

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144729.7 - phase: 0
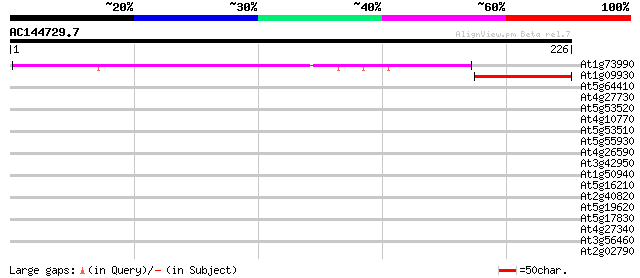
(226 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g73990 putative protease SppA (SppA) 160 5e-40
At1g09930 unknown protein 44 9e-05
At5g64410 Isp4-like protein 40 0.001
At4g27730 unknown protein 38 0.004
At5g53520 isp4 protein 37 0.011
At4g10770 oligopeptide transporter like protein 33 0.094
At5g53510 isp4 protein 33 0.16
At5g55930 sexual differentiation process protein ISP4-like 32 0.27
At4g26590 isp4 like protein 30 0.79
At3g42950 polygalacturonase -like protein 30 0.79
At1g50940 unknown protein 28 3.9
At5g16210 unknown protein 28 5.1
At2g40820 hypothetical protein 28 5.1
At5g19620 unknown protein 27 6.7
At5g17830 putative protein 27 8.8
At4g27340 putative protein 27 8.8
At3g56460 quinone reductase-like protein 27 8.8
At2g02790 unknown protein 27 8.8
>At1g73990 putative protease SppA (SppA)
Length = 677
Score = 160 bits (405), Expect = 5e-40
Identities = 100/226 (44%), Positives = 130/226 (57%), Gaps = 42/226 (18%)
Query: 2 ISFRKYSKVRKWTVGISEAKEQIAIIRASGTMD----------SDIVTSNFIEKIGMVKD 51
+ ++KYS V+KWT+G++ ++QIAIIRA G++ S I+ IEKI V++
Sbjct: 354 VDYKKYSGVKKWTLGLTGGRDQIAIIRAGGSISRVKGPLSTPGSAIIAEQLIEKIRSVRE 413
Query: 52 SKKFKAVIVRIDSVGGDFHASQSTWEAIRSLASKKPVIASMSDAATSAGYYMAMGAGAIV 111
SKK+KA I+RIDS GGD AS W I+ LA KPVIASMSD A S GYYMAM A AIV
Sbjct: 414 SKKYKAAIIRIDSPGGDALASDLMWREIKLLAETKPVIASMSDVAASGGYYMAMAANAIV 473
Query: 112 AENLTLTGSIRGTVSQNFNL---------DNELPSFIPY-------------DDAKVLDN 149
AENLTLTGSI G V+ F L + E S Y ++A++ +
Sbjct: 474 AENLTLTGSI-GVVTARFTLAKLYEKIGFNKETISRGKYAELLGAEERPLKPEEAELFEK 532
Query: 150 VV---------RLRLMTLGKLDKMKKVAHGRIWTGKDAASHGLVDA 186
+ L +DKM++VA GR+WTGKDA S GL+DA
Sbjct: 533 SAQHAYQLFRDKAALSRSMPVDKMEEVAQGRVWTGKDAHSRGLIDA 578
>At1g09930 unknown protein
Length = 734
Score = 43.5 bits (101), Expect = 9e-05
Identities = 19/39 (48%), Positives = 25/39 (63%)
Query: 188 MPVISYGFAGMPPATPTNFARWPFTRMIFNYFVFRFHKR 226
+PV+ A MPPAT NF W +IFNYFVF++ K+
Sbjct: 631 LPVLLGATAAMPPATSVNFNCWIIVGVIFNYFVFKYCKK 669
>At5g64410 Isp4-like protein
Length = 729
Score = 39.7 bits (91), Expect = 0.001
Identities = 18/38 (47%), Positives = 22/38 (57%)
Query: 188 MPVISYGFAGMPPATPTNFARWPFTRMIFNYFVFRFHK 225
+PV+ A MPPAT N+ W IFN FVFR+ K
Sbjct: 627 LPVLLGATAMMPPATAVNYNSWILVGTIFNLFVFRYRK 664
>At4g27730 unknown protein
Length = 736
Score = 38.1 bits (87), Expect = 0.004
Identities = 15/38 (39%), Positives = 22/38 (57%)
Query: 188 MPVISYGFAGMPPATPTNFARWPFTRMIFNYFVFRFHK 225
+PV+ A MPPAT NF W IF +F+F++ +
Sbjct: 632 IPVLVGATAMMPPATAVNFTSWLIVAFIFGHFIFKYRR 669
>At5g53520 isp4 protein
Length = 733
Score = 36.6 bits (83), Expect = 0.011
Identities = 15/41 (36%), Positives = 23/41 (55%)
Query: 185 DAGMPVISYGFAGMPPATPTNFARWPFTRMIFNYFVFRFHK 225
D +PV+ A MPPA+ NF W +F +FVF++ +
Sbjct: 628 DIHIPVLIGATAIMPPASAVNFTSWLVMAFVFGHFVFKYRR 668
>At4g10770 oligopeptide transporter like protein
Length = 766
Score = 33.5 bits (75), Expect = 0.094
Identities = 13/36 (36%), Positives = 19/36 (52%)
Query: 188 MPVISYGFAGMPPATPTNFARWPFTRMIFNYFVFRF 223
MPV+ + MPPAT N+ W + + VFR+
Sbjct: 663 MPVLISATSSMPPATAVNYTTWVLAGFLSGFVVFRY 698
>At5g53510 isp4 protein
Length = 741
Score = 32.7 bits (73), Expect = 0.16
Identities = 12/38 (31%), Positives = 21/38 (54%)
Query: 189 PVISYGFAGMPPATPTNFARWPFTRMIFNYFVFRFHKR 226
PVI + MPPA NF W +F +F++++ ++
Sbjct: 638 PVILGATSMMPPAMAVNFTSWCIVAFVFGHFLYKYKRQ 675
>At5g55930 sexual differentiation process protein ISP4-like
Length = 755
Score = 32.0 bits (71), Expect = 0.27
Identities = 12/38 (31%), Positives = 21/38 (54%)
Query: 188 MPVISYGFAGMPPATPTNFARWPFTRMIFNYFVFRFHK 225
+P+I + MP A ++ W ++FNY++FR K
Sbjct: 651 VPLIFSAVSAMPQAKAVHYWSWAIVGVVFNYYIFRRFK 688
>At4g26590 isp4 like protein
Length = 753
Score = 30.4 bits (67), Expect = 0.79
Identities = 13/38 (34%), Positives = 22/38 (57%)
Query: 188 MPVISYGFAGMPPATPTNFARWPFTRMIFNYFVFRFHK 225
+P+I G MP A ++ W ++FNY++FR +K
Sbjct: 649 IPLIFSGANVMPMAKAVHYWSWFAVGIVFNYYIFRRYK 686
>At3g42950 polygalacturonase -like protein
Length = 484
Score = 30.4 bits (67), Expect = 0.79
Identities = 21/68 (30%), Positives = 31/68 (44%), Gaps = 8/68 (11%)
Query: 64 SVGGDFHASQSTWEAI--------RSLASKKPVIASMSDAATSAGYYMAMGAGAIVAENL 115
SVG D A +S W+ +++ + +I SM A S G M+ G I EN+
Sbjct: 272 SVGDDGIAIKSGWDQYGTTYGKPSKNILIRNLIIRSMVSAGISIGSEMSGGVSNITVENI 331
Query: 116 TLTGSIRG 123
+ S RG
Sbjct: 332 LIWSSRRG 339
>At1g50940 unknown protein
Length = 363
Score = 28.1 bits (61), Expect = 3.9
Identities = 31/130 (23%), Positives = 52/130 (39%), Gaps = 7/130 (5%)
Query: 1 MISFRKYS-KVRKWTVGISEAKEQIAIIRASGTMDSDIVTSNFIEKIGMVKDSKKFKAVI 59
M++ R S V T K ++ I S D + S ++ + +D+++
Sbjct: 186 MLTIRSTSFPVTPITANSESKKATVSQIDLSNFEDDSVSKSRYVGR--STQDTERPDLGS 243
Query: 60 VRIDSVGGDFHASQSTWEAIRSLASKKPVIASMSDAATSAGYY---MAMG-AGAIVAENL 115
R+ GG S ++ I LA K + AA AGY + +G G IVA L
Sbjct: 244 ARVVITGGRALKSVENFKMIEKLAEKLGGAVGATRAAVDAGYVPNDLQVGQTGKIVAPEL 303
Query: 116 TLTGSIRGTV 125
+ + G +
Sbjct: 304 YMAFGVSGAI 313
>At5g16210 unknown protein
Length = 1180
Score = 27.7 bits (60), Expect = 5.1
Identities = 21/65 (32%), Positives = 27/65 (41%), Gaps = 1/65 (1%)
Query: 121 IRGTVSQNFNLDNELPSFIPYDDAKVLDNVVRLRLMTLGKLDKMKKVAHGRIWTGKDAAS 180
+ T N L + +PY DAKV V L+TLG D+ V + I A
Sbjct: 903 VDSTAELKINAAKLLKTIVPYIDAKVASANVLPALITLGS-DQNLNVKYASIDAFGSVAQ 961
Query: 181 HGLVD 185
H VD
Sbjct: 962 HFKVD 966
>At2g40820 hypothetical protein
Length = 945
Score = 27.7 bits (60), Expect = 5.1
Identities = 16/52 (30%), Positives = 24/52 (45%), Gaps = 7/52 (13%)
Query: 95 AATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQNFNLDNELPSFIPYDDAKV 146
A T G+ + +GAGA+V L GS F +LPS +P+ +
Sbjct: 895 ARTGPGFAVGVGAGAVVGAGAALYGS-------EFMSGIDLPSSLPHPSVPI 939
>At5g19620 unknown protein
Length = 732
Score = 27.3 bits (59), Expect = 6.7
Identities = 31/110 (28%), Positives = 51/110 (46%), Gaps = 9/110 (8%)
Query: 17 ISEAKEQIAIIRASGTMDS----DIVTSNFIEKIGMVKDSKKFKAVIVRIDSVGGDFHAS 72
I E +E + I SG S + T + I + V+ +++F+ ++ +V S
Sbjct: 201 IREVQEDVHRIIESGYFCSCTPVAVDTRDGIRLMFQVEPNQEFRGLVCENANV----LPS 256
Query: 73 QSTWEAIRSLASKKPVIASMSDAATSA-GYYMAMGAGAIVAENLTLTGSI 121
+ EA R K I + +A TS G+YM G IV++ TL+G I
Sbjct: 257 KFIHEAFRDGFGKVINIKRLEEAITSINGWYMERGLFGIVSDIDTLSGGI 306
>At5g17830 putative protein
Length = 474
Score = 26.9 bits (58), Expect = 8.8
Identities = 15/42 (35%), Positives = 22/42 (51%), Gaps = 2/42 (4%)
Query: 71 ASQSTWEAIRSLASKKPVIASMSDAATSAGYYMAMGAGAIVA 112
AS TW R++ + +I D +S ++ AMG GAI A
Sbjct: 343 ASSDTWRRFRTIPEFEQLIDF--DITSSICFFSAMGIGAIAA 382
>At4g27340 putative protein
Length = 562
Score = 26.9 bits (58), Expect = 8.8
Identities = 14/57 (24%), Positives = 24/57 (41%)
Query: 17 ISEAKEQIAIIRASGTMDSDIVTSNFIEKIGMVKDSKKFKAVIVRIDSVGGDFHASQ 73
+ A E + I D + I K+ + K+ K + V+ +ID + DF Q
Sbjct: 340 VPSAFEMVGHIAHLNLRDEHLPYKRLIAKVVLDKNQPKIQTVVNKIDPIHNDFRTMQ 396
>At3g56460 quinone reductase-like protein
Length = 348
Score = 26.9 bits (58), Expect = 8.8
Identities = 15/54 (27%), Positives = 27/54 (49%), Gaps = 1/54 (1%)
Query: 15 VGISEAKEQIAIIRASGTMDS-DIVTSNFIEKIGMVKDSKKFKAVIVRIDSVGG 67
+ ++ E+I ++++ G D+ T N I + ++K K V V D VGG
Sbjct: 179 IAVARGTEKIQLLKSMGVDHVVDLGTENVISSVKEFIKTRKLKGVDVLYDPVGG 232
>At2g02790 unknown protein
Length = 650
Score = 26.9 bits (58), Expect = 8.8
Identities = 17/55 (30%), Positives = 28/55 (50%), Gaps = 5/55 (9%)
Query: 79 IRSLASKKPVIASMSDAATSAGYYMAMGAGAIVAENLTLTGSIRGTVSQNFNLDN 133
+ L ++ PV++S AAT + ++AE L+G I G S N NL++
Sbjct: 53 VSKLPTEPPVVSSQEVAATQT----VVVPDVVIAEK-QLSGDIEGDESSNVNLES 102
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.134 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 4,692,038
Number of Sequences: 26719
Number of extensions: 179973
Number of successful extensions: 478
Number of sequences better than 10.0: 18
Number of HSP's better than 10.0 without gapping: 10
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 466
Number of HSP's gapped (non-prelim): 19
length of query: 226
length of database: 11,318,596
effective HSP length: 96
effective length of query: 130
effective length of database: 8,753,572
effective search space: 1137964360
effective search space used: 1137964360
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144729.7


