

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144730.8 - phase: 0
(840 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
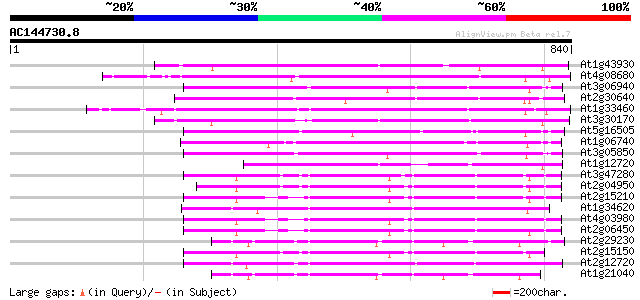
Score E
Sequences producing significant alignments: (bits) Value
At1g43930 hypothetical protein 271 1e-72
At4g08680 putative MuDR-A-like transposon protein 265 6e-71
At3g06940 putative mudrA protein 247 2e-65
At2g30640 Mutator-like transposase 246 4e-65
At1g33460 mutator transposase MUDRA, putative 244 1e-64
At3g30170 unknown protein 233 4e-61
At5g16505 mutator-like transposase-like protein (MQK4.25) 228 1e-59
At1g06740 mudrA-like protein 217 2e-56
At3g05850 unknown protein 216 3e-56
At1g12720 hypothetical protein 197 3e-50
At3g47280 putative protein 195 1e-49
At2g04950 putative transposase of transposon FARE2.9 (cds1) 190 3e-48
At2g15210 putaive transposase of transposon FARE2.7 (cds1) 184 1e-46
At1g34620 Mutator-like protein 181 2e-45
At4g03980 putative transposon protein 179 4e-45
At2g06450 putative transposase of FARE2.11 179 4e-45
At2g29230 Mutator-like transposase 179 6e-45
At2g15150 putative transposase of FARE2.2 (CDS1) 178 1e-44
At2g12720 putative protein 172 9e-43
At1g21040 hypothetical protein 165 1e-40
>At1g43930 hypothetical protein
Length = 946
Score = 271 bits (692), Expect = 1e-72
Identities = 172/641 (26%), Positives = 301/641 (46%), Gaps = 34/641 (5%)
Query: 218 DEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIE 277
D+ D + + + L SD + NDD Q P+ + ++ FN +FK A++
Sbjct: 200 DDIWDDERIPDPLVHSDDEGDNDDEDGAPQTDDPEES----LRLAKTFNNPEDFKIAVLR 255
Query: 278 WNVLNGYQIKVLKNESYRVRVECRD---QCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVL 334
+++ Y IK+ +++S RV +C D C ++ CS ++ Q+K + H+C
Sbjct: 256 YSLKTRYDIKLYRSQSLRVGAKCTDTDVNCQWRCYCSYDKKKQKMQVKVYKDGHSCVRSG 315
Query: 335 ENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGD 394
+K R +A +++ K++ QE+ +E+ Y + +T D+ +A+ + +
Sbjct: 316 YSKMLKQRTIAWLFRERLRKNPKITKQEMVDEIKREYKLTVTEDQCSKAKTMVMRERQAT 375
Query: 395 ADKQYAMIRRYAAELKRVCRDNNVKIN-VAGPSVTIQLRFGSFYFCFDGCKKGFISACRP 453
+ +A I Y A++ R +I + GP++ RF + CF K+ + CRP
Sbjct: 376 HQEHFARIWDYQAKIFRSNPGTTFEIETILGPTIGSLQRFLRLFICFKSQKESWKQTCRP 435
Query: 454 FVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLME--DIGE 511
+G+DG LK G LL A GRD +++ P+A+ VVE E + W WF ++L D+ +
Sbjct: 436 IIGIDGAFLKWDIKGHLLAATGRDGDNRIVPIAWAVVEIENDDNWDWFVRMLSRTLDLQD 495
Query: 512 DKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATY 571
+ ISD+Q GL+ + +HR C RH+ N+K+ +++ L ++
Sbjct: 496 GRNVAIISDKQSGLVKAIHSVIPQAEHRQCARHIMENWKRN-SHDMELQRLFWKIVRSYT 554
Query: 572 NQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSAR 631
+ M LK + A+E L+ P W + F S C+ +NN+SESFN TI AR
Sbjct: 555 EGEFGAHMRALKSYNASAFELLLKTLPVTWSRAFFKIGSCCNDNLNNLSESFNRTIREAR 614
Query: 632 DQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGE 691
+P++ M E IR M R + ++ + R R K S + +
Sbjct: 615 RKPLLDMLEDIRRQCMVR--------NEKRYVIADRLRTRFTKRAHAEIEKMIAGSQVCQ 666
Query: 692 QFQVMHTYNR------QQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDD 745
++ H + F VD+ +C C W++ GIPC H S + ++Q+ E +V D
Sbjct: 667 RWMARHNKHEIKVGPVDSFTVDMNNNTCGCMKWQMTGIPCIHAASVIIGKRQKVEDYVSD 726
Query: 746 YYSRSKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNG-PGRPRKLRIRE-------- 796
+Y+ S + Y+ I P+ G +WP V +LPP ++ G PGRP+ R+
Sbjct: 727 WYTTSMWKQTYNDGIGPVQGKLLWPTVNKVGVLPPPWRRGNPGRPKNHARRKGVFESSTA 786
Query: 797 FDENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDVQNPEAAK 837
+ + + R C+ C GHN++ CK++ P A +
Sbjct: 787 SSSSTTELSRLHRVMTCSNCQGEGHNKQGCKNETVAPPAKR 827
>At4g08680 putative MuDR-A-like transposon protein
Length = 761
Score = 265 bits (678), Expect = 6e-71
Identities = 205/726 (28%), Positives = 332/726 (45%), Gaps = 46/726 (6%)
Query: 139 FDDSEDESTTALEDGFEDVEVEAPANGNNRVTVNNKTLRIKKCGSKTPKKKSPKKNNSGR 198
FDD D DG DVE + + + +R KK K K+K K
Sbjct: 2 FDDDVD-MFYKYSDG--DVEYNGGNESESEERIEEEIVRSKKNTKKNKKEKKKKDEEDPL 58
Query: 199 MNVIAPVSLLKNGKGKYVVDEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFK 258
V +G + + I D + E SSD D+ D I R+ + D K
Sbjct: 59 ERFDFEVD---DGVANFEDEIPIGRDIIPE---SSDEDE---DPILVRDRRIRRAMGD-K 108
Query: 259 FKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECR--DQCGYKVLCSKVGDR 316
F IG F T +EFKEAI+E + +G+ + + E ++ +C +C ++V CS DR
Sbjct: 109 FVIGSTFFTGIEFKEAILEVTLKHGHNVVQDRWEKEKISFKCGMGGKCKWRVYCSYDPDR 168
Query: 317 RTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGIT 376
+ F +KT H+C+P + + S +A+ + K+++ +K ++ + + + T
Sbjct: 169 QLFVVKTSCTWHSCSPNGKCQILKSPVIARLFLDKLRLNEKFMPMDIQEYIKERWKMVST 228
Query: 377 MDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKI----NVAGPSVTIQLR 432
+ + R R +A +++ + ++Q+A IR Y E+ + I N G V
Sbjct: 229 IPQCQRGRLLALKMLKKEYEEQFAHIRGYVEEIHSQNPGSVAFIDTYRNEKGEDV----- 283
Query: 433 FGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVET 492
F FY CF+ + + +CRP +G+DG LK G LL AVG DPN+Q +P+A+ VV++
Sbjct: 284 FNRFYVCFNILRTQWAGSCRPIIGLDGTFLKVVVKGVLLTAVGHDPNNQIYPIAWAVVQS 343
Query: 493 ECKETWKWFTQLLMEDIG--EDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFK 550
E E W WF Q + +D+ + R+V +SD+ KGLLS + + +HR+C++H+ N K
Sbjct: 344 ENAENWLWFVQQIKKDLNLEDGSRFVILSDRSKGLLSAVKQELPNAEHRMCVKHIVENLK 403
Query: 551 KKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYS 610
K ++ L+ + + + + +N L+ D + ++ EP W + + S
Sbjct: 404 KNHAKKDMLKTLVWKLAWSYNEKEYGKNLNNLRCYDEALYNDVLNEEPHTWSRCFYKLGS 463
Query: 611 KCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRK 670
C+ + NN +ESFN TI AR + +I M E IR M R+ K + + K
Sbjct: 464 CCEDVDNNATESFNSTITKARAKSLIPMLETIRRQGMTRIVKRNKKSLRHEGRFTKYALK 523
Query: 671 RLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVS 730
L E T A K + F+V N + VDI K CSC W+I GIPC H
Sbjct: 524 MLALEKTDADRSK-VYRCTHGVFEVYIDENGHR--VDIPKTQCSCGKWQISGIPCEHSYG 580
Query: 731 ALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPE-----VEAP--ELLPPNYK 783
A+ + E ++ +++S + Y A P+ G W V AP +LP K
Sbjct: 581 AMIEAGLDAENYISEFFSTDLWRDSYETATMPLRGPKYWLNSSYGLVTAPPEPILPGRKK 640
Query: 784 NGPGRPRKLRIREFDENGSRMRR----------QGVAYRCTKCDQFGHNQRRCKSDVQNP 833
+ + RI+ +E+ + +R +G C C + GHN RCK +
Sbjct: 641 EKSKKEKFARIKGKNESPKKKKRKKNEVEKLGKKGKIIHCKSCGEAGHNALRCKKFPKEK 700
Query: 834 EAAKRK 839
KRK
Sbjct: 701 VPRKRK 706
>At3g06940 putative mudrA protein
Length = 749
Score = 247 bits (631), Expect = 2e-65
Identities = 156/582 (26%), Positives = 276/582 (46%), Gaps = 29/582 (4%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTF 319
+ FNT +EF++A+ +++V +G+ K KN+S+RV V+C+ Q C +++ S++ +
Sbjct: 182 VDQRFNTFLEFRDALHKYSVAHGFAYKYKKNDSHRVSVKCKAQGCPWRITASRLSTTQLI 241
Query: 320 QIKTLEGPHTC--APVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITM 377
IK + HTC A + A WV K++ +++ ++ YG+ +
Sbjct: 242 CIKKMNPRHTCERAVIKPGYRATRGWVRTILKEKLKAFPDYKPKDIAEDIKKEYGIQLNY 301
Query: 378 DRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFY 437
+AWRA++IA+ ++G + Y+ + ++K ++ + + F SFY
Sbjct: 302 SQAWRAKEIAREQLQGSYKEAYSQLPLICEKIKET-NPGSIATFMTKEDSSFHRLFISFY 360
Query: 438 FCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKET 497
G K+G RP + +D L ++Y G +L+A D D FP+AF +V++E +E
Sbjct: 361 ASISGFKQG----SRPLLFLDNAILNSKYQGVMLVATASDAEDGIFPVAFAIVDSETEEN 416
Query: 498 WKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFD-SIDHRLCLRHLYANFKKKFGG- 555
W WF + L + E + F++D Q GL + ++F+ H CL L G
Sbjct: 417 WLWFLEQLKTALSESRIITFVADFQNGLKNAIAQVFEKDAHHAYCLGQLAEKLNVDLKGQ 476
Query: 556 -GSQIRDLMM----GATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYS 610
+ R M+ AT G+ + +K I A+ W++ EP W F
Sbjct: 477 FSHEARRYMLNDFYSVAYATTPVGYYLALENIKSISPDAYNWVIESEPHHWANALFQ-GE 535
Query: 611 KCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRK 670
+ + + +N + F + A + PI M + R +M+ + T K ++W + P +
Sbjct: 536 RYNKMNSNFGKDFYSWVSEAHEFPITQMIDEFRAKMMQSIYTRQVKSREWVTTLTPSNEE 595
Query: 671 RLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVS 730
+L KEI LA + + S G FQV + IVDI + C C W + G+PC H ++
Sbjct: 596 KLQKEIELAQLLQVS-SPEGSLFQV-NGGESSVSIVDINQCDCDCKTWRLTGLPCSHAIA 653
Query: 731 ALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPE----LLPPNYKNGP 786
+G ++ P + Y + + L Y+ +I P+ MD + PE + PP + P
Sbjct: 654 VIGCIEKSPYEYCSTYLTVESHRLMYAESIQPVPNMDRMMLDDPPEGLVCVTPPPTRRTP 713
Query: 787 GRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRCKS 828
GRP+ ++ D M+RQ +C+KC GHN++ CK+
Sbjct: 714 GRPKIKKVEPLD----MMKRQ---LQCSKCKGLGHNKKTCKA 748
>At2g30640 Mutator-like transposase
Length = 754
Score = 246 bits (628), Expect = 4e-65
Identities = 160/601 (26%), Positives = 284/601 (46%), Gaps = 31/601 (5%)
Query: 247 QFRMPQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CG 305
Q R Q + +GMEF+ ++ + A+ + + ++++ +K++ R +C + C
Sbjct: 168 QCRSLQTAPEHNLIVGMEFSDVLACRRALRDAAIALRFEMQTVKSDKTRFTAKCNSEGCP 227
Query: 306 YKVLCSKVGDRRTFQIKTLEGPHTCAPV--LENKSANSRWVAKKAVPKMQVTKKMSVQEV 363
+++ C+K+ TF I+T+ G HTC + L + A+ +WVA +++V +E+
Sbjct: 228 WRIHCAKLPGLPTFTIRTIHGSHTCGGISHLGHHQASVQWVADAVTERLKVNPHCKPKEI 287
Query: 364 FNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVA 423
++ +G+ +T +AWR ++ + + G ++ Y ++ RY +++R +V +
Sbjct: 288 LEQIHQVHGITLTYKQAWRGKERIMAAVRGSYEEDYRLLPRYCDQIRRT-NPGSVAVVHG 346
Query: 424 GPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYF 483
P + F F+ F GF++ACRP +G+D LK++Y G LL+A G D F
Sbjct: 347 SP---VDGSFQQFFISFQASICGFLNACRPLIGLDRTVLKSKYVGTLLLATGFDGEGAVF 403
Query: 484 PLAFGVVETECKETWKWF----TQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHR 539
PLAF ++ E +W+WF QLL + + +S + + ++ D F + H
Sbjct: 404 PLAFAIISEENDSSWQWFLSELRQLLEVNSENMPKLTILSSRDQSIVDGVDTNFPTAFHG 463
Query: 540 LCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPK 599
LC+ L + + +F S + +L+ A K + + KM E+ +I A W+ ++
Sbjct: 464 LCVHCLTESVRTQF-NNSILVNLVWEAAKCLTDFEFEGKMGEIAQISPEAASWIRNIQHS 522
Query: 600 KWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQK 659
+W + F ++ L N+SES N + A PII M E IR LM +
Sbjct: 523 QWATYCFE-GTRFGHLTANVSESLNSWVQDASGLPIIQMLESIRRQLMTLFNERRETSMQ 581
Query: 660 WQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWE 719
W ++P + + + I QF+VM + + +IVDI R+C C WE
Sbjct: 582 WSGMLVPSAERHVLEAIE-ECRLYPVHKANEAQFEVMTSEGK--WIVDIRCRTCYCRGWE 638
Query: 720 IVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMW----PEVEAP 775
+ G+PC H V+AL +Q F + Y++ + Y Y+ I P+ W P E+
Sbjct: 639 LYGLPCSHAVAALLACRQNVYRFTESYFTVANYRRTYAETIHPVPDKTEWKTTEPAGESG 698
Query: 776 E------LLPPNYKNGPGRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRCKSD 829
E + PP P RP+K R G RQ RC++C+Q GH + C +
Sbjct: 699 EDGEEIVIRPPRDLRQPPRPKKRR-----SQGEDRGRQKRVVRCSRCNQAGHFRTTCTAP 753
Query: 830 V 830
+
Sbjct: 754 I 754
>At1g33460 mutator transposase MUDRA, putative
Length = 826
Score = 244 bits (624), Expect = 1e-64
Identities = 186/747 (24%), Positives = 334/747 (43%), Gaps = 42/747 (5%)
Query: 115 DLDVGYDEEDVMVSTDDDSGKFVGFDDSEDESTTALEDGFEDVEVEAPANGNNRVTVNNK 174
D G + +DV + D +S G ++ ED+ A DG ++E + + + + +
Sbjct: 43 DEATGVEGDDVGIDGDRESD---GEENREDDGDIA-SDGDINLEDDGDRDESQKKRTREE 98
Query: 175 TLRIKKCGSKTPKKKSPKKNNSGRMNVIAPVSLLKNGKGKYVVDEEIDGDY-----LSEE 229
+ ++ K++ KK+ P L + ++ ++E + Y E
Sbjct: 99 KRKEEEAEEGQKKRRKQKKDE--------PEEDLAE-RFEFEIEEAVAMWYDELKIRRNE 149
Query: 230 LGSSDPDDSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVL 289
+ SD ++ D I D R +L D + IG F T EFKE ++ + + + K
Sbjct: 150 IPESD-NEEEDHVITRD--RKIRLASDDRLAIGRTFFTGFEFKEVVLHYAMKHRINAKQN 206
Query: 290 KNESYRVRVEC--RDQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKK 347
+ E ++ C R +C + V S +R+ + +KT H+C + K R + +
Sbjct: 207 RWEKDKISFRCAQRKECEWYVYASYSHERQLWVLKTKCLDHSCTSNGKCKLLKRRVIGRL 266
Query: 348 AVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAA 407
+ K+++ ++ + + + T+ + R +A ++ + +Q+A +R Y A
Sbjct: 267 FMDKLRLQPNFMPLDIQRHIKEQWKLVSTIGQVQDGRLLALKWLKEEYAQQFAHLRGYVA 326
Query: 408 ELKRVCRDNNVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYG 467
E+ + + ++ + + F Y CF K F CRP +G+DG LK
Sbjct: 327 EILSTNKGSTAIVDTIRDANENDV-FNRIYVCFGAMKNAFYF-CRPLIGIDGTFLKHAVK 384
Query: 468 GQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIG--EDKRYVFISDQQKGL 525
G LL A+ D N+Q +P+A+ V+ E E W WF L D+ + YV ISD+ KG+
Sbjct: 385 GCLLTAIAHDANNQIYPVAWATVQFENAENWLWFLNQLKHDLELKDGSGYVVISDRCKGI 444
Query: 526 LSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKI 585
+S + +HR C++H+ N KK+ G ++ + + + + +NE++
Sbjct: 445 ISAVKNALPNAEHRPCVKHIVENLKKRHGSLDLLKKFVWNLAWSYSDTQYKANLNEMRAY 504
Query: 586 DLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHY 645
+ +E +M EP WC+ F S C+ + NN +ESFN TI+ AR + ++ M E IR
Sbjct: 505 IMSLYEDVMKEEPNTWCRSWFRIGSYCEDVDNNATESFNATIVKARAKALVPMMETIRRQ 564
Query: 646 LMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFI 705
M R++ K+ KW+ + + L +E LA + T G E+F+V N
Sbjct: 565 AMARISKRKAKIGKWKKKISEYVSEILKEEWELAVKCEVT-KGTHEKFEVWFDGNSNAVS 623
Query: 706 VDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPING 765
+ + CSC W++ GIPC H +A+ + E FV + + Y P+ G
Sbjct: 624 LKTEEWDCSCCKWQVTGIPCEHAYAAINDVGKDVEDFVIPMFYTIAWREQYDTGPDPVRG 683
Query: 766 MDMWPE----VEAP--ELLPPNYKNGPGRPRKLRIREFDENGS-------RMRRQGVAYR 812
WP + AP + +PP K G + RI+ +E+ ++ ++GV
Sbjct: 684 QMYWPTGLGLITAPLQDPVPPGRKKGE-KKNYHRIKGPNESPKKKGPQRLKVGKKGVVMH 742
Query: 813 CTKCDQFGHNQRRCKSDVQNPEAAKRK 839
C C + GHN CK + + K+K
Sbjct: 743 CKSCGEAGHNAAGCKKFPKEKKGRKKK 769
>At3g30170 unknown protein
Length = 995
Score = 233 bits (593), Expect = 4e-61
Identities = 167/637 (26%), Positives = 289/637 (45%), Gaps = 42/637 (6%)
Query: 218 DEEIDGDYLSEELGSSDPDDSNDDTIKYDQFRMPQLNKDFKFKIGMEFNTLVEFKEAIIE 277
D+ + D + + S D D+S + + N++ ++ +NT +FK A++
Sbjct: 188 DDVWEEDKIPDPFSSDDEDESRTREVHGHR---DAANEEVLLELKKTYNTPDDFKLAVLR 244
Query: 278 WNVLNGYQIKVLKNESYRVRVEC----RD--QCGYKVLCSKVGDRRTFQIKTLEGPHTCA 331
+++ Y IK+ ++E+ V +C +D QC +KV CS QI+T H C
Sbjct: 245 YSLKTRYDIKLFRSEARIVAAKCSYVDKDGVQCPWKVYCSYEKTMHKMQIRTYVNNHICV 304
Query: 332 PVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSII 391
+K +A +++V K++ E+ E+ Y + +T D+ +A+
Sbjct: 305 RSGHSKMLKRSSIAYLFEERLRVNPKLTKYEMAAEILREYNLEVTPDQCAKAKTKVVKAR 364
Query: 392 EGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISAC 451
+ +A + Y AE+ + +I +V G K+
Sbjct: 365 NASHEAHFARVWDYQAEVIKQNPWTEFEIETTAGAVI-------------GAKQ------ 405
Query: 452 RPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIG- 510
RPF+G+DG LK G LL AVGRD +++ P+A+ VVE E + W WF + L +G
Sbjct: 406 RPFIGLDGAFLKWDIKGHLLAAVGRDGDNKIVPIAWAVVEIENDDNWDWFLRHLSASLGL 465
Query: 511 -EDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKA 569
+ +SD+Q GL+ + +HR C +H+ N+K+ +++ L ++
Sbjct: 466 CQMANIAILSDKQSGLVKAIHTILPHAEHRQCSKHIMDNWKRD-SHDLELQRLFWKIARS 524
Query: 570 TYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILS 629
+ + M EL++ + A+E L P W + F + C+ +NN+SESFN TI
Sbjct: 525 YTVEEFNNHMMELQQYNRQAYESLQLTSPVTWSRAFFRIGTCCNDNLNNLSESFNRTIRQ 584
Query: 630 ARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGI 689
AR +PI+ M E IR M R +K Q + ++K I + +
Sbjct: 585 ARRKPILDMLEDIRRQCMVRSAKRFLIAEKLQTRFTKRAHEEIEKMIVGLRQCERYMAR- 643
Query: 690 GEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSR 749
E ++ N + VD+ +++C C ++VGIPC H + +K++ E +V DYY++
Sbjct: 644 -ENLHEIYV-NNVSYFVDMEQKTCDCRKCQMVGIPCIHATCVIIGKKEKVEDYVSDYYTK 701
Query: 750 SKYALCYSFAISPINGMDMWPEVEAPELLPPNYKNG-PGRPRKLRIRE------FDENGS 802
++ Y I P+ GM +W +LPP ++ G GRP R+ N +
Sbjct: 702 VRWRETYLKGIRPVQGMPLWCRTNRLPVLPPPWRRGNAGRPSNYARRKGRNEAAAPSNPN 761
Query: 803 RMRRQGVAYRCTKCDQFGHNQRRCK-SDVQNPEAAKR 838
+M R+ C+ C Q GHN++RC V NP R
Sbjct: 762 KMSREKRIMTCSNCHQEGHNKQRCNIPTVLNPTKRPR 798
>At5g16505 mutator-like transposase-like protein (MQK4.25)
Length = 597
Score = 228 bits (581), Expect = 1e-59
Identities = 148/587 (25%), Positives = 271/587 (45%), Gaps = 33/587 (5%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVEC-RDQCGYKVLCSKVGDRRTF 319
+G EF + + + + + + ++++K++ R +C ++ C +++ +K +TF
Sbjct: 27 VGKEFPDVETCRRTLKDLAIALHFDLRIVKSDRSRFIAKCSKEGCPWRIHAAKCPGVQTF 86
Query: 320 QIKTLEGPHTCAPV--LENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITM 377
++TL HTC V L ++ A+ WVA+ +++ + +E+ ++ +GV ++
Sbjct: 87 TVRTLNSEHTCEGVRDLHHQQASVGWVARSVEARIRDNPQYKPKEILQDIRDEHGVAVSY 146
Query: 378 DRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFY 437
+AWR ++ + + + G ++ Y + Y ++K V + ++ GP Q F
Sbjct: 147 MQAWRGKERSMAALHGTYEEGYRFLPAYCEQIKLVNPGSFASVSALGPENCFQRLF---- 202
Query: 438 FCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKET 497
+ C GF S+CRP + +D HLK +Y G +L A D +D FPLA +V+ E E
Sbjct: 203 IAYRACISGFFSSCRPLLELDRAHLKGKYLGAILCAAAVDADDGLFPLAIAIVDNESDEN 262
Query: 498 WKWFTQLLMEDIGED----KRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKF 553
W WF L + +G + + +S++Q ++ + F + H CLR++ NF+ F
Sbjct: 263 WSWFLSELRKLLGMNTDSMPKLTILSERQSAVVEAVETHFPTAFHGFCLRYVSENFRDTF 322
Query: 554 GGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAF--TFYSK 611
+++ ++ A A + K NE+ +I +W P W F Y
Sbjct: 323 -KNTKLVNIFWSAVYALTPAEFETKSNEMIEISQDVVQWFELYLPHLWAVAYFQGVRYGH 381
Query: 612 CDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKR 671
+ I+E L + PII M E IRH + W ++P +R
Sbjct: 382 FGL---GITEVLYNWALECHELPIIQMMEHIRHQISSWFDNRRELSMGWNSILVPSAERR 438
Query: 672 LDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSA 731
+ + + A ++ + E F+++ T + IVDI R CSC W+I G+PC H +A
Sbjct: 439 ITEAVADARCYQVLRANEVE-FEIVST--ERTNIVDIRTRDCSCRRWQIYGLPCAHAAAA 495
Query: 732 LGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPE--------VEAPELLPPNYK 783
L + +F + ++ S Y YS I I +W + VE+ + PP +
Sbjct: 496 LISCGRNVHLFAEPCFTVSSYQQTYSQMIGEIPDRSLWKDEGEEAGGGVESLRIRPPKTR 555
Query: 784 NGPGRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRCKSDV 830
PGRP+K +R EN R +R +C +C GH+Q++C +
Sbjct: 556 RPPGRPKKKVVRV--ENLKRPKR---IVQCGRCHLLGHSQKKCTQPI 597
>At1g06740 mudrA-like protein
Length = 726
Score = 217 bits (552), Expect = 2e-56
Identities = 148/584 (25%), Positives = 272/584 (46%), Gaps = 27/584 (4%)
Query: 256 DFKFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVG 314
D + +GMEF+ + AI + ++++ +K++ R +C + C +++ C+KV
Sbjct: 153 DHEMVVGMEFSDAYACRRAIKNAAISLRFEMRTIKSDKTRFTAKCNSKGCPWRIHCAKVS 212
Query: 315 DRRTFQIKTLEGPHTCAPV--LENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYG 372
+ TF I+T+ G HTC + L ++ A+ +WVA K++ +E+ E+ +G
Sbjct: 213 NAPTFTIRTIHGSHTCGGISHLGHQQASVQWVADVVAEKLKENPHFKPKEILEEIYRVHG 272
Query: 373 VGITMDRAWRARK-----IAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSV 427
+ ++ +AWR ++ + S + G +++Y ++ +Y E++R +V + P
Sbjct: 273 ISLSYKQAWRGKERIMATLRGSTLRGSFEEEYRLLPQYCDEIRR-SNPGSVAVVHVNP-- 329
Query: 428 TIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAF 487
I F + F GF++ACRP + +D LK++Y G LL+A G D + FPLAF
Sbjct: 330 -IDGCFQHLFISFQASISGFLNACRPLIALDSTVLKSKYPGTLLLATGFDGDGAVFPLAF 388
Query: 488 GVVETECKETWKWFTQLLMEDIGED-KRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLY 546
+V E + W F L + + E+ + +S ++ ++ + F + H CL +L
Sbjct: 389 AIVNEENDDNWHRFLSELRKILDENMPKLTILSSGERPVVDGVEANFPAAFHGFCLHYLT 448
Query: 547 ANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAF 606
F+++F S + DL A + K+N++++I A W+ P +W F
Sbjct: 449 ERFQREF-QSSVLVDLFWEAAHCLTVLEFKSKINKIEQISPEASLWIQNKSPARWASSYF 507
Query: 607 TFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMP 666
+ N I+ES + + PII E I +L+ + W + ++P
Sbjct: 508 EGTRFGQLTANVITESLSNWVEDTSGLPIIQTMECIHRHLINMLKERRETSLHWSNVLVP 567
Query: 667 MPRKRLDKEITLA-AHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPC 725
K++ I + AH + +F+VM +V+I SC C W++ G+PC
Sbjct: 568 SAEKQMLAAIEQSRAH--RVYRANEAEFEVMTC--EGNVVVNIENCSCLCGRWQVYGLPC 623
Query: 726 RHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEA---PELLPPNY 782
H V AL ++ + + ++ Y Y+ + PI+ W E ++ E +
Sbjct: 624 SHAVGALLSCEEDVYRYTESCFTVENYRRAYAETLEPISDKVQWKENDSERDSENVIKTP 683
Query: 783 KNGPGRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRC 826
K G PRK R+R E+ R+RR C +C+Q GH + C
Sbjct: 684 KAMKGAPRKRRVRA--EDRDRVRR---VVHCGRCNQTGHFRTTC 722
>At3g05850 unknown protein
Length = 777
Score = 216 bits (551), Expect = 3e-56
Identities = 150/583 (25%), Positives = 269/583 (45%), Gaps = 33/583 (5%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTF 319
+G F + EF+EA+ ++ + N + + KN+S+RV V+C+ + C +++ S++ +
Sbjct: 206 VGQRFKNVGEFREALRKYAIANQFGFRYKKNDSHRVTVKCKAEGCPWRIHASRLSTTQLI 265
Query: 320 QIKTLEGPHTC--APVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAVNYGVGITM 377
IK + HTC A + + WVA K++V +++ +++ YG+ +
Sbjct: 266 CIKKMNPTHTCEGAGGINGLQTSRSWVASIIKEKLKVFPNYKPKDIVSDIKEEYGIQLNY 325
Query: 378 DRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFY 437
+AWR ++IA+ ++G Y + + ++ + S F +
Sbjct: 326 FQAWRGKEIAREQLQGSYKDGYKQLPLFCEKIMETNPGSLATFTTKEDS-----SFHRVF 380
Query: 438 FCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKET 497
F GF+ ACRP V +D LK++Y G LL A D +D+ FPLAF VV+ E +
Sbjct: 381 VSFHASVHGFLEACRPLVFLDSMQLKSKYQGTLLAATSVDGDDEVFPLAFAVVDAETDDN 440
Query: 498 WKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGG-- 555
W+WF L + F++D+QK L ++F+ H CLR+L K G
Sbjct: 441 WEWFLLQLRSLLSTPCYITFVADRQKNLQESIPKVFEKSFHAYCLRYLTDELIKDLKGPF 500
Query: 556 GSQIRDLMM----GATKATYNQGWLEKMNELKKIDLGAWEWLM-AVEPKKWCKHAFTFYS 610
+I+ L++ A A + + +K + A++W++ +P W +A+ +
Sbjct: 501 SHEIKRLIVDDFYSAAYAPRADSFERHVENIKGLSPEAYDWIVQKSQPDHWA-NAYFRGA 559
Query: 611 KCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRK 670
+ + + ++ E F A D PI M + IR +M + + N+ P
Sbjct: 560 RYNHMTSHSGEPFFSWASDANDLPITQMVDVIRGKIMGLIHVRRISANEANGNLTPSMEV 619
Query: 671 RLDKEITLAAHWKSTWSGIGEQFQVM-HTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVV 729
+L+KE A S FQV TY +V++A+ CSC W++ G+PC H V
Sbjct: 620 KLEKESLRAQTVHVAPSADNNLFQVRGETYE----LVNMAECDCSCKGWQLTGLPCHHAV 675
Query: 730 SALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGM--DMWPEV---EAPELLPPNYKN 784
+ + + P + Y++ + Y Y+ +I+P+ + +M E A + PP +
Sbjct: 676 AVINYYGRNPYDYCSKYFTVAYYRSTYAQSINPVPLLEGEMCRESSGGSAVTVTPPPTRR 735
Query: 785 GPGRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRCK 827
PGRP K + + M+RQ +C++C GHN+ CK
Sbjct: 736 PPGRPPKKKTPAEEV----MKRQ---LQCSRCKGLGHNKSTCK 771
>At1g12720 hypothetical protein
Length = 585
Score = 197 bits (500), Expect = 3e-50
Identities = 133/489 (27%), Positives = 221/489 (44%), Gaps = 40/489 (8%)
Query: 351 KMQVTKKMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELK 410
+++V KM+ E+ E+ Y + +T D+ +A+ D ++ I Y AE+
Sbjct: 14 RLRVNPKMTKYEMVAEIKREYKLEVTPDQCAKAKTKVLKARNASHDTHFSRIWDYQAEVL 73
Query: 411 RVCRDNNVKINVAGPS-VTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQ 469
+++ I + + + RF Y CF+ K + CRP +G+DG LK G
Sbjct: 74 NRNPNSDFDIETTARTFIGSKQRFFRLYICFNSQKVSWKQHCRPVIGIDGAFLKWDIKGH 133
Query: 470 LLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIG--EDKRYVFISDQQKGLLS 527
LL AVGRD +++ PLA+ VVE E + W WF + L E +G E ISD+Q GL+
Sbjct: 134 LLAAVGRDGDNRIVPLAWAVVEIENDDNWDWFLKKLSESLGLCEMVNLALISDKQSGLVK 193
Query: 528 VFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDL 587
+ +HR C +H+ N+K+ +++ L +++ + + M LK +
Sbjct: 194 AIHNVLPQAEHRQCSKHIMDNWKRD-SHDMELQRLFWKISRSYTIEEFNTHMANLKSYNP 252
Query: 588 GAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLM 647
A+ L P W TI AR +P++ M E IR M
Sbjct: 253 QAYASLQLTSPMTW------------------------TIRQARRKPLLDMLEDIRRQCM 288
Query: 648 RRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTY-NRQQFIV 706
R ++ + P ++K I +A + + + + Y N + V
Sbjct: 289 VRTAKRFIIAERLKSRFTPRAHAEIEKMIAGSAGCERHLA----RNNLHEIYVNDVGYFV 344
Query: 707 DIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISPINGM 766
D+ K++C C WE+VGIPC H + RK++ E +V DYY++ ++ Y I P+ GM
Sbjct: 345 DMDKKTCGCRKWEMVGIPCVHTPCVIIGRKEKVEDYVSDYYTKVRWRETYRDGIRPVQGM 404
Query: 767 DMWPEVEAPELLPPNYKNG-PGRPRKLRIRE------FDENGSRMRRQGVAYRCTKCDQF 819
+WP + +LPP ++ G PGR ++ N ++M R C+ C Q
Sbjct: 405 PLWPRMSRLPVLPPPWRRGNPGRQSNYARKKGRYETASSSNKNKMSRANRIMTCSNCKQE 464
Query: 820 GHNQRRCKS 828
GHN+ CK+
Sbjct: 465 GHNKSSCKN 473
>At3g47280 putative protein
Length = 739
Score = 195 bits (495), Expect = 1e-49
Identities = 141/580 (24%), Positives = 270/580 (46%), Gaps = 31/580 (5%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTF 319
IG F E + ++ V + ++ +K++ R + C D+ C +++ ++ G ++
Sbjct: 173 IGKIFKDKDEMVFTLRKFAVKHSFKFHTVKSDLTRYVLHCIDENCSWRLRATRAGGSESY 232
Query: 320 QIKTLEGPHTCAPVLENKS---ANSRWVAKKAVPKMQVTK-KMSVQEVFNEMAVNYGVGI 375
I+ H+C L N S A++R + + + K + +++ ++GVGI
Sbjct: 233 VIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGI 292
Query: 376 TMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGS 435
+AWR +K A + G D + ++ R+ RV N I + ++
Sbjct: 293 NYSKAWRVQKHAAELARGLPDDSFEVLPRW---FHRVQVTNPGSITFFKKDSANKFKYA- 348
Query: 436 FYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECK 495
+ F +G+ R + +DG HL +++ G LL A +D N +P+AF +V++E
Sbjct: 349 -FLAFGASIRGY-KLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSEND 406
Query: 496 ETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGG 555
+W WF + L+ I ++ VF+SD+ + S + H LC HL N + F G
Sbjct: 407 ASWDWFLKCLLNIIPDENDLVFVSDRAASIASRLSGNYPLAHHGLCTFHLQKNLETHFRG 466
Query: 556 GSQIRDLMMGA---TKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKC 612
S I + TK ++ + E N KK+ ++L V+ +KW + A++ ++
Sbjct: 467 SSLIPVYYAASRIYTKTEFDSLFWEITNSDKKLA----QYLCEVDVRKWSR-AYSPSNRY 521
Query: 613 DVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRL 672
+++ +N++ES N + R+ P++ + E IR + R + + V K++
Sbjct: 522 NIMTSNLAESVNALLKQNREYPVVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 581
Query: 673 DKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSAL 732
+ W E+F+V + + +V++ KR+C+C ++I PC H +++
Sbjct: 582 KASYDTSTRWLEVCQVNQEEFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 639
Query: 733 GKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPE------LLPPNYKNGP 786
MFVD+++S ++ YS +I P M+ W E PE LPP+ +
Sbjct: 640 KHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYW---EIPETISEVICLPPSTRVPT 696
Query: 787 GRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRC 826
G+ +K RI E+G R + + ++C++C Q GHN+ C
Sbjct: 697 GKRKKKRIPSVWEHG-RSQPKPKLHKCSRCGQSGHNKSTC 735
>At2g04950 putative transposase of transposon FARE2.9 (cds1)
Length = 616
Score = 190 bits (482), Expect = 3e-48
Identities = 137/561 (24%), Positives = 261/561 (46%), Gaps = 31/561 (5%)
Query: 280 VLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKS 338
V + ++ +K++ + + C D+ C +++ ++ G ++ I+ H+C L N S
Sbjct: 69 VKHSFEFHTVKSDLTKYVLHCIDENCSWRLRATRAGGSESYVIRKYVSHHSCDSSLRNVS 128
Query: 339 ---ANSRWVAKKAVPKMQVTK-KMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGD 394
A++R + + + K + +++ ++GVGI +AWR ++ A + G
Sbjct: 129 HRQASARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGINYSKAWRVQEHAAELARGL 188
Query: 395 ADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPF 454
D + ++ R+ RV N I + ++ + F +G+ +
Sbjct: 189 PDDSFEVLPRW---FHRVQVTNPGSITFFKKDSANKFKYA--FLAFGASIRGY-KLMKKV 242
Query: 455 VGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKR 514
+ +DG HL +++ G LL A +D N +P+AF +V++E +W WF + L+ I ++
Sbjct: 243 ISIDGAHLISKFKGTLLGASAQDGNFNLYPIAFAIVDSENDASWDWFLKCLLNIIPDEND 302
Query: 515 YVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGGGSQIRDLMMGA---TKATY 571
VF+SD+ + S + + LC HL N + F G S I + TK +
Sbjct: 303 LVFVSDRAASIASGLSGNYPLAHNGLCTFHLQKNLETHFRGSSLIPVYYAASRVYTKTEF 362
Query: 572 NQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSAR 631
+ + E N KK+ WE V+ +KW + A++ ++ +++ +N++ES N + R
Sbjct: 363 DSLFWEITNSDKKLAQYLWE----VDVRKWSR-AYSPSNRYNIMTSNLAESVNALLKQNR 417
Query: 632 DQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGE 691
+ PI+ + E IR + R + + V K++ + W E
Sbjct: 418 EYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKMKASYDTSTRWLEVCQVNQE 477
Query: 692 QFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSK 751
+F+V + + +V++ KR+C+C ++I PC H +++ MFVD+++S K
Sbjct: 478 EFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASAKHINLNENMFVDEFHSTYK 535
Query: 752 YALCYSFAISPINGMDMWPEVEAPE------LLPPNYKNGPGRPRKLRIREFDENGSRMR 805
+ YS +I P M+ W E PE LPP+ + GR +K RI E+G R +
Sbjct: 536 WRQAYSKSIHPNGDMEYW---EIPETISEVICLPPSTRVPSGRRKKKRIPSVWEHG-RSQ 591
Query: 806 RQGVAYRCTKCDQFGHNQRRC 826
+ ++C++C Q GHN C
Sbjct: 592 PKPKLHKCSRCGQSGHNNSTC 612
>At2g15210 putaive transposase of transposon FARE2.7 (cds1)
Length = 580
Score = 184 bits (468), Expect = 1e-46
Identities = 140/580 (24%), Positives = 260/580 (44%), Gaps = 67/580 (11%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTF 319
IG F E + + V + ++ +K++ R + C D+ C +++ ++ G ++
Sbjct: 50 IGKNFKDKDEMVFTLRMFAVKHNFEFHTVKSDLTRYVLHCIDENCSWRLRATRAGGSESY 109
Query: 320 QIKTLEGPHTCAPVLENKS---ANSRWVAKKAVPKMQVTK-KMSVQEVFNEMAVNYGVGI 375
I+ H+C L N S A++R + + + K + +++ ++GVGI
Sbjct: 110 VIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGI 169
Query: 376 TMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGS 435
+AWR ++ +AAEL R D++
Sbjct: 170 NYSKAWR-------------------VQEHAAELARGLPDDS------------------ 192
Query: 436 FYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECK 495
F+ +G+ R + +DG HL +++ G LL A +D N +P+AF +V++E
Sbjct: 193 ----FEVLPRGY-KLMRKVISIDGAHLTSKFKGTLLGASAQDRNFNLYPIAFAIVDSEND 247
Query: 496 ETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGG 555
+W WF + L+ I ++ VF+S++ + S + H LC HL N + F G
Sbjct: 248 ASWDWFLKCLLNIIPDENDLVFVSERAASIASGLSGNYPLAHHGLCTFHLQKNLETHFRG 307
Query: 556 GSQIRDLMMGA---TKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKC 612
S I + TK ++ + E N KK+ WE V +KW + A++ ++
Sbjct: 308 SSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWE----VHVRKWSR-AYSPSNRY 362
Query: 613 DVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRL 672
+++ +N++ES N + R+ PI+ + E IR + R + + V K++
Sbjct: 363 NIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 422
Query: 673 DKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSAL 732
+ W E+F+V + + +V++ KR+C+C ++I PC H +++
Sbjct: 423 KASYDTSTRWLEVCQVNQEEFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 480
Query: 733 GKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPE------LLPPNYKNGP 786
MFVD+++S ++ YS +I P M+ W E PE LPP+ +
Sbjct: 481 NHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYW---EIPETISEVICLPPSTRVPS 537
Query: 787 GRPRKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRC 826
GR +K RI E+G R + + ++C++C Q GHN+ C
Sbjct: 538 GRRKKKRIPSVWEHG-RSQPKPKLHKCSRCGQSGHNKSTC 576
>At1g34620 Mutator-like protein
Length = 1034
Score = 181 bits (458), Expect = 2e-45
Identities = 146/563 (25%), Positives = 264/563 (45%), Gaps = 20/563 (3%)
Query: 258 KFKIGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDR 316
K +G F + + K I + + + ++ + ++C + C ++V SKV
Sbjct: 364 KLFVGRVFKSKSDCKIKIAIHAINRKFHFRTARSTPKFMVLKCISKTCPWRVYASKVDTS 423
Query: 317 RTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTKKMSVQEVFNEMAV------N 370
+FQ++ HTC ++ + R + + ++ ++ + ++ N + +
Sbjct: 424 DSFQVRQANQRHTCT--IDQRRRYHRLATTQVIGELMQSRFLGIKRGPNAAVIRKFLLDD 481
Query: 371 YGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQ 430
Y V I+ +AWRAR++A G YA+I YA L++ ++ T
Sbjct: 482 YHVSISYWKAWRAREVAMEKSLGSMAGSYALIPAYAGLLQQA-NPGSLCFTEYDDDPTGP 540
Query: 431 LRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVV 490
RF + F KG+ + R + VDG +K RYGG L+ A +D N Q FPLAFG+V
Sbjct: 541 RRFKYQFIAFAASIKGY-AFMRKVIVVDGTSMKGRYGGCLISACCQDGNFQIFPLAFGIV 599
Query: 491 ETECKETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFK 550
+E ++WF Q L ++ +FISD+ + + + +++ +H C HL+ N +
Sbjct: 600 NSENDSAYEWFFQRLSIIAPDNPDLMFISDRHESIYTGLSKVYTQANHAACTVHLWRNIR 659
Query: 551 KKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYS 610
+ S R LM A +A + + +++K++ G +L+ + +W + +
Sbjct: 660 HLYKPKSLCR-LMSEAAQAFHVTDFNRIFLKIQKLNPGCAAYLVDLGFSEWTR-VHSKGR 717
Query: 611 KCDVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRK 670
+ +++ +NI ES+N I AR+ P+I M E+IR LM T + + + P ++
Sbjct: 718 RFNIMDSNICESWNNVIREAREYPLICMLEYIRTTLMDWFATRRAQAEDCPTTLAPRVQE 777
Query: 671 RLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVS 730
R+++ +A S +FQV + FIV + + +CSC ++ +GIPC H ++
Sbjct: 778 RVEENYQ-SAMSMSVKPICNFEFQVQER-TGECFIVKLDESTCSCLEFQGLGIPCAHAIA 835
Query: 731 ALGKRKQRPEMFVDDYYSRSKYALCYSFAISPIN--GMDMWPEVEA---PELLPPNYKNG 785
A + + + Y L Y I PI+ G ++ PE+ A EL PP +
Sbjct: 836 AAARIGVPTDSLAANGYFNELVKLSYEEKIYPIHSVGGEVAPEIAAGTTGELQPPFVRRP 895
Query: 786 PGRPRKLRIREFDENGSRMRRQG 808
PGRPRK+RI E +R +G
Sbjct: 896 PGRPRKIRILSRGEFKARQGPRG 918
>At4g03980 putative transposon protein
Length = 580
Score = 179 bits (455), Expect = 4e-45
Identities = 138/577 (23%), Positives = 260/577 (44%), Gaps = 61/577 (10%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTF 319
IG +F E + + V + ++ +K++ R + C D+ C +++ +K G ++
Sbjct: 50 IGKKFKDKDEMVFTLRMFAVKHSFEFHTVKSDLTRYVLHCIDENCSWRLRATKAGGSESY 109
Query: 320 QIKTLEGPHTCAPVLENKS---ANSRWVAKKAVPKMQVTK-KMSVQEVFNEMAVNYGVGI 375
I+ H+C L N S A +R + + ++ K + +++ ++GVGI
Sbjct: 110 VIRKYVSHHSCDSSLRNVSHRQAFARTLGRLISNHLEGGKLPLGPKQLIEIFRKDHGVGI 169
Query: 376 TMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGS 435
+AWR ++ + EL R D++
Sbjct: 170 NYSKAWR-------------------VQEHVVELARGLPDDS------------------ 192
Query: 436 FYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECK 495
F+ +G+ R + +DG HL +++ G LL A +D N +P AF +V+TE
Sbjct: 193 ----FEVLPRGY-KLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPSAFAIVDTEND 247
Query: 496 ETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGG 555
+W WF + L+ I ++ VF+SD+ + S E + H LC HL N + F G
Sbjct: 248 ASWDWFLKCLLNIIPDENDLVFVSDRAASIASGLSENYPLAHHGLCTFHLQKNLETHFRG 307
Query: 556 GSQIRDLMMGA---TKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKC 612
S I + TK ++ + + N KK+ WE V+ +KW + A++ ++
Sbjct: 308 SSLIPVNYAASRVYTKTEFDSLFWKITNSDKKLAQYLWE----VDVRKWSR-AYSPSNRY 362
Query: 613 DVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRL 672
+++ +N++ES N + R+ PI+ + E IR + R + + V K++
Sbjct: 363 NIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 422
Query: 673 DKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSAL 732
+ W E+F+V + + +V++ KR+C+C ++I P H +++
Sbjct: 423 KASYDTSTRWLEVCQVNQEEFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPYAHGIASA 480
Query: 733 GKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMW--PEVEAPEL-LPPNYKNGPGRP 789
MFVD+++S ++ YS +I P M+ W P+ + + LPP+ + GR
Sbjct: 481 KHINLNKNMFVDEFHSTYRWRQAYSESIHPNGDMEYWEIPDTVSEVICLPPSTRVPSGRR 540
Query: 790 RKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRC 826
+K RI E+G R + + ++C++C Q GHN+ C
Sbjct: 541 KKKRIPSVWEHG-RSQPKPKLHKCSRCGQSGHNKSTC 576
>At2g06450 putative transposase of FARE2.11
Length = 580
Score = 179 bits (455), Expect = 4e-45
Identities = 137/577 (23%), Positives = 258/577 (43%), Gaps = 61/577 (10%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTF 319
IG F E + + V + ++ +K++ R + C D+ C +++ ++ G ++
Sbjct: 50 IGKIFKDKDEMVFTLRMFAVKHNFKFHTVKSDLTRYVLHCIDENCSWRLRATRAGGSESY 109
Query: 320 QIKTLEGPHTCAPVLENKS---ANSRWVAKKAVPKMQVTK-KMSVQEVFNEMAVNYGVGI 375
I+ H+C L N S A++R + + + K + +++ ++GVGI
Sbjct: 110 VIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGI 169
Query: 376 TMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGS 435
+AWR ++ +AAEL R D++
Sbjct: 170 NYSKAWR-------------------VQEHAAELARGLPDDS------------------ 192
Query: 436 FYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECK 495
F+ +G+ R + +DG HL +++ G LL A +D N +P+AF +V++E
Sbjct: 193 ----FEVLPRGY-KLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSEND 247
Query: 496 ETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGG 555
+W WF + L+ I ++ VF+SD+ + S + H LC HL N + F G
Sbjct: 248 ASWDWFLKCLLNIIPDENDLVFVSDRAAFIASGLSGNYPLAHHGLCTFHLQKNLETHFRG 307
Query: 556 GSQIRDLMMGA---TKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKC 612
S I + TK ++ + E N KK+ WE V+ +KW + ++ ++
Sbjct: 308 SSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWE----VDVRKWSR-TYSPSNRY 362
Query: 613 DVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRL 672
+++ +N++ES N + R+ PI+ + E IR + R + + V K++
Sbjct: 363 NIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 422
Query: 673 DKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSAL 732
+ W E+F+V + + +V++ KR+C+C ++I PC H +++
Sbjct: 423 KASYDTSTRWLEVCQVNQEEFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 480
Query: 733 GKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPE---LLPPNYKNGPGRP 789
MFVD+++S ++ YS +I P M+ W +E LPP+ + GR
Sbjct: 481 KHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYWEILETISEVICLPPSTRVPSGRR 540
Query: 790 RKLRIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRC 826
K RI E+G R + + ++C++C + GHN+ C
Sbjct: 541 EKKRIPSVWEHG-RSQPKPKLHKCSRCGRSGHNKSTC 576
>At2g29230 Mutator-like transposase
Length = 915
Score = 179 bits (454), Expect = 6e-45
Identities = 138/552 (25%), Positives = 249/552 (45%), Gaps = 47/552 (8%)
Query: 302 DQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTK----- 356
+ C ++V K+ D +QI++ HTC +E +S R + + + +K
Sbjct: 388 ETCPWRVYIVKLEDSDNYQIRSANLEHTCT--VEERSNYHRAATTRVIGSIIQSKYAGNS 445
Query: 357 ----KMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRV 412
+ +Q + + +Y V I+ +AW++R+IA +G A + ++ Y L+
Sbjct: 446 RGPRAIDLQRI---LLTDYSVRISYWKAWKSREIAMDSAQGSAANSFTLLPAYLHVLREA 502
Query: 413 CRDN--NVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQL 470
+ ++K V G RF + F +GF S + + +DG HLK +YGG L
Sbjct: 503 NPGSIVDLKTEVDGKG---NHRFKYMFLAFAASIQGF-SCMKRVIVIDGAHLKGKYGGCL 558
Query: 471 LIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFD 530
L A G+D N Q FP+AFGVV++E + W+WF ++L I + F+SD+ + +
Sbjct: 559 LTASGQDANFQVFPIAFGVVDSENDDAWEWFFRVLSTAIPDGDNLTFVSDRHSSIYTGLR 618
Query: 531 EMFDSIDHRLCLRHLYAN----FKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKID 586
++ H C+ HL N +KKK + + A +A + NE+ ++D
Sbjct: 619 RVYPKAKHGACIVHLQRNIATSYKKK-----HLLFHVSRAARAFRICEFHTYFNEVIRLD 673
Query: 587 LGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYL 646
+L +V W + A+ + +V+ +N++ES N + AR+ PIIS+ E+IR L
Sbjct: 674 PACARYLESVGFCHWTR-AYFLGKRYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTL 732
Query: 647 M----RRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQ 702
+ R + T+ + + + +K + A H + +
Sbjct: 733 ISWFAMRREAARTEASPLPPKMREVVHRNFEKSVRFAVHRLDRYD------YEIREEGAS 786
Query: 703 QFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISP 762
+ V + +R+CSC ++++ +PC H ++A + + YS + + Y I P
Sbjct: 787 VYHVKLMERTCSCRAFDLLHLPCPHAIAAAVAEGVPIQGLMAPEYSVESWRMSYLGTIKP 846
Query: 763 ---INGMDMWPE-VEAPELLPPNYKNGPGRPRKLRIREFDENGSRMRRQGVAYRCTKCDQ 818
+ + PE + + L PP + GRP+K RI E R+ RC++C
Sbjct: 847 VPEVGDVFALPEPIASLRLFPPATRRPSGRPKKKRITSRGEFTCPQRQ---VTRCSRCTG 903
Query: 819 FGHNQRRCKSDV 830
GHN+ CK +
Sbjct: 904 AGHNRATCKMPI 915
>At2g15150 putative transposase of FARE2.2 (CDS1)
Length = 607
Score = 178 bits (452), Expect = 1e-44
Identities = 134/555 (24%), Positives = 252/555 (45%), Gaps = 30/555 (5%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECRDQ-CGYKVLCSKVGDRRTF 319
IG F E + + V + ++ +K++ R + C D+ C +++ ++ G ++
Sbjct: 50 IGKIFKDKDEMVFTLRMFAVKHNFEFHTVKSDLTRYVLHCIDENCSWRLRATRAGGSESY 109
Query: 320 QIKTLEGPHTCAPVLENKS---ANSRWVAKKAVPKMQVTK-KMSVQEVFNEMAVNYGVGI 375
I+ H+C L N S A++R + + + K + +++ ++GVGI
Sbjct: 110 AIRKYVSHHSCDSSLRNVSHRQASARTLGRLISNHFEGGKLPLRPKQLIEIFRKDHGVGI 169
Query: 376 TMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRFGS 435
+AWR ++ A + G D + ++ R+ RV N I + ++
Sbjct: 170 NYSKAWRVQEHAAELARGLPDDSFEVLPRW---FHRVQVTNPGSITFFKKDSANKFKYA- 225
Query: 436 FYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETECK 495
+ F +G+ R + +DG HL +++ G LL A +D N +P+AF +V++E
Sbjct: 226 -FLAFGASIRGY-KLMRKVISIDGAHLTSKFKGTLLGASAQDGNFNLYPIAFAIVDSEND 283
Query: 496 ETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKFGG 555
+W WF + L+ I ++ VF+S++ + S + H LC HL N + F G
Sbjct: 284 ASWDWFLKCLLNIIPDENDLVFVSERAASIASGLSGNYPLAHHGLCTFHLQKNLETHFRG 343
Query: 556 GSQIRDLMMGA---TKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKC 612
S I + TK ++ + E N KK+ WE V +KW + A++ ++
Sbjct: 344 SSLIPVYYAASRVYTKTEFDSLFWEITNSDKKLAQYLWE----VHVRKWSR-AYSPSNRY 398
Query: 613 DVLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRL 672
+++ +N++ES N + R+ PI+ + E IR + R + + V K++
Sbjct: 399 NIMTSNLAESVNALLKQNREYPIVCLFESIRSIMTRWFNERREESSQHPSAVTINVGKKM 458
Query: 673 DKEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSAL 732
+ W E+F+V + + +V++ KR+C+C ++I PC H +++
Sbjct: 459 KASYDTSTRWLEVCQVNQEEFEVKG--DTKTHLVNLDKRTCTCCMFDIDKFPCAHGIASA 516
Query: 733 GKRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPE------LLPPNYKNGP 786
MFVD+++S ++ YS +I P M+ W E PE LPP+ +
Sbjct: 517 NHINLNENMFVDEFHSTYRWRQAYSESIHPNGDMEYW---EIPETISEVICLPPSTRVPS 573
Query: 787 GRPRKLRIREFDENG 801
GR +K RI E+G
Sbjct: 574 GRRKKKRIPSVWEHG 588
>At2g12720 putative protein
Length = 819
Score = 172 bits (435), Expect = 9e-43
Identities = 133/576 (23%), Positives = 243/576 (42%), Gaps = 21/576 (3%)
Query: 261 IGMEFNTLVEFKEAIIEWNVLNGYQIKVLKNESYRVRVECR-DQCGYKVLCSKVGDRRTF 319
+G ++ + E + + + +G+ V + V EC D+C ++V + G+ F
Sbjct: 257 VGQQYRSKFELEYRLKLLAIRDGFDFDVPTSNKTTVSYECWVDRCLWRVRACRQGNDPNF 316
Query: 320 QIKTLEGPHTCAPVLENKSANSRWVAKKAVPKM------QVTKKMSVQEVFNEMAVNYGV 373
+ + HTC+ + +S SR + + V + + + + ++ V
Sbjct: 317 YVYIYDSEHTCS--VRERSGRSRQATPDVLGVLYRDYLGDVGPDVKPKSIGIIITKHFRV 374
Query: 374 GITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRVCRDNNVKINVAGPSVTIQLRF 433
++ ++++ + A+ + G D + + Y ++R ++ + RF
Sbjct: 375 KMSYSKSYKTLRFARELTLGTPDSSFEELPSYLYMIRRANPGTVARLQIDESG-----RF 429
Query: 434 GSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQLLIAVGRDPNDQYFPLAFGVVETE 493
+ F GF R V VDG L Y G LL A+ +D N Q FPLAFGVV+TE
Sbjct: 430 NYMFIVFGASIAGF-HYMRRVVVVDGTFLHGSYKGTLLTALAQDGNFQIFPLAFGVVDTE 488
Query: 494 CKETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFDEMFDSIDHRLCLRHLYANFKKKF 553
++W+W L I + ISD+ K + E++ +C HLY N KF
Sbjct: 489 NDDSWRWLFTQLKVVIPDATDLAIISDRHKSIGKAIGEVYPLAARGICTYHLYKNILVKF 548
Query: 554 GGGSQIRDLMMGATKATYNQGWLEKMNELKKIDLGAWEWLMAVEPKKWCKHAFTFYSKCD 613
+ L+ A + + NE++++D +L + W + F + +
Sbjct: 549 -KRKDLFPLVKKAARCYRLNDFTNAFNEIEELDPLLHAYLQRAGVEMWARAHFP-GDRYN 606
Query: 614 VLMNNISESFNVTILSARDQPIISMAEWIRHYLMRRMTTSATKLQKWQHNVMPMPRKRLD 673
++ NI+ES N + A++ PI+ M E IR + R K + P K L
Sbjct: 607 LMTTNIAESMNRALSQAKNLPIVRMLEAIRQMMTRWFAERRDDASKQHTQLTPGVEKLLQ 666
Query: 674 KEITLAAHWKSTWSGIGEQFQVMHTYNRQQFIVDIAKRSCSCNFWEIVGIPCRHVVSALG 733
+T ++ + + QV Y +V++ ++ C+C + +PC H ++A
Sbjct: 667 TRVT-SSRLLDVQTIDASRVQV--AYEASLHVVNVDEKQCTCRLFNKEKLPCIHAIAAAE 723
Query: 734 KRKQRPEMFVDDYYSRSKYALCYSFAISPINGMDMWPEVEAPE-LLPPNYKNGPGRPRKL 792
YY S Y+ AI P + P++ + LPP N PGRP+KL
Sbjct: 724 HMGVSRISLCSPYYKSSHLVNVYACAIMPSDTEVPVPQIVIDQPCLPPIVANQPGRPKKL 783
Query: 793 RIREFDENGSRMRRQGVAYRCTKCDQFGHNQRRCKS 828
R++ E +R + C++C + GHN + C++
Sbjct: 784 RMKSALEVAVETKRPRKEHACSRCKETGHNVKTCRA 819
>At1g21040 hypothetical protein
Length = 904
Score = 165 bits (417), Expect = 1e-40
Identities = 128/516 (24%), Positives = 234/516 (44%), Gaps = 44/516 (8%)
Query: 302 DQCGYKVLCSKVGDRRTFQIKTLEGPHTCAPVLENKSANSRWVAKKAVPKMQVTK----- 356
+ C ++V K+ D +QI++ HTC +E +S R + + + +K
Sbjct: 388 ETCPWRVYIVKLEDSDNYQIRSANLEHTCT--VEERSNYHRAATTRVIGSIIQSKYAGNS 445
Query: 357 ----KMSVQEVFNEMAVNYGVGITMDRAWRARKIAKSIIEGDADKQYAMIRRYAAELKRV 412
+ +Q + + +Y V I+ +AW++R+IA +G A + ++ Y L+
Sbjct: 446 RGPRAIDLQRI---LLTDYSVRISYWKAWKSREIAMDSAQGSAANSFTLLPAYLHVLREA 502
Query: 413 CRDN--NVKINVAGPSVTIQLRFGSFYFCFDGCKKGFISACRPFVGVDGCHLKTRYGGQL 470
+ ++K V G RF + F +GF S + + +DG HLK +YGG L
Sbjct: 503 NPGSIVDLKTEVDGKG---NHRFKYMFLAFAASIQGF-SCMKRVIVIDGAHLKGKYGGCL 558
Query: 471 LIAVGRDPNDQYFPLAFGVVETECKETWKWFTQLLMEDIGEDKRYVFISDQQKGLLSVFD 530
L A G+D N Q FP+AFGVV++E + W+WF ++L I + F+SD+ + +
Sbjct: 559 LTASGQDANFQVFPIAFGVVDSENDDAWEWFFRVLSTAIPDGDNLTFVSDRHSSIYTGLR 618
Query: 531 EMFDSIDHRLCLRHLYAN----FKKKFGGGSQIRDLMMGATKATYNQGWLEKMNELKKID 586
++ H C+ HL N +KKK + + A +A + NE+ ++D
Sbjct: 619 RVYPKAKHGACIVHLQRNIATSYKKK-----HLLFHVSRAARAFRICEFHTYFNEVIRLD 673
Query: 587 LGAWEWLMAVEPKKWCKHAFTFYSKCDVLMNNISESFNVTILSARDQPIISMAEWIRHYL 646
+L +V W + A+ + +V+ +N++ES N + AR+ PIIS+ E+IR L
Sbjct: 674 PACARYLESVGFCHWTR-AYFLGERYNVMTSNVAESLNAVLKEARELPIISLLEFIRTTL 732
Query: 647 M----RRMTTSATKLQKWQHNVMPMPRKRLDKEITLAAHWKSTWSGIGEQFQVMHTYNRQ 702
+ R + T+ + + + +K + A H + +
Sbjct: 733 ISWFAMRREAARTEASPLPPKMREVVHRNFEKSVRFAVHRLDRYD------YEIREEGAS 786
Query: 703 QFIVDIAKRSCSCNFWEIVGIPCRHVVSALGKRKQRPEMFVDDYYSRSKYALCYSFAISP 762
+ V + +R+CSC ++++ +PC H ++A + + YS + + Y I P
Sbjct: 787 VYHVKLMERTCSCRAFDLLHLPCPHAIAAAVAEGVPIQGLMAPEYSVESWRMSYLGTIKP 846
Query: 763 ---INGMDMWPE-VEAPELLPPNYKNGPGRPRKLRI 794
+ + PE + + L PP + GRP+K RI
Sbjct: 847 VPEVGDVFALPEPIASLRLFPPATRRPSGRPKKKRI 882
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 20,338,284
Number of Sequences: 26719
Number of extensions: 939686
Number of successful extensions: 3197
Number of sequences better than 10.0: 139
Number of HSP's better than 10.0 without gapping: 93
Number of HSP's successfully gapped in prelim test: 46
Number of HSP's that attempted gapping in prelim test: 2782
Number of HSP's gapped (non-prelim): 233
length of query: 840
length of database: 11,318,596
effective HSP length: 108
effective length of query: 732
effective length of database: 8,432,944
effective search space: 6172915008
effective search space used: 6172915008
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144730.8


