

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.5 - phase: 0
(497 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
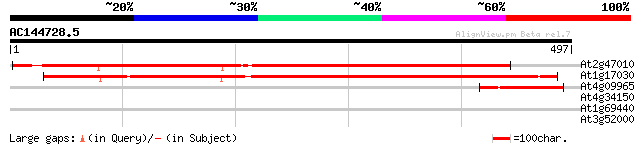
At2g47010 unknown protein 639 0.0
At1g17030 unknown protein 601 e-172
At4g09965 putative protein 101 8e-22
At4g34150 unknown protein 29 6.9
At1g69440 hypothetical protein 29 6.9
At3g52000 serine-type carboxypeptidase like protein 28 8.9
>At2g47010 unknown protein
Length = 451
Score = 639 bits (1648), Expect = 0.0
Identities = 303/462 (65%), Positives = 354/462 (76%), Gaps = 33/462 (7%)
Query: 3 KQLQKFFVLSNVMLLCASLVSTTQSQTNVSAVGDPGMQRDGLRVAFEAWNFCNEVGQEAP 62
KQL + N++LL SAVGDPGM+RDGLRVAFEAWNFCNEVG EAP
Sbjct: 2 KQLAPILAVVNLILLVHG--------DERSAVGDPGMKRDGLRVAFEAWNFCNEVGFEAP 53
Query: 63 YMGSPRAAQCFDLSQ-----------------GSLIHKVTEKDNKLGVGESLPGLRPED- 104
+MGSPRAA CFDLS SL+HKV++ DN+LG+G+ PG+ E
Sbjct: 54 HMGSPRAADCFDLSSKCIKAYTEDQSNKTTSGSSLVHKVSDSDNELGIGKPKPGIISESA 113
Query: 105 INNIDLYAVQKELYLGSLCEVKDTPRPWQFWMIMLKNGNYDSRSGLCPQDGKRVPPF-KP 163
++N DLYAV+KELYLGSLC+V D P PW FWM+MLKNGNYD++S LCP++GK++PPF +P
Sbjct: 114 LHNPDLYAVEKELYLGSLCQVSDKPNPWSFWMVMLKNGNYDTKSALCPKNGKKIPPFNQP 173
Query: 164 GRFPCFGTGCMNQPILCHQWTKLE--GDVMRGGFNGTYDLDSSCRDSGLDDNNNLSFYEV 221
G FPCFG+GCMNQP L H T+L+ G M+G FNGTY+ + +GLD +S+YEV
Sbjct: 174 GLFPCFGSGCMNQPTLNHGKTELQRDGQTMKGWFNGTYEQGADF-GNGLD---GISYYEV 229
Query: 222 VWEKKVNVGSWLFKHKLKTSKKYPWLMLYLRADTVSGFSGGYHYDTRGMLKAPLESPNFK 281
VWEK+V VG W+FKHKLKTS KYPWLMLYLRAD GFSGGYHYDTRGMLK ESPNFK
Sbjct: 230 VWEKRVGVGGWVFKHKLKTSAKYPWLMLYLRADATKGFSGGYHYDTRGMLKTLPESPNFK 289
Query: 282 VRLTLDVKKGGGSKSQFYLLDMGSCWKNNGAPCDGDVLTDVTRYSEMIINPETPAWCSPT 341
VRLTL+VK+GGG+KSQFYLLD+GSCWKNNG PCDGDV TDVTRYSEMIINPETP WC+P
Sbjct: 290 VRLTLNVKQGGGAKSQFYLLDIGSCWKNNGKPCDGDVTTDVTRYSEMIINPETPLWCNPK 349
Query: 342 GLGNCPPFHITPDNKKIFRNDTANFPYSAYHYYCAPGNAQHLEQPVSTCDPYSNPQAQEI 401
L NCPP+H + ++ R D +FPY AYH YCAPGNA+HLE PV TCD YSNPQAQEI
Sbjct: 350 SLHNCPPYHTFRNGTRVHRTDHRSFPYEAYHVYCAPGNAEHLELPVGTCDAYSNPQAQEI 409
Query: 402 VQLLPHPIWGEYGYPINKGDGWVGDARTWELDVGGLASRLYF 443
+QLLPHP+WGEYGYP GDGWVGD RTW+LDVGGL+SRL+F
Sbjct: 410 LQLLPHPVWGEYGYPTRLGDGWVGDPRTWDLDVGGLSSRLFF 451
>At1g17030 unknown protein
Length = 502
Score = 601 bits (1549), Expect = e-172
Identities = 282/465 (60%), Positives = 343/465 (73%), Gaps = 17/465 (3%)
Query: 31 VSAVGDPGMQRDGLRVAFEAWNFCNEVGQEAPYMGSPRAAQCFDLSQGS----LIHKVTE 86
VSAVGDPGM+ D LRVA EAWN CNEVG+EA MGSPR A CFD+ S +IHKV E
Sbjct: 40 VSAVGDPGMRNDNLRVAIEAWNQCNEVGEEATNMGSPRMADCFDIDNSSFPVKIIHKVDE 99
Query: 87 KDNKLGVGE-SLPGLRPEDINNIDLYAVQKELYLGSLCEVKDTPRPWQFWMIMLKNGNYD 145
+DN+LGVG + G+ D N D+YA QKE+YLG+ C+V D P PWQFWMIMLKNGN D
Sbjct: 100 RDNRLGVGNGTYGGISAGD--NADIYAAQKEVYLGNKCQVVDKPNPWQFWMIMLKNGNTD 157
Query: 146 SRSGLCPQDGKRVPPFKP-GRFPCFGTGCMNQPILCHQWTKL---EGDVMRGGFNGTYDL 201
+ + +CP++GK+ PF P GRFPCFG GCMN P + H++T L E M G F GT+DL
Sbjct: 158 TLAAICPENGKKAKPFPPTGRFPCFGKGCMNMPSMHHEYTSLVDNEEGHMSGSFYGTWDL 217
Query: 202 DSSCRDSGLDDNNNLSFYEVVWEKKVNVG-SWLFKHKLKTSKKYPWLMLYLRADTVSGFS 260
D+ +D N S+Y+V WEKK+ SW+F H LKTS KYPWLMLYLRAD GFS
Sbjct: 218 DNDQKDPV----GNNSYYKVKWEKKIGGNESWVFHHLLKTSSKYPWLMLYLRADASRGFS 273
Query: 261 GGYHYDTRGMLKAPLESPNFKVRLTLDVKKGGGSKSQFYLLDMGSCWKNNGAPCDGDVLT 320
GGYHYDTRGM+K L+SP+FKV+ L++ KGGGS SQFYL+DMGSCWKN+G CDGDV T
Sbjct: 274 GGYHYDTRGMMKMTLKSPDFKVKFKLEIIKGGGSGSQFYLMDMGSCWKNDGRDCDGDVTT 333
Query: 321 DVTRYSEMIINPETPAWCSPTGLGNCPPFHITPDNKKIFRNDTANFPYSAYHYYCAPGNA 380
DVTRYSEMIINP A C+ LG CPP H P+ K+ R D FP+ AYHYYC PGNA
Sbjct: 334 DVTRYSEMIINPGATAVCTRNRLGACPPEHTFPNGTKVHRTDKEKFPFEAYHYYCVPGNA 393
Query: 381 QHLEQPVSTCDPYSNPQAQEIVQLLPHPIWGEYGYPINKGDGWVGDARTWELDVGGLASR 440
+ E P CDPYSNPQ QEI+Q+LPHP+W ++GYP KG GW+GD RTWELDVG L+
Sbjct: 394 RFAESPYEVCDPYSNPQPQEILQILPHPVWEQFGYPTKKGQGWIGDPRTWELDVGKLSQS 453
Query: 441 LYFYQDPGTSPAKRIWTSIDTGTEIFVSDKDEVAEWTISDFDVIL 485
L+FYQDPGT P +R W+SID GTEI++S K+++AEWT++DFD+++
Sbjct: 454 LFFYQDPGTKPVERHWSSIDLGTEIYMS-KNQIAEWTVTDFDIVI 497
>At4g09965 putative protein
Length = 223
Score = 101 bits (252), Expect = 8e-22
Identities = 45/75 (60%), Positives = 64/75 (85%), Gaps = 2/75 (2%)
Query: 417 INKGDGWVGDARTWELDVGGLASRLYFYQD-PGTSPAKRIWTSIDTGTEIFVSDKDEVAE 475
I +G+GW+GD+RTWE++ G L+SRLYFYQ+ PGT PAKR+WTSI+ T+I+VS++ E AE
Sbjct: 147 IKQGNGWIGDSRTWEVN-GALSSRLYFYQEYPGTKPAKRMWTSINVVTDIYVSNRQETAE 205
Query: 476 WTISDFDVILIQPKS 490
WT+SDFDV++ Q ++
Sbjct: 206 WTVSDFDVLVQQKET 220
>At4g34150 unknown protein
Length = 247
Score = 28.9 bits (63), Expect = 6.9
Identities = 21/55 (38%), Positives = 24/55 (43%), Gaps = 9/55 (16%)
Query: 367 PYSAY-HYYCAPGNAQHLEQPVSTCDPYSNPQAQEIVQLLPHPIWGEYGYPINKG 420
PY+ + Y AP +A P ST PYS P VQ P P GYP G
Sbjct: 143 PYAPHVPQYSAPPSAS----PYSTAPPYSGPSLYPQVQQYPQP----SGYPPASG 189
>At1g69440 hypothetical protein
Length = 990
Score = 28.9 bits (63), Expect = 6.9
Identities = 18/64 (28%), Positives = 27/64 (42%)
Query: 345 NCPPFHITPDNKKIFRNDTANFPYSAYHYYCAPGNAQHLEQPVSTCDPYSNPQAQEIVQL 404
N PPF + P + + +N P S Y+YY +Q P+ P + + L
Sbjct: 37 NPPPFLLHPSSHQNLNLVASNLPSSYYYYYYCYFYSQFHNSLPPPPPPHLLPLSPPLPPL 96
Query: 405 LPHP 408
LP P
Sbjct: 97 LPLP 100
>At3g52000 serine-type carboxypeptidase like protein
Length = 482
Score = 28.5 bits (62), Expect = 8.9
Identities = 19/48 (39%), Positives = 22/48 (45%), Gaps = 9/48 (18%)
Query: 348 PFHITPDNKKIFRNDTANFPYSAYHYYCAPGNAQHLEQPVSTCDPYSN 395
PF + D K +FRN PYS + N LE PV T YSN
Sbjct: 131 PFRVHSDGKTLFRN-----PYS----WNNEANVLFLETPVGTGFSYSN 169
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.138 0.446
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,108,150
Number of Sequences: 26719
Number of extensions: 641572
Number of successful extensions: 1114
Number of sequences better than 10.0: 6
Number of HSP's better than 10.0 without gapping: 3
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 1098
Number of HSP's gapped (non-prelim): 6
length of query: 497
length of database: 11,318,596
effective HSP length: 103
effective length of query: 394
effective length of database: 8,566,539
effective search space: 3375216366
effective search space used: 3375216366
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144728.5


