

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144730.2 - phase: 0 /pseudo
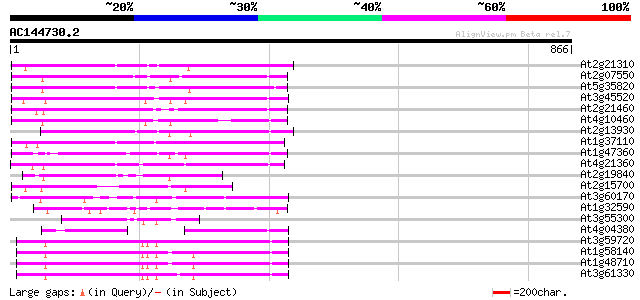
(866 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g21310 putative retroelement pol polyprotein 263 4e-70
At2g07550 putative retroelement pol polyprotein 253 4e-67
At5g35820 copia-like retrotransposable element 251 9e-67
At3g45520 copia-like polyprotein 248 1e-65
At2g21460 putative retroelement pol polyprotein 235 7e-62
At4g10460 putative retrotransposon 229 4e-60
At2g13930 putative retroelement pol polyprotein 214 2e-55
At1g37110 197 3e-50
At1g47360 polyprotein, putative 195 8e-50
At4g21360 putative transposable element 188 1e-47
At2g19840 copia-like retroelement pol polyprotein 158 1e-38
At2g15700 copia-like retroelement pol polyprotein 119 1e-26
At3g60170 putative protein 109 6e-24
At1g32590 hypothetical protein, 5' partial 106 5e-23
At3g55300 putative protein 104 2e-22
At4g04380 putative polyprotein 95 2e-19
At3g59720 copia-type reverse transcriptase-like protein 93 7e-19
At1g58140 hypothetical protein 93 7e-19
At1g48710 hypothetical protein 92 1e-18
At3g61330 copia-type polyprotein 92 2e-18
>At2g21310 putative retroelement pol polyprotein
Length = 838
Score = 263 bits (671), Expect = 4e-70
Identities = 168/458 (36%), Positives = 260/458 (56%), Gaps = 30/458 (6%)
Query: 3 DIEKFTGSNDFGLWKVKMRAI---------LIQQKCVEALKGEAQMDVHLTPAEKT---E 50
++EK G D+ LWK K+ A L + + +E + A+ D LT E E
Sbjct: 7 EVEKLDGEGDYVLWKEKLLAHIELLGLLEGLEEDEAIEEEESTAETDSLLTKTEDKVLKE 66
Query: 51 MNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVE 110
KA S +IL LG+ VLR+V +E TA M LD L+M KSL +R LKQ+LY Y+M +
Sbjct: 67 KRGKARSTVILSLGNHVLRKVIKEKTAAGMIRVLDKLFMAKSLPNRIYLKQRLYGYKMSD 126
Query: 111 SKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLE 170
S I E + +F K+I DL N+ V++ DED+A+ L +LP+ F+ KDT+ YGK T+ L+
Sbjct: 127 SMTIEENVNDFFKLISDLENVKVSVPDEDQAIVLLMSLPKQFDQLKDTLKYGK-TTLALD 185
Query: 171 EVQAALRTKELTKFKELK-VEDSGEGLNV-SRERSQNRGKGKGKNSRSKSRSKGDGNKTQ 228
E+ A+R+K L K +++S + L V R RS+ R K +N +S+SRSK K
Sbjct: 186 EITGAIRSKVLELGASGKMLKNSSDALFVQDRGRSEKRDKSSERN-KSQSRSKSREKKV- 243
Query: 229 YKCFICHNPGHFKKDCP--ERKGNGGGNPSVQIASNEEGYES-AGALTV------TSWEP 279
C++C GHFKK C + K G N +SN G + A AL V + E
Sbjct: 244 --CWVCGKEGHFKKQCYVWKEKNKKGNNSEKGESSNVIGQAADAAALAVREESNADNQEV 301
Query: 280 EKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLL 339
+ W++D+GCS+H+ PR+++F + + G V + N +I+ IG+IR++ D+ LL
Sbjct: 302 DNEWIMDTGCSFHMTPRRDWFVEFDESQTGRVKMANQTYSEIKGIGSIRIQNDDNTTVLL 361
Query: 340 KDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVI 399
K+VRY+P + +NLIS+ + G + + G +++ G + + KG K+ LY+L+G V
Sbjct: 362 KNVRYVPSMSKNLISMGTLEDQGCWFQSKAGTLKVVKGCMTLLKGKKVGTLYLLQGVVVT 421
Query: 400 ADASVASVDTLDVTKLWHLRLGHVSERGIWLN*LNKGC 437
+A+ A + D +K+WH RL H+S+R I + + KGC
Sbjct: 422 GNAN-AVTSSKDESKIWHSRLCHMSQRNIDVL-IKKGC 457
>At2g07550 putative retroelement pol polyprotein
Length = 1356
Score = 253 bits (645), Expect = 4e-67
Identities = 163/457 (35%), Positives = 255/457 (55%), Gaps = 41/457 (8%)
Query: 3 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKT------------- 49
++EKF G D+ +WK K+ A + ALK + +++
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHMDILGLNTALKESESTGEKKSVLDESDEDYEEKLEKFEA 66
Query: 50 --EMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYR 107
E KA SAI+L + D+VLR++ +ESTA +M LD LYM+K+L +R KQ+LY ++
Sbjct: 67 LEEKKKKARSAIVLSVTDRVLRKIKKESTAAAMLLALDKLYMSKALPNRIYPKQKLYSFK 126
Query: 108 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTI 167
M E+ + + EF +II DL N++V + DED+A+ L ALP++F+ KDT+ Y +I
Sbjct: 127 MSENLSVEGNIDEFLQIITDLENMNVIISDEDQAILLLTALPKAFDQLKDTLKYSSGKSI 186
Query: 168 -TLEEVQAALRTKEL---TKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKGD 223
TL+EV AA+ +KEL + K +KV+ EGL V +++++N+GKG+ K K + K
Sbjct: 187 LTLDEVAAAIYSKELELGSVKKSIKVQ--AEGLYV-KDKNENKGKGEQKG---KGKGKKG 240
Query: 224 GNKTQYKCFICHNPGHFKKDCPER-----------KG-NGGGNPSVQIASNEEGYESAGA 271
+K + C+ C GHF+ CP + KG + GG ++ A+ GY + A
Sbjct: 241 KSKKKPGCWTCGEEGHFRSSCPNQNKPQFKQSQVVKGESSGGKGNLAEAA---GYYVSEA 297
Query: 272 LTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKM 331
L+ T E W+LD+GCSYH+ ++E+F + GG V +GN +++ +GTIR+K
Sbjct: 298 LSSTEVHLEDEWILDTGCSYHMTYKREWFHEFNEDAGGSVRMGNKTVSRVRGVGTIRVKN 357
Query: 332 FDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLY 391
D +L +VRYIP + RNL+S+ F+ GY E G++RI G ++ G + LY
Sbjct: 358 SDGLTIVLTNVRYIPDMDRNLLSLGTFEKAGYKFESEDGILRIKAGNQVLLTGRRYDTLY 417
Query: 392 ILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGI 428
+L V A S+A V D T LWH RL H+S++ +
Sbjct: 418 LLNWKPV-ASESLAVVKRADDTVLWHQRLCHMSQKNM 453
>At5g35820 copia-like retrotransposable element
Length = 1342
Score = 251 bits (642), Expect = 9e-67
Identities = 164/457 (35%), Positives = 248/457 (53%), Gaps = 38/457 (8%)
Query: 3 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKT------------- 49
++EKF G D+ LWK K+ A + +E L E + V + E +
Sbjct: 7 EVEKFDGDGDYILWKEKLLAHMEMLGLLEGLGEEEEAVVEDSTTEISDGGNQDPETATSK 66
Query: 50 -------EMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQ 102
E KA S IIL LG+ VLR+V ++ TA M LD L+M KSL +R LKQ+
Sbjct: 67 LEDKILKEKRGKARSTIILSLGNNVLRKVIKQKTAAGMIKVLDQLFMAKSLPNRIYLKQR 126
Query: 103 LYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYG 162
LY Y+M E+ + E + +F K+I DL N+ V + DED+A+ L +LPR F+ K+T+ Y
Sbjct: 127 LYGYKMSENMTMEENVNDFFKLISDLENVKVVVPDEDQAIVLLMSLPRQFDQLKETLKYC 186
Query: 163 K*GTITLEEVQAALRTKELTKFKELK-VEDSGEGLNV-SRERSQNRGKGKGKNSRSKSRS 220
K T+ LEE+ +A+R+K L K ++++ +GL V R RS+ RGKG KN +S+S+S
Sbjct: 187 K-TTLHLEEITSAIRSKILELGASGKLLKNNSDGLFVQDRGRSETRGKGPNKN-KSRSKS 244
Query: 221 KGDGNKTQYKCFICHNPGHFKKDC---PERKGNGGGNPSVQIASNEEGYESAGALTVT-- 275
KG G C+IC GHFKK C ER G + + ++ A AL V+
Sbjct: 245 KGAGK----TCWICGKEGHFKKQCYVWKERNKQGSTSERGEASTVTARVTDAAALVVSRA 300
Query: 276 ----SWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKM 331
+ W+LD+GCS+H+ RK++ + G V +GN+ +++ IG +R+K
Sbjct: 301 LLGFAEVTPDTWILDTGCSFHMTCRKDWIIDFKETASGKVRMGNDTYSEVKGIGDVRIKN 360
Query: 332 FDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLY 391
D LL DVRYIP++ +NLIS+ + G ++G++ I L + G K LY
Sbjct: 361 EDGSTILLTDVRYIPEMSKNLISLGTLEDKGCWFESKKGILTIFKNDLTVLTGKKESTLY 420
Query: 392 ILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGI 428
L+G+T+ +A+V + D T LWH RLGH+ +G+
Sbjct: 421 FLQGTTLAGEANVIDKEK-DETSLWHSRLGHIGAKGL 456
>At3g45520 copia-like polyprotein
Length = 1363
Score = 248 bits (632), Expect = 1e-65
Identities = 158/466 (33%), Positives = 254/466 (53%), Gaps = 48/466 (10%)
Query: 3 DIEKFTGSNDFGLWKVKM---------RAILIQQKCVEALKGEAQMDVHLTPAEKTEMND 53
++EKF G D+ +WK K+ A+L + + + +++ E+ +M
Sbjct: 7 EVEKFDGRGDYTMWKEKLLAHIDMLGLSAVLRESETPMGKERDSEKSDEDEKEEREKMEA 66
Query: 54 ------KAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYR 107
KA S I+L + D+VLR++ +E++A +M LD LYM+K+L +R LKQ+LY ++
Sbjct: 67 FEEKKRKARSTIVLSVSDRVLRKIKKETSAAAMLEALDRLYMSKALPNRIYLKQKLYSFK 126
Query: 108 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTI 167
M E+ I + EF I+ DL N++V + DED+A+ L +LP+ F+ KDT+ Y T+
Sbjct: 127 MSENLSIEGNIDEFLHIVADLENLNVLVSDEDQAILLLMSLPKPFDQLKDTLKYSSGKTV 186
Query: 168 -TLEEVQAALRTKELTKFKELK--VEDSGEGLNVSRERSQNRG----KGKGKNSRSKSRS 220
+L+EV AA+ ++EL +F +K ++ EGL V +++++NRG K KGK RSKS+S
Sbjct: 187 LSLDEVAAAIYSREL-EFGSVKKSIKGQAEGLYV-KDKAENRGRSEQKDKGKGKRSKSKS 244
Query: 221 KGDGNKTQYKCFICHNPGHFKKDCPER-----------KGNGGGNPSVQIASNEEGYESA 269
K C+IC GH K CP + KG G + + ESA
Sbjct: 245 KRG-------CWICGEDGHLKSTCPNKNKPQFKNQGSNKGESSGGKGNLVEGSVNFVESA 297
Query: 270 G-----ALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVI 324
G AL+ T E W++D+GC YH+ ++E+ E + E GG V +GN +++ +
Sbjct: 298 GMFVSEALSSTDIHLEDEWIMDTGCIYHMTHKREWLEDFDEEAGGSVRMGNKSISRVKGV 357
Query: 325 GTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKG 384
GT+R+ + L++VRYIP + RNL+S+ F+ G+ E G++RI G ++ +G
Sbjct: 358 GTVRIVNDNGLTVTLQNVRYIPDMDRNLLSLGTFEKAGHKFESENGMLRIKSGNQVLLEG 417
Query: 385 SKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWL 430
+ LYIL G D S+A D T LWH RL H+S++ + L
Sbjct: 418 RRYDTLYILHGKPA-TDESLAVARANDDTVLWHRRLCHMSQKNMSL 462
>At2g21460 putative retroelement pol polyprotein
Length = 1333
Score = 235 bits (600), Expect = 7e-62
Identities = 158/446 (35%), Positives = 247/446 (54%), Gaps = 41/446 (9%)
Query: 3 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQM-----DVHLTPAEKTE------- 50
++EKF G D+ +WK K+ A L ALK E + ++ LT E+ E
Sbjct: 7 EVEKFDGRGDYTMWKEKLMAHLDILGLSVALKEEDDLVEKVAEMQLTEEEEKEEVLRREL 66
Query: 51 ---MNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYR 107
KA SAI+L + D+VLR++ +E +A +M LD LYM+K+L +R KQ+LY ++
Sbjct: 67 LEEKRRKARSAIVLSVTDRVLRKIKKEQSAAAMLGVLDKLYMSKALPNRIYQKQKLYSFK 126
Query: 108 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*G-T 166
M E+ I + EF +II DL N +V + DED+A+ L +LP+ F+ +DT+ YG T
Sbjct: 127 MSENLSIEGNIDEFLRIIADLENTNVLVSDEDQAILLLMSLPKPFDQLRDTLKYGLGRVT 186
Query: 167 ITLEEVQAALRTKELTKFKELK-VEDSGEGLNVSRERSQNRGKGK---GKNSRSKSRSKG 222
++L+EV AA+ +KEL K ++ EGL V +E+++ RG+ + N+ KSRSK
Sbjct: 187 LSLDEVVAAIYSKELELGSNKKSIKGQAEGLFV-KEKTETRGRTEQRGNNNNNKKSRSK- 244
Query: 223 DGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGYESAGALTVTSWEPEKG 282
++++ C+IC NG N S G + AL+ T E
Sbjct: 245 --SRSKKGCWIC-----------GESSNGSSN-----YSEANGLYVSEALSSTDIHLEDE 286
Query: 283 WVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDV 342
WV+D+GCSYH+ ++E+FE L + GG V +GN K++ IGTIR+K L +V
Sbjct: 287 WVMDTGCSYHMTYKREWFEDLNEDAGGSVRMGNKTVSKVRGIGTIRVKNEAGMVVRLTNV 346
Query: 343 RYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVIADA 402
RYIP++ RNL+S+ F+ GY ++E G + I G ++ + + LY+L+ V +
Sbjct: 347 RYIPEMDRNLLSLGTFEKSGYSFKLENGTLSIIAGDSVLLTVRRCYTLYLLQWRPV-TEE 405
Query: 403 SVASVDTLDVTKLWHLRLGHVSERGI 428
S++ V D T LWH RLGH+S++ +
Sbjct: 406 SLSVVKRQDDTILWHRRLGHMSQKNM 431
>At4g10460 putative retrotransposon
Length = 1230
Score = 229 bits (585), Expect = 4e-60
Identities = 155/457 (33%), Positives = 232/457 (49%), Gaps = 57/457 (12%)
Query: 3 DIEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEK-------------T 49
++EKF G D+ LWK K+ A + AL+ + L E+
Sbjct: 7 EMEKFDGHGDYTLWKEKLMAHMDLLGLTVALRETQSVSDPLESEEEGKESEKGDKEALME 66
Query: 50 EMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMV 109
E KA S I+L + D+VLR+ +E TA SM LD LYM+K+L +R LKQ+LY Y+M
Sbjct: 67 EKRQKARSTIVLSVSDQVLRKSKKEKTAPSMLEALDKLYMSKALPNRIYLKQKLYSYKMQ 126
Query: 110 ESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK-*GTIT 168
E+ + + EF ++I DL N +V + DED+A+ L +LP+ F+ KDT+ YG T++
Sbjct: 127 ENLSVEGNIDEFLRLIADLENTNVLVSDEDQAILLLMSLPKQFDQLKDTLKYGSGRTTLS 186
Query: 169 LEEVQAALRTKELTKFKELK-VEDSGEGLNVSRERSQNRG----KGKGKNSRSKSRSKGD 223
++EV AA+ +KEL K + EGL V +++ + RG K KG RS+SRSKG
Sbjct: 187 VDEVVAAIYSKELELGSNKKSIRGQAEGLYV-KDKPETRGMSEQKEKGNKGRSRSRSKG- 244
Query: 224 GNKTQYKCFICHNPGHFKKDCPER---------KGNGGGNPSVQIASNE---EGYESAGA 271
C+IC GHFK CP + + +G + I N GY + A
Sbjct: 245 ----WKGCWICGEEGHFKTSCPNKGKQQNKGKDQASGSKGEAATIKGNTSEGSGYYVSEA 300
Query: 272 LTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKM 331
L T WV+D+GC+YH+ +KE+FE L + GG V +GN K +
Sbjct: 301 LHSTDVNLGNEWVMDTGCNYHMTHKKEWFEELSEDAGGTVRMGNKSTSKFR--------- 351
Query: 332 FDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLY 391
V+YIP + RNL+S+ + GY + GV+ + G + GS+ LY
Sbjct: 352 ----------VKYIPDMDRNLLSMGTLEEHGYSFESKNGVLVVKEGTRTLLIGSRHEKLY 401
Query: 392 ILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGI 428
+L+G ++ S+ D T LWH RLGH+S++ +
Sbjct: 402 LLQGKPEVSH-SMTVERRNDDTVLWHRRLGHISQKNM 437
>At2g13930 putative retroelement pol polyprotein
Length = 1335
Score = 214 bits (545), Expect = 2e-55
Identities = 135/404 (33%), Positives = 213/404 (52%), Gaps = 22/404 (5%)
Query: 48 KTEMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYR 107
+ E DKA + I L + DKVLR++ TA LD L+M +SL HR + Y ++
Sbjct: 42 RLERCDKAKNVIFLNVADKVLRKIELCKTAAEAWETLDRLFMIRSLPHRVYTQLSFYTFK 101
Query: 108 MVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GT- 166
M E+K I E + +F KI+ DL ++ +++ DE +A+ L +LP ++ +TM Y
Sbjct: 102 MQENKKIDENIDDFLKIVADLNHLQIDVTDEVQAILLLSSLPARYDGLVETMKYSNSREK 161
Query: 167 ITLEEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKG-DGN 225
+ L++V A R KE + + G + +R R + +G +++SRSK DG
Sbjct: 162 LRLDDVMVAARDKERELSQNNRPVVEG---HFARGRPDGKNNNQGNKGKNRSRSKSADGK 218
Query: 226 KTQYKCFICHNPGHFKKDC------PERKGNGGGNPSVQIASNEEGYESAGALTVTSW-- 277
+ C+IC GHFKK C + K G N +A + E + A L T
Sbjct: 219 RV---CWICGKEGHFKKQCYKWIERNKSKQQGSDNGESSLAKSTEAFNPAMVLLATDETL 275
Query: 278 ----EPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFD 333
WVLD+GCS+H+ PRK++F+ + G V +GN+ ++ IG+I+++ D
Sbjct: 276 VVTDSIANEWVLDTGCSFHMTPRKDWFKDFKELSSGYVKMGNDTYSPVKGIGSIKIRNSD 335
Query: 334 DRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYIL 393
+L DVRY+P + RNLIS+ + G + + G+++I G I KG K LYIL
Sbjct: 336 GSQVILTDVRYMPNMTRNLISLGTLEDRGCWFKSQDGILKIVKGCSTILKGQKRDTLYIL 395
Query: 394 EGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWLN*LNKGC 437
+G T + S +S + D T LWH RLGH+S++G+ + + KGC
Sbjct: 396 DGVTEEGE-SHSSAEVKDETALWHSRLGHMSQKGMEIL-VKKGC 437
>At1g37110
Length = 1356
Score = 197 bits (500), Expect = 3e-50
Identities = 129/443 (29%), Positives = 226/443 (50%), Gaps = 24/443 (5%)
Query: 3 DIEKFTGSNDFGLWKVKMRAIL------------IQQKCVEALKGEAQMDV--------- 41
+I+ F G DF LWK++++A L K V K EA+ +
Sbjct: 9 EIKVFNGDRDFSLWKIRIQAQLGVLGLKDTLTDFSLTKTVPLTKSEAKQESGDGESSGTK 68
Query: 42 HLTPAEKTEMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQ 101
+ K E +++A + II + D VL +V+ +T + L+ YM SL +R +
Sbjct: 69 EVPDPVKIEQSEQAKNIIINHISDVVLLKVNHYATTADLWATLNKKYMETSLPNRIYTQL 128
Query: 102 QLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLY 161
+LY ++MV + I + + EF +I+ +L ++++ +++E +A+ + +LP S K T+ Y
Sbjct: 129 KLYSFKMVSTMTIDQNVDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLKHTLKY 188
Query: 162 GK*GTITLEEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNSRSKSRSK 221
G T+T+++V ++ ++ E + + ++ + + ER + + K + K RS+
Sbjct: 189 GN-KTLTVQDVTSSAKSLERELAEAVDLDKGQAAVLYTTERGRPLVRNNQKGGQGKGRSR 247
Query: 222 GDGNKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGYESAGALTVTSWEPEK 281
+ +KT+ C+ C GH KKDC RK + E + AL+V +
Sbjct: 248 SN-SKTKVPCWYCKKEGHVKKDCYSRKKKMESEGQGEAGVITEKLVFSEALSVNEQMVKD 306
Query: 282 GWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKD 341
W+LDSGC+ H+ R+++F + + + LG++ + + Q GTIR+ +L++
Sbjct: 307 LWILDSGCTSHMTSRRDWFISFQEKGNTTILLGDDHSVESQGQGTIRIDTHGGTIKILEN 366
Query: 342 VRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVIAD 401
V+Y+P LRRNLIS D LGY G +R +GS +GLY+L+GSTV+++
Sbjct: 367 VKYVPHLRRNLISTGTLDKLGYRHEGGEGKVRYFKNNKTALRGSLSNGLYVLDGSTVMSE 426
Query: 402 ASVASVDTLDVTKLWHLRLGHVS 424
A D + T LWH RLGH+S
Sbjct: 427 LCNAETDKVK-TALWHSRLGHMS 448
>At1g47360 polyprotein, putative
Length = 1182
Score = 195 bits (496), Expect = 8e-50
Identities = 141/444 (31%), Positives = 223/444 (49%), Gaps = 49/444 (11%)
Query: 4 IEKFTGSNDFGLWKVKMRAILIQQKCVEALKGEAQMDVHLTPAEKTEMNDKAVSAIILCL 63
I F S DF LWK ++ A L + + G+ +TP E D I LC
Sbjct: 6 IAVFDESGDFSLWKTRIMAHLSVIGLKDVVIGKT-----ITPLTAEEEEDPE-KKIELC- 58
Query: 64 GDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNK 123
TA LD L+M +SL HR + Y ++M E+K I E + +F K
Sbjct: 59 -----------QTAQEAWETLDRLFMIRSLPHRVYTQLSFYTFKMQENKKIDENIDDFLK 107
Query: 124 IIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GT-ITLEEVQAALRTKELT 182
I+ DL ++ + + DE +A+ L +LP ++ +TM Y + L++V R KE
Sbjct: 108 IVADLNHLQIEVTDEVQAILLLSSLPARYDGLVETMKYSNSREKLRLDDVMVGARDKE-- 165
Query: 183 KFKELKVEDS--GEGLNVSRERSQNRGKGKGKNSRSKSRSKG-DGNKTQYKCFICHNPGH 239
+EL + EG + +R R + + +G +++SRSK DG + C+IC GH
Sbjct: 166 --RELSQNNRPVAEG-HYARGRPEGKNNNQGNKGKNRSRSKSADGKRV---CWICGKEGH 219
Query: 240 FKKDC--------PERKGNGGGNPSVQIASNEEGYESA-------GALTVTSWEPEKGWV 284
FKK C +++G+ G S +A + E ++ A L VT + ++ W
Sbjct: 220 FKKQCYKWLERNKSKQQGSDVGESS--LAKSSEIFDPAMVIMATGETLLVTGRDADE-WF 276
Query: 285 LDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRY 344
LD+GCS+H+ PR++ F+ + G V +GN+ ++ IG+I+++ D +L DVRY
Sbjct: 277 LDTGCSFHMTPRRDLFKDFKELSSGFVKMGNDTYSPVKGIGSIKIRNNDGTQVILTDVRY 336
Query: 345 IPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLYILEGSTVIADASV 404
+P + RNLIS+ + G + + GV++I G I KG K LYILEG + S
Sbjct: 337 VPNMARNLISLGTLEDKGCWFKSQDGVLKIVKGCSTILKGQKRETLYILEGLAENGE-SH 395
Query: 405 ASVDTLDVTKLWHLRLGHVSERGI 428
+S + D T LWH RLGH+S++G+
Sbjct: 396 SSAELKDETSLWHSRLGHMSQKGM 419
>At4g21360 putative transposable element
Length = 1308
Score = 188 bits (477), Expect = 1e-47
Identities = 132/453 (29%), Positives = 234/453 (51%), Gaps = 38/453 (8%)
Query: 1 KWDIEKFTGSNDFGLWKVKMRAILIQQKCVEAL---------------KGEAQMDVHLTP 45
K +I+ F G DF LWK+++ A L AL K E++ + T
Sbjct: 6 KVEIKTFNGDRDFSLWKIRIEAQLGVLGLKPALSDFTLTKTILVVKSEKKESESEDDETD 65
Query: 46 AEKTE---------MNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHR 96
++KTE +D+A + II + D VL +V TA + L+ L+M SL +R
Sbjct: 66 SKKTEEVPDPIKFEQSDQAKNFIINHITDTVLLKVQHCVTAAELWATLNKLFMETSLPNR 125
Query: 97 QCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFK 156
+ +LY ++MV++ I + EF +I+ +L ++ + + +E +A+ + +LP S+ K
Sbjct: 126 IYTQLRLYSFKMVDNLSIDQNTDEFLRIVAELGSLQIQVGEEVQAILILNSLPPSYIQLK 185
Query: 157 DTMLYGK*GTITLEEVQAALRT--KELTKFKELKVEDSGEGLNVSRERSQNRGKGKGKNS 214
T+ YG T+++++V ++ ++ +EL++ KE + L + RG+ + KN+
Sbjct: 186 HTLKYGN-KTLSVQDVVSSAKSLERELSEQKETIRAPASTALYTAE-----RGRPQTKNT 239
Query: 215 RSKSRSKGDGN-KTQYKCFICHNPGHFKKDC--PERKGNGGGNPSVQIASNEEGYESAGA 271
+ + + +G N K++ C+ C GH KKDC +RK G + + + Y A
Sbjct: 240 QGQGKGRGRSNSKSRLTCWFCKKEGHVKKDCYAGKRKLENEGQGKAGVITEKLVYSEA-- 297
Query: 272 LTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLKM 331
L++ E + WV+DSGC+YH+ R ++F E ++ LG++ + + GT+++
Sbjct: 298 LSMYDQEAKDKWVIDSGCTYHMTSRMDWFSEFNENETTMILLGDDHTVESKGSGTVKVNT 357
Query: 332 FDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGLY 391
+LK+VR++P LRRNLIS D LGY G +R G+ ++GLY
Sbjct: 358 HGGSIRVLKNVRFVPNLRRNLISTGTLDKLGYKHEGGDGKVRFYKENKTALCGNLVNGLY 417
Query: 392 ILEGSTVIADASVASVDTLDVTKLWHLRLGHVS 424
+L+G TV+ + + + + T+LWH RLGH+S
Sbjct: 418 VLDGHTVV-NENCNVEGSNEKTELWHCRLGHMS 449
>At2g19840 copia-like retroelement pol polyprotein
Length = 1137
Score = 158 bits (400), Expect = 1e-38
Identities = 106/329 (32%), Positives = 176/329 (53%), Gaps = 40/329 (12%)
Query: 20 MRAILIQQKCVEALKGEAQMDVHLTPAEKTE----------MNDKAVSAIILCLGDKVLR 69
++ L+++ + LK E + D PA+K + ++KA+ I + +GDKVLR
Sbjct: 3 LKDALVERAPLPPLKEEDESD----PAKKKQRIEEEKARIDQDEKAMDMIFINVGDKVLR 58
Query: 70 EVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLA 129
+ TA LD LY+ KSL +R L+ ++Y YRM +SK + E + EF K+I DL
Sbjct: 59 NIENSKTAAEAWATLDKLYLVKSLPNRVYLQLKVYNYRMQDSKTLEENVDEFQKMISDLN 118
Query: 130 NIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKELTKFKELKV 189
N+ + + DE +A+ + ALP S++ K+T+ YG+ G I L++V +A K KEL++
Sbjct: 119 NLQIQVPDEVQAILILSALPDSYDMLKETLKYGREG-IKLDDVISA------AKSKELEL 171
Query: 190 EDSGEGLNVSRERSQNRGKGKGKNSRSKSRSKG-DGNKT---QYKCFICHNPGHFKKDC- 244
DS G +R G+G R KS+++G DG K+ + C+IC GHFK+ C
Sbjct: 172 RDSSGG---------SRPVGEGLYVRGKSQARGSDGPKSTEGKKVCWICGKEGHFKRQCY 222
Query: 245 ---PERKGNGGGNPSV--QIASNEEGYESAGALTVTSWEPEKGWVLDSGCSYHICPRKEY 299
+ K NG G ++ A + G ++ + ++ W++D+GCS+H+ PRKEY
Sbjct: 223 KWLEKNKANGAGETALVKDDAQDLVGLVASEVNMSEGKDDQEEWIMDTGCSFHMTPRKEY 282
Query: 300 FEMLELEEGGVVCLGNNKACKIQVIGTIR 328
+ G V + NN +++ IG ++
Sbjct: 283 LMDFVEAKSGKVRMANNSFSEVKGIGKVK 311
>At2g15700 copia-like retroelement pol polyprotein
Length = 1166
Score = 119 bits (297), Expect = 1e-26
Identities = 110/374 (29%), Positives = 165/374 (43%), Gaps = 70/374 (18%)
Query: 3 DIEKFTGSNDFGLWKVKMRA---------------ILIQQKCVEALKGEAQMDVHLTPAE 47
++++F G+ DF LWKV+M A +L +E A D H+ E
Sbjct: 8 EVDRFDGTGDFSLWKVRMLAHYGVLGLKGILNDEQLLRDPPVIEEEAAVAGRDYHVGDFE 67
Query: 48 --------KTEMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCL 99
K E ++KA I+L +G++VLR++ TA +M + L LYM SL +R L
Sbjct: 68 LPSNVDLIKFEKSEKAKDLIVLNVGNQVLRKIKNCETAAAMWSTLKRLYMETSLPNRIYL 127
Query: 100 KQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTM 159
+ + Y Y+M +S+ I + +F K++ DL NI V++
Sbjct: 128 QLKFYTYKMTDSRSIDGNVDDFLKLVTDLNNIGVDV------------------------ 163
Query: 160 LYGK*GTITLEEVQAALRTKELTKFKE--LKVEDS-GEGLNVSRERSQNRG--KGKGKNS 214
EEVQA L L KE KVE S EGL V +E Q RG K K +
Sbjct: 164 ---------TEEVQAILLLSSLYDRKENSQKVESSQSEGLYVEQEADQRRGLKKEKANHG 214
Query: 215 RSKSRSKGDGNKTQYKCFICHNPGH-FKKDCPERKGNGGGNPSVQIASNEEGYESAG--- 270
R+K D N+ Q KKD G G S+ S+ +
Sbjct: 215 EDVLRAKADQNQGQSTIKTTRVVSSVVKKDI----GRGSVLKSLHKPSSSANIATEPKQP 270
Query: 271 -ALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRL 329
LT + + ++ WV+DSGCS+HI K+ L+ +GG V +GN +++ IG I++
Sbjct: 271 LVLTASPQDTKEEWVMDSGCSFHITLDKDSLFDLQEFDGGKVLMGNMTHSEVKGIGKIKI 330
Query: 330 KMFDDRDFLLKDVR 343
DD +L +VR
Sbjct: 331 LNPDDYVVILTNVR 344
>At3g60170 putative protein
Length = 1339
Score = 109 bits (273), Expect = 6e-24
Identities = 108/468 (23%), Positives = 198/468 (42%), Gaps = 70/468 (14%)
Query: 4 IEKFTGSNDFGLWKVKMRAILIQQKCVEALK-GEAQMDVHLTPA--------EKTEMND- 53
I +F G DF W + M L ++ ++ G + V TP E+ ++ D
Sbjct: 12 IPRFDGYYDF--WSMTMENFLRSRELWRLVEEGIPAIVVGTTPVSEAQRSAVEEAKLKDL 69
Query: 54 KAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHR---QCLKQQLYFYRMVE 110
K + + + ++L + +ST+ ++ + Y + R Q L+++ M E
Sbjct: 70 KVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAMKE 129
Query: 111 SKPI--------------------MEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPR 150
+ I MEQ T +KI+ L K ++ C++
Sbjct: 130 GEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLT---------PKFNYVVCSIEE 180
Query: 151 SFENFKDTMLYGK*GTITLEEVQAALRTKELTKFKELKVEDSGEGLNVSRERSQNRGKGK 210
S + T++++E+ +L E ++ E + L V+ E ++G+G+
Sbjct: 181 SNDL----------STLSIDELHGSLLVHEQRLNGHVQEE---QALKVTHEERPSQGRGR 227
Query: 211 GKNSRSKSRSKGDG----NKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASNEEGY 266
G S+ R +G G N+ +C+ CHN GHF+ +CPE + N + EE
Sbjct: 228 GVFRGSRGRGRGRGRSGTNRAIVECYKCHNLGHFQYECPEWEKNA----NYAELEEEEEL 283
Query: 267 ESAGALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGT 326
+ ++ W LDSGCS H+ KE+F LE V LGN+ + G+
Sbjct: 284 LLMAYVEQNQANRDEVWFLDSGCSNHMTGSKEWFSELEEGFNRTVKLGNDTRMSVVGKGS 343
Query: 327 IRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISH---GALIIAK 383
+++K+ + ++ +V Y+P+LR NL+S+ G I G ++ H GA++
Sbjct: 344 VKVKV-NGVTQVIPEVYYVPELRNNLLSLGQLQERGLAILIRDGTCKVYHPSKGAIMETN 402
Query: 384 GSKIHGLYILEGSTVIADASVASVDTLD-VTKLWHLRLGHVSERGIWL 430
S ++L + + + +D LWH R GH+++ G+ L
Sbjct: 403 MSGNRMFFLLASKPQKNSLCLQTEEVMDKENHLWHCRFGHLNQEGLKL 450
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 106 bits (265), Expect = 5e-23
Identities = 106/430 (24%), Positives = 189/430 (43%), Gaps = 65/430 (15%)
Query: 38 QMDVHLTPAEKTEMNDKAV------SAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTK 91
+ +V LT A++TE+ +K V + + + +L+ + ++ T+ + + Y
Sbjct: 5 ERNVILTGAQRTELAEKTVKDHKVKNYLFASIDKTILKTILQKETSKDLWESMKRKYQGN 64
Query: 92 SLAHRQCLKQQLYFYRMVESKPIMEQLTEF----NKIIDDLANIDVNLEDE--------- 138
L++ + ++E K I E +T + +I +D+ N+ ++ D
Sbjct: 65 DRVQSAQLQRLRRSFEVLEMK-IGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRT 123
Query: 139 --DKALHLPCALPRSFENFKDTMLYGK*GTITLEEVQAALRTKELTKFKELKVE-----D 191
+K ++ CA+ S N K+ + G ++ + E + E + LK E D
Sbjct: 124 LVEKFTYVVCAIEES-NNIKELTVDGLQSSLMVHEQNLSRHDVEE---RVLKAETQWRPD 179
Query: 192 SGEGLNVSRERSQNRGKGKGKNSRSKSRSKGDGNKTQYKCFICHNPGHFKKDCP--ERKG 249
G G R S +RG+G+G + R +G N+ +CF CH GH+K +CP E++
Sbjct: 180 GGRG----RGGSPSRGRGRGGY---QGRGRGYVNRDTVECFKCHKMGHYKAECPSWEKEA 232
Query: 250 NGGGNPSVQIASNEEGYESAGALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEG- 308
N EE + E ++ W LDSGCS H+C +E+F LEL+ G
Sbjct: 233 N--------YVEMEEDLLLMAHVEQIGDEEKQIWFLDSGCSNHMCGTREWF--LELDSGF 282
Query: 309 -GVVCLGNNKACKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRI 367
V LG+++ ++ G +RL++ D R ++ DV ++P L+ NL S+ G I
Sbjct: 283 KQNVRLGDDRRMAVEGKGKLRLEV-DGRIQVISDVYFVPGLKNNLFSVGQLQQKGLRFII 341
Query: 368 ERGVMRISHGALIIAKGSKIHGLYILEGSTVIADASVASVDTLD---------VTKLWHL 418
E V + H K +H V+ A S +T + +WH
Sbjct: 342 EGDVCEVWHKT---EKRMVMHSTMTKNRMFVVFAAVKKSKETEETRCLQVIGKANNMWHK 398
Query: 419 RLGHVSERGI 428
R GH++ +G+
Sbjct: 399 RFGHLNHQGL 408
>At3g55300 putative protein
Length = 262
Score = 104 bits (260), Expect = 2e-22
Identities = 73/225 (32%), Positives = 112/225 (49%), Gaps = 26/225 (11%)
Query: 81 RNKLDSLYMTKSLAHRQCLKQQLYFYRMVESKPIMEQLTEFNKIIDDLANIDVNLEDEDK 140
R + Y TK+LA+ LK + ++MVE+K I E L K++ DLA++D+N DED+
Sbjct: 33 REGYEGNYQTKTLANMIYLKHKFTSFKMVENKSIEENLDTSLKLVADLASLDINTSDEDQ 92
Query: 141 ALHLPCALPRSFENFKDTMLYGK-*GTITLEEVQAALRTKELTKFKELKVEDSGEGLNVS 199
AL LP +++ DT+ YG T+TL V ++ +KE+ ELK + LNVS
Sbjct: 93 ALQFMAGLPPQYDSLVDTLKYGSGKDTLTLNAVISSAYSKEI----ELKEKGL---LNVS 145
Query: 200 RERSQ-------NRGKGKGKNSRSKSRSKGDGN-----KTQYKCFICHNPGHFKKDCPER 247
+ S+ R KG S G+G CFIC H+K++CP+R
Sbjct: 146 KPDSEALLGDFKGRSNNKGNKRPCTGSSSGNGKFQSRPSNNTSCFICGKEDHWKRECPQR 205
Query: 248 KGNGGGNPSVQIASNEEGYESAGALTVTSWEPEKGWVLDSGCSYH 292
K + ++A+ LTV++ K WV+DSG ++H
Sbjct: 206 KKH------AELANIATEPPQPLVLTVSTQSTCKEWVMDSGRTFH 244
>At4g04380 putative polyprotein
Length = 778
Score = 94.7 bits (234), Expect = 2e-19
Identities = 51/160 (31%), Positives = 87/160 (53%), Gaps = 1/160 (0%)
Query: 271 ALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQVIGTIRLK 330
AL+ T E W++D+GCSYH+ ++E+ E + E G V +GN +++ +GT+R+
Sbjct: 187 ALSSTDIHLEDEWIMDTGCSYHMTHKREWLEDFDEEAGCSVRMGNKTISRVKGVGTVRIA 246
Query: 331 MFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRISHGALIIAKGSKIHGL 390
+ L++VRYI + R +S+ F+ G E ++R++ G ++ +G + L
Sbjct: 247 NDNGLTVTLQNVRYISDMDRIFLSLGTFEKAGRKFESENDMLRVNAGEQVLLEGRRYDTL 306
Query: 391 YILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWL 430
YIL G D S+A D LWH RL H+S++ + L
Sbjct: 307 YILHGKPA-TDESLAVAKANDDIVLWHRRLCHMSQKNMSL 345
Score = 81.3 bits (199), Expect = 2e-15
Identities = 52/133 (39%), Positives = 79/133 (59%), Gaps = 14/133 (10%)
Query: 50 EMNDKAVSAIILCLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHRQCLKQQLYFYRMV 109
E K S I+L + D+VL E S D LYM+K+L ++ LKQ+LY ++M
Sbjct: 51 EKKRKTRSTIVLSVSDRVL-EAS------------DRLYMSKALPNQIYLKQKLYRFKMS 97
Query: 110 ESKPIMEQLTEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*GTI-T 168
E+ + + EF I+ DL N++V + DED+ + L +LP+SF+ KDT+ Y T+ T
Sbjct: 98 ENLSMEGNIDEFLHIVADLENLNVLVSDEDQTILLLMSLPKSFDQLKDTLQYSSGKTVLT 157
Query: 169 LEEVQAALRTKEL 181
L+EV AA+ +KEL
Sbjct: 158 LDEVTAAIYSKEL 170
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 92.8 bits (229), Expect = 7e-19
Identities = 104/469 (22%), Positives = 194/469 (41%), Gaps = 51/469 (10%)
Query: 11 NDFGLWKVKMRAILIQQKCVEAL-KG--EAQMDVHLTPAEKTEMND------KAVSAIIL 61
+++ W ++M+AIL E + KG E + + L+ +K + D KA+ I
Sbjct: 16 SNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQ 75
Query: 62 CLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHR---QCLKQQLYFYRMVESKPIMEQL 118
L + +V ++A KL + Y + Q L+ + +M E + + +
Sbjct: 76 GLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYF 135
Query: 119 TEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*-GTITLEEVQAALR 177
+ + ++L L+D + +L FE+ + K +T+E++ +L+
Sbjct: 136 SRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQ 195
Query: 178 TKELTKFKELKVEDSGEGLNVSRERS----------QNRGKGKG--------------KN 213
E K K+ + + + +++E + Q RG+G+G N
Sbjct: 196 AYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTN 255
Query: 214 SRSKSRSKGDG--------NKTQYKCFICHNPGHFKKDC--PERKGNGGGNPSVQIASNE 263
R ++ S+G G +K+ KC+ C GH+ +C P K V+ E
Sbjct: 256 QRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFKEKANYVEEKIQE 315
Query: 264 EGYESAGALTVTSWEPEKGWVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACKIQV 323
E + E W LDSG S H+C RK F L+ G V LG+ +++
Sbjct: 316 EDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKMEVKG 375
Query: 324 IGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRI--SHGALII 381
G I +++ + + +V YIP ++ N++S+ GY R++ + I LI
Sbjct: 376 KGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDKESNLIT 435
Query: 382 AKGSKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWL 430
+ +++L IA + + + LWHLR GH++ G+ L
Sbjct: 436 KVPMSKNRMFVLNIRNDIAQC--LKMCYKEESWLWHLRFGHLNFGGLEL 482
>At1g58140 hypothetical protein
Length = 1320
Score = 92.8 bits (229), Expect = 7e-19
Identities = 104/474 (21%), Positives = 200/474 (41%), Gaps = 61/474 (12%)
Query: 11 NDFGLWKVKMRAILIQQKCVEAL-KG--EAQMDVHLTPAEKTEMND------KAVSAIIL 61
+++ W ++M+AIL E + KG E + + L+ +K + D KA+ I
Sbjct: 16 SNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQ 75
Query: 62 CLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHR---QCLKQQLYFYRMVESKPIMEQL 118
L + +V ++A KL + Y + Q L+ + +M E + + +
Sbjct: 76 GLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYF 135
Query: 119 TEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*-GTITLEEVQAALR 177
+ + ++L L+D + +L FE+ + K +T+E++ +L+
Sbjct: 136 SRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQ 195
Query: 178 TKELTKFKELKVEDSGEGLNVSRERS----------QNRGKGKG--------------KN 213
E K K+ + + + +++E + Q RG+G+G N
Sbjct: 196 AYEEKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTN 255
Query: 214 SRSKSRSKGDG--------NKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASN--E 263
R ++ S+G G +K+ KC+ C GH+ +C N + +N E
Sbjct: 256 QRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASEC-----KAPSNKKFEEKANYVE 310
Query: 264 EGYESAGALTVTSWEPEKG-----WVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKA 318
E + L + S++ ++ W LDSG S H+C RK F L+ G V LG+
Sbjct: 311 EKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESK 370
Query: 319 CKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRI--SH 376
+++ G I +++ + + +V YIP ++ N++S+ GY R++ + I
Sbjct: 371 MEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQE 430
Query: 377 GALIIAKGSKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWL 430
LI + +++L IA + + + LWHLR GH++ G+ L
Sbjct: 431 SNLITKVPMSKNRMFVLNIRNDIAQC--LKMCYKEESWLWHLRFGHLNFGGLEL 482
>At1g48710 hypothetical protein
Length = 1352
Score = 92.4 bits (228), Expect = 1e-18
Identities = 104/474 (21%), Positives = 200/474 (41%), Gaps = 61/474 (12%)
Query: 11 NDFGLWKVKMRAILIQQKCVEAL-KG--EAQMDVHLTPAEKTEMND------KAVSAIIL 61
+++ W ++M+AIL E + KG E + + L+ +K + D KA+ I
Sbjct: 16 SNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQ 75
Query: 62 CLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHR---QCLKQQLYFYRMVESKPIMEQL 118
L + +V ++A KL + Y + Q L+ + +M E + + +
Sbjct: 76 GLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYF 135
Query: 119 TEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*-GTITLEEVQAALR 177
+ + ++L L+D + +L FE+ + K +T+E++ +L+
Sbjct: 136 SRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQ 195
Query: 178 TKELTKFKELKVEDSGEGLNVSRERS----------QNRGKGKG--------------KN 213
E K K+ + + + +++E + Q RG+G+G N
Sbjct: 196 AYEEKKKKKEDIIEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTN 255
Query: 214 SRSKSRSKGDG--------NKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASN--E 263
R ++ S+G G +K+ KC+ C GH+ +C N + +N E
Sbjct: 256 QRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASEC-----KAPSNKKFEEKANYVE 310
Query: 264 EGYESAGALTVTSWEPEKG-----WVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKA 318
E + L + S++ ++ W LDSG S H+C RK F L+ G V LG+
Sbjct: 311 EKIQEEDMLLMASYKKDEQEENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESK 370
Query: 319 CKIQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRI--SH 376
+++ G I +++ + + +V YIP ++ N++S+ GY R++ + I
Sbjct: 371 MEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQE 430
Query: 377 GALIIAKGSKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWL 430
LI + +++L IA + + + LWHLR GH++ G+ L
Sbjct: 431 SNLITKVPMSKNRMFVLNIRNDIAQC--LKMCYKEESWLWHLRFGHLNFGGLEL 482
>At3g61330 copia-type polyprotein
Length = 1352
Score = 91.7 bits (226), Expect = 2e-18
Identities = 102/472 (21%), Positives = 198/472 (41%), Gaps = 57/472 (12%)
Query: 11 NDFGLWKVKMRAILIQQKCVEAL-KG--EAQMDVHLTPAEKTEMND------KAVSAIIL 61
+++ W ++M+AIL E + KG E + + L+ +K + D KA+ I
Sbjct: 16 SNYDNWSLRMKAILGAHDVWEIVEKGFIEPENEGSLSQTQKDGLRDSRKRDKKALCLIYQ 75
Query: 62 CLGDKVLREVSRESTAVSMRNKLDSLYMTKSLAHR---QCLKQQLYFYRMVESKPIMEQL 118
L + +V ++A KL + Y + Q L+ + +M E + + +
Sbjct: 76 GLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLRGEFEALQMKEGELVSDYF 135
Query: 119 TEFNKIIDDLANIDVNLEDEDKALHLPCALPRSFENFKDTMLYGK*-GTITLEEVQAALR 177
+ + ++L L+D + +L FE+ + K +T+E++ +L+
Sbjct: 136 SRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEETKDLEAMTIEQLLGSLQ 195
Query: 178 TKELTKFKELKVEDSGEGLNVSRERS----------QNRGKGKG--------------KN 213
E K K+ + + + +++E + Q RG+G+G N
Sbjct: 196 AYEEKKKKKEDIAEQVLNMQITKEENGQSYQRRGGGQVRGRGRGGYGNGRGWRPHEDNTN 255
Query: 214 SRSKSRSKGDG--------NKTQYKCFICHNPGHFKKDCPERKGNGGGNPSVQIASNEEG 265
R ++ S+G G +K+ KC+ C GH+ +C + + EE
Sbjct: 256 QRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYASECKAPSNKKFEEKAHYV---EEK 312
Query: 266 YESAGALTVTSWEPEKG-----WVLDSGCSYHICPRKEYFEMLELEEGGVVCLGNNKACK 320
+ L + S++ ++ W LDSG S H+C RK F L+ G V LG+ +
Sbjct: 313 IQEEDMLLMASYKKDEQKENHKWYLDSGASNHMCGRKSMFAELDESVRGNVALGDESKME 372
Query: 321 IQVIGTIRLKMFDDRDFLLKDVRYIPKLRRNLISISMFDGLGYCTRIERGVMRI--SHGA 378
++ G I +++ + + +V YIP ++ N++S+ GY R++ + I
Sbjct: 373 VKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSIRDQESN 432
Query: 379 LIIAKGSKIHGLYILEGSTVIADASVASVDTLDVTKLWHLRLGHVSERGIWL 430
LI + +++L IA + + + LWHLR GH++ G+ L
Sbjct: 433 LITKVPMSKNRMFVLNIRNDIAQC--LKMCYKEESWLWHLRFGHLNFGGLEL 482
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.344 0.151 0.505
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,223,233
Number of Sequences: 26719
Number of extensions: 685579
Number of successful extensions: 3931
Number of sequences better than 10.0: 143
Number of HSP's better than 10.0 without gapping: 74
Number of HSP's successfully gapped in prelim test: 69
Number of HSP's that attempted gapping in prelim test: 3525
Number of HSP's gapped (non-prelim): 258
length of query: 866
length of database: 11,318,596
effective HSP length: 108
effective length of query: 758
effective length of database: 8,432,944
effective search space: 6392171552
effective search space used: 6392171552
T: 11
A: 40
X1: 15 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.6 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144730.2


