL---VIQHAATR 169
L + WW VD + E KK +E+ ++L + + AA+R
Sbjct: 124 ALAETRVVNKDVWWK--VDPNDVKDHEKAKKMMEKYQELYDKLCEQAASR 171
>At1g72350 putative SRF-type transcription factor
Length = 224
Score = 87.0 bits (214), Expect = 7e-18
Identities = 49/153 (32%), Positives = 80/153 (52%), Gaps = 33/153 (21%)
Query: 8 RGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHP 67
+GRQKIE+K+I E+ QVTFSK +GLFKKA+EL LCGA + ++ FS +++SFG
Sbjct: 42 KGRQKIEIKEIMLETRRQVTFSKRRSGLFKKAAELSVLCGAQIGIITFSRCDRIYSFG-- 99
Query: 68 NLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLH 127
N++++ID++L P + +H NV +N
Sbjct: 100 NVNSLIDKYLRKAPV-------MLRSHPGGNV------------------------ANGE 128
Query: 128 KETEAKFWWACVVDGMNRDQLEIFKKALEELKK 160
++ + WW V+ + + +E +K AL L++
Sbjct: 129 EDNDGLMWWERAVESVPEEHMEEYKNALSVLRE 161
>At4g09960 MADS-box protein AGL11
Length = 230
Score = 83.6 bits (205), Expect = 8e-17
Identities = 50/121 (41%), Positives = 75/121 (61%), Gaps = 6/121 (4%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS G+++ + + N
Sbjct: 2 GRGKIEIKRIENSTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLYEYANNN 61
Query: 69 LDTVIDRF-LSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKK--IGDELSN 125
+ + I+R+ + + N T+Q I A A ++ +A+L Q T+ + +GD LS+
Sbjct: 62 IRSTIERYKKACSDSTNTSTVQEINA---AYYQQESAKLRQQIQTIQNSNRNLMGDSLSS 118
Query: 126 L 126
L
Sbjct: 119 L 119
>At2g45660 MADS-box protein (AGL20)
Length = 214
Score = 82.4 bits (202), Expect = 2e-16
Identities = 56/161 (34%), Positives = 89/161 (54%), Gaps = 18/161 (11%)
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R K +MK+I N ++ QVTFSK NGL KKA EL LC A+V+L++FSP GK++ F N+
Sbjct: 3 RGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVSLIIFSPKGKLYEFASSNM 62
Query: 70 DTVIDRFL----SLIPTQ--NDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIG--- 120
IDR+L + T+ ++ MQ ++ + AN+ + QL L E IG
Sbjct: 63 QDTIDRYLRHTKDRVSTKPVSEENMQHLK-YEAANMMKKIEQLEASKRKLLGE-GIGTCS 120
Query: 121 -DELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKK 160
+EL + ++ E V + + ++FK+ +E+LK+
Sbjct: 121 IEELQQIEQQLEKS------VKCIRARKTQVFKEQIEQLKQ 155
>At1g22130 hypothetical protein
Length = 335
Score = 82.4 bits (202), Expect = 2e-16
Identities = 45/118 (38%), Positives = 70/118 (59%), Gaps = 2/118 (1%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF-GHP 67
GR K+E+K+I N +N QVTFSK NGL KKA EL LC D+AL++FSPS ++ F G
Sbjct: 2 GRVKLEIKRIENTTNRQVTFSKRRNGLIKKAYELSILCDIDIALIMFSPSDRLSLFSGKT 61
Query: 68 NLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSN 125
++ V RF++L + + + F + +R +++ L +I L E I +++N
Sbjct: 62 RIEDVFSRFINLPKQERESALYFPDQNRRPDIQNKEC-LLRILQQLKTENDIALQVTN 118
>At1g17310 transcription factor, putative
Length = 217
Score = 81.3 bits (199), Expect = 4e-16
Identities = 51/153 (33%), Positives = 75/153 (48%), Gaps = 36/153 (23%)
Query: 8 RGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHP 67
RGRQKIE+KKI E+ QVTFSK GLFKK++EL L GA +A++ FS +++ FGH
Sbjct: 47 RGRQKIEIKKIEEETKRQVTFSKRRRGLFKKSAELSVLTGAKIAVITFSKCDRIYRFGH- 105
Query: 68 NLDTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLH 127
+D +ID++L P + +G GD N
Sbjct: 106 -VDALIDKYLRKSPVKLEGYS-------------------------------GD---NAA 130
Query: 128 KETEAKFWWACVVDGMNRDQLEIFKKALEELKK 160
E + WW V+ + ++LE + AL L++
Sbjct: 131 DEESRRPWWERPVESVPEEELEEYMAALSMLRE 163
>At2g45650 MADS-box protein (AGL6)
Length = 252
Score = 80.1 bits (196), Expect = 9e-16
Identities = 36/68 (52%), Positives = 50/68 (72%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR ++EMK+I N+ N QVTFSK NGL KKA EL LC A+VAL++FS GK++ FG
Sbjct: 2 GRGRVEMKRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEFGSVG 61
Query: 69 LDTVIDRF 76
+++ I+R+
Sbjct: 62 IESTIERY 69
>At4g24540 MADS-box protein AGL24
Length = 220
Score = 79.7 bits (195), Expect = 1e-15
Identities = 52/147 (35%), Positives = 81/147 (54%), Gaps = 17/147 (11%)
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R+KI +KKI N + QVTFSK G+FKKA EL LC ADVAL++FS +GK+F F +
Sbjct: 3 REKIRIKKIDNITARQVTFSKRRRGIFKKADELSVLCDADVALIIFSATGKLFEFSSSRM 62
Query: 70 DTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLHKE 129
++ R+ SL + + M H +R N L++++ ++ + K +L L E
Sbjct: 63 RDILGRY-SLHASNINKLMDPPSTH----LRLENCNLSRLSKEVEDKTK---QLRKLRGE 114
Query: 130 TEAKFWWACVVDGMNRDQLEIFKKALE 156
+DG+N ++L+ +K LE
Sbjct: 115 D---------LDGLNLEELQRLEKLLE 132
>At4g18960 floral homeotic protein agamous
Length = 284
Score = 78.6 bits (192), Expect = 3e-15
Identities = 70/238 (29%), Positives = 106/238 (44%), Gaps = 29/238 (12%)
Query: 2 SSVRKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKV 61
SS + GR KIE+K+I N +N QVTF K NGL KKA EL LC A+VAL+VFS G++
Sbjct: 43 SSPLRKSGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRL 102
Query: 62 FSFGHPNLDTVIDRFLSLI-PTQNDGTMQFIEA----HRNANVRELNAQLTQINNTLDAE 116
+ + + ++ I+R+ I N G++ I A +A +R+ + N L E
Sbjct: 103 YEYSNNSVKGTIERYKKAISDNSNTGSVAEINAQYYQQESAKLRQQIISIQNSNRQLMGE 162
Query: 117 KKIGD----ELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQ-HAATRTL 171
IG EL NL E + + + E+ ++ ++K + H + L
Sbjct: 163 -TIGSMSPKELRNLEGRLERS------ITRIRSKKNELLFSEIDYMQKREVDLHNDNQIL 215
Query: 172 PFFVGNTSSSNIYLHHQPNPQQAE--MFPPQ----------IFQNAMLQLQTHFFDGS 217
+ +N + P E M PPQ FQ A LQ H + +
Sbjct: 216 RAKIAENERNNPSISLMPGGSNYEQLMPPPQTQSQPFDSRNYFQVAALQPNNHHYSSA 273
>At1g28450 hypothetical protein
Length = 185
Score = 77.0 bits (188), Expect = 8e-15
Identities = 47/171 (27%), Positives = 87/171 (50%), Gaps = 6/171 (3%)
Query: 5 RKGRGRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSF 64
+K +G+QKI +KKI + + VT SK N ++ EL LCG +VA + +S SGK ++F
Sbjct: 4 KKTKGKQKINIKKIEKDEDRSVTLSKRLNAIYTMIIELSILCGVEVAFIGYSCSGKPYTF 63
Query: 65 GHPNLDTVIDRFLSLIPTQNDGT---MQFIEAHRNANVRELNAQLTQINNTLDAEK---K 118
G P+ V++RFL+ + + + AH+ A ++EL + ++ L ++ K
Sbjct: 64 GSPSFQAVVERFLNGEASSSSSSSLQRSVKNAHKQAKIQELCKRYNRLVEELKVDEVKVK 123
Query: 119 IGDELSNLHKETEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQHAATR 169
L+ + +W A D + ++ + + +ELK+ + + A R
Sbjct: 124 KAAALAETRAVNKDAWWKADPNDVKDHEKAKKMMEKYQELKEKLREEVALR 174
>At2g22540 putative MADS-box protein
Length = 210
Score = 75.9 bits (185), Expect = 2e-14
Identities = 62/225 (27%), Positives = 109/225 (47%), Gaps = 24/225 (10%)
Query: 10 RQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPNL 69
R+KI+++KI N + QVTFSK GLFKKA EL LC ADVAL++FS +GK+F F ++
Sbjct: 3 REKIQIRKIDNATARQVTFSKRRRGLFKKAEELSVLCDADVALIIFSSTGKLFEFCSSSM 62
Query: 70 DTVIDRFLSLIPTQNDGTMQFIEAHRNANVRELNAQLTQINNTLDAEKKIGDELSNLHKE 129
V++R + ++N + L QL + ++ K+I D+ L +
Sbjct: 63 KEVLER--HNLQSKN---------LEKLDQPSLELQLVENSDHARMSKEIADKSHRLRQM 111
Query: 130 TEAKFWWACVVDGMNRDQLEIFKKALEELKKLVIQHAATRTLPFFVGNTSSSNIYL--HH 187
+ + G++ ++L+ +KALE VI+ + + + + + L +
Sbjct: 112 RGEE------LQGLDIEELQQLEKALETGLTRVIETKSDKIMS-EISELQKKGMQLMDEN 164
Query: 188 QPNPQQAEMFPPQIFQNAML----QLQTHFFDGSMIPHHVFNNMV 228
+ QQ + P + N L + T F+ + H+F++ V
Sbjct: 165 KRLRQQVCVLPSLLITNPFLLSTINVHTPKFNPQLSTTHMFDHTV 209
>At3g57390 MADS transcription factor-like protein
Length = 256
Score = 75.1 bits (183), Expect = 3e-14
Identities = 34/68 (50%), Positives = 49/68 (72%)
Query: 9 GRQKIEMKKISNESNLQVTFSKHHNGLFKKASELCTLCGADVALVVFSPSGKVFSFGHPN 68
GR +IE+KKI N ++ QVTFSK NGL KKA EL LC A+VAL++FS +GK++ F
Sbjct: 2 GRGRIEIKKIENINSRQVTFSKRRNGLIKKAKELSILCDAEVALIIFSSTGKIYDFSSVC 61
Query: 69 LDTVIDRF 76
++ ++ R+
Sbjct: 62 MEQILSRY 69
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.133 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,007,216
Number of Sequences: 26719
Number of extensions: 207220
Number of successful extensions: 799
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 103
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 677
Number of HSP's gapped (non-prelim): 113
length of query: 228
length of database: 11,318,596
effective HSP length: 96
effective length of query: 132
effective length of database: 8,753,572
effective search space: 1155471504
effective search space used: 1155471504
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144731.7


