

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.4 + phase: 0
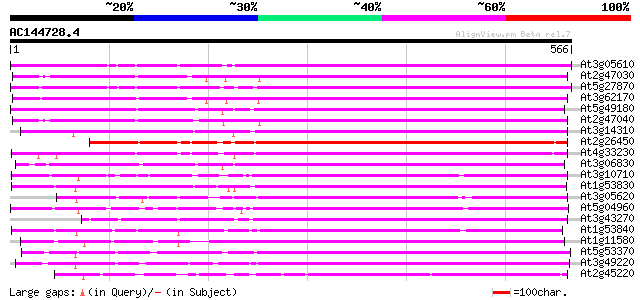
(566 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g05610 putative pectinesterase 416 e-116
At2g47030 putative pectinesterase 408 e-114
At5g27870 pectin methyl-esterase - like protein 405 e-113
At3g62170 PECTINESTERASE-like protein 399 e-111
At5g49180 pectin methylesterase 389 e-108
At2g47040 putative pectinesterase 389 e-108
At3g14310 pectin methylesterase like protein 382 e-106
At2g26450 putative pectinesterase 375 e-104
At4g33230 pectinesterase - like protein 373 e-103
At3g06830 pectin methylesterase like protein 370 e-103
At3g10710 putative pectinesterase 367 e-102
At1g53830 unknown protein 355 5e-98
At3g05620 putative pectinesterase 354 6e-98
At5g04960 pectinesterase 352 3e-97
At3g43270 pectinesterase like protein 343 1e-94
At1g53840 unknown protein 343 2e-94
At1g11580 unknown protein 341 6e-94
At5g53370 pectinesterase 339 3e-93
At3g49220 pectinesterase - like protein 321 8e-88
At2g45220 pectinesterase like protein 317 1e-86
>At3g05610 putative pectinesterase
Length = 669
Score = 416 bits (1069), Expect = e-116
Identities = 225/569 (39%), Positives = 336/569 (58%), Gaps = 16/569 (2%)
Query: 2 ERKIFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFC 61
+R+ V +S +L++ + + V V N D+ E+ + KAV VC +D K C
Sbjct: 12 KRRYIVITISSVLLISMVVAVTVGVSLNKHDGDSKGKAEVNASVKAVKDVCAPTDYRKTC 71
Query: 62 ADTL--GSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDL 119
DTL NT+DP + +K +++ + A + + +E +K+++ T+MALD CK+L
Sbjct: 72 EDTLIKNGKNTTDPMELVKTAFNVTMKQITDAAKKSQTI-MELQKDSR-TRMALDQCKEL 129
Query: 120 LEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQ 179
+++A+DEL ++S H +++ +L+ WL A +++++CL+GF G + +
Sbjct: 130 MDYALDEL-SNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGFQ--GTQGNAGE 186
Query: 180 LQTGSLDHVGKLTALALDVVTAITKVLAALDLD-LNVKPSSRRLFEVDEDGNPEWMSGAD 238
+L +LT L +++ ++ + + + LN SRRL +G P W+
Sbjct: 187 TMKKALKTAIELTHNGLAIISEMSNFVGQMQIPGLN----SRRLLA---EGFPSWVDQRG 239
Query: 239 RKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQI 298
RKLL + V P+ VVA+DGSG++KT+ +A+ PK +V+++KAG+Y EY+Q+
Sbjct: 240 RKLLQAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVHIKAGLYKEYVQV 299
Query: 299 DKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKH 358
+KT +++ GDGP KTII+G KN+ DG+ T +TAT + V FIAK + FENTAGA KH
Sbjct: 300 NKTMSHLVFIGDGPDKTIISGNKNYKDGITTYRTATVAIVGNYFIAKNIGFENTAGAIKH 359
Query: 359 QAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKI 418
QAVA+RVQ D+S FF+C GYQDTLY H+HRQF+R+C ISGT+DF+FG A+ V QN +
Sbjct: 360 QAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFLFGDAAAVFQNCTL 419
Query: 419 VVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSR 478
+VRKP NQ I A G TG V Q C I EP + +++L RPWK YSR
Sbjct: 420 LVRKPLPNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSKAYLGRPWKEYSR 479
Query: 479 AIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADA 537
I M I D +QP G+ PW G L T F++E NTGPGS + RV W G LS+ D
Sbjct: 480 TIIMNTFIPDFVQPQGWQPWLGDFGLKTLFYSEVQNTGPGSALANRVTWAGIKTLSEEDI 539
Query: 538 TKYTAAQWIEGGVWLPATGIPFDLGFTKG 566
K+T AQ+I+G W+P G+P+ G G
Sbjct: 540 LKFTPAQYIQGDDWIPGKGVPYTTGLLAG 568
>At2g47030 putative pectinesterase
Length = 588
Score = 408 bits (1048), Expect = e-114
Identities = 234/597 (39%), Positives = 336/597 (56%), Gaps = 51/597 (8%)
Query: 4 KIFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFCAD 63
K+ VS S++L+VGVA+GVVA + N DAN L+ KAV +CQ++ D C
Sbjct: 4 KVVVSVASILLIVGVAIGVVAFINKNG---DAN----LSPQMKAVQGICQSTSDKASCVK 56
Query: 64 TLGSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKN-NQSTKMALDDCKDLLEF 122
TL V + DPN IKA + + + + K+ N T + V + + + K LD CK + +
Sbjct: 57 TLEPVKSEDPNKLIKAFMLATKDELTKSSNFTGQTEVNMGSSISPNNKAVLDYCKRVFMY 116
Query: 123 AIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQT 182
A+++L ++I+ + + + LK WL V+ YQ CLD + D ++ +
Sbjct: 117 ALEDL--ATIIEEMGEDLSQIGSKIDQLKQWLIGVYNYQTDCLDDIEEDDLRKAIGE--- 171
Query: 183 GSLDHVGKLTALALD----VVTAITKVLAALDLDLNVK---------------------- 216
+ + LT A+D VV+A+ K+ +D N+
Sbjct: 172 -GIANSKILTTNAIDIFHTVVSAMAKINNKVDDLKNMTGGIPTPGAPPVVDESPVADPDG 230
Query: 217 PSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMS--------VTPNAVVAKDGSGKFKTV 268
P+ R L ++DE G P W+SGADRKL+A G V N VVAKDGSG+FKTV
Sbjct: 231 PARRLLEDIDETGIPTWVSGADRKLMAKAGRGRRGGRGGGARVRTNFVVAKDGSGQFKTV 290
Query: 269 LDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFV--DG 326
A+++ P+N++GR +IY+KAG+Y E + I K K NI ++GDG KT+I+ ++ G
Sbjct: 291 QQAVDACPENNRGRCIIYIKAGLYREQVIIPKKKNNIFMFGDGARKTVISYNRSVALSRG 350
Query: 327 VKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYA 386
T +AT +EGF+AK M F+NTAG HQA A+RV GD++ F+C GYQDTLY
Sbjct: 351 TTTSLSATVQVESEGFMAKWMGFKNTAGPMGHQAAAIRVNGDRAVIFNCRFDGYQDTLYV 410
Query: 387 HAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGT-VQKNMPTGV 445
+ RQFYRNC +SGTVDFIFG ++TVIQN+ IVVRK Q N + ADG + M G+
Sbjct: 411 NNGRQFYRNCVVSGTVDFIFGKSATVIQNTLIVVRKGSKGQYNTVTADGNELGLGMKIGI 470
Query: 446 VLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLD 505
VLQNC I+P+ L P+RL V ++L RPWK +S + M +GDLI+P+G+ W G F
Sbjct: 471 VLQNCRIVPDRKLTPERLTVATYLGRPWKKFSTTVIMSTEMGDLIRPEGWKIWDGESFHK 530
Query: 506 TCFFAEYANTGPGSNVQARVKWGKGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
+C + EY N GPG+ RV W K S A+ +TAA W+ W+ +P +G
Sbjct: 531 SCRYVEYNNRGPGAFANRRVNWAKVARSAAEVNGFTAANWLGPINWIQEANVPVTIG 587
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 405 bits (1040), Expect = e-113
Identities = 223/569 (39%), Positives = 340/569 (59%), Gaps = 20/569 (3%)
Query: 2 ERKIFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFC 61
+++ + +S +L++ + + V V N +D E+T+ KA+ VC +D + C
Sbjct: 13 KKRYVIISISSVLLISMVVAVTIGVSVNK--SDNAGDEEITTSVKAIKDVCAPTDYKETC 70
Query: 62 ADTL--GSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDL 119
DTL + +TSDP + +K +++ + + + +E +K+ ++ KMALD CK+L
Sbjct: 71 EDTLRKDAKDTSDPLELVKTAFNATMKQISDVAKKSQTM-IELQKDPRA-KMALDQCKEL 128
Query: 120 LEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQ 179
+++AI EL + S H V++ L+ WL A +++Q+CLDGF G + +
Sbjct: 129 MDYAIGEL-SKSFEELGKFEFHKVDEALVKLRIWLSATISHEQTCLDGFQ--GTQGNAGE 185
Query: 180 LQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWMSGADR 239
+L +LT L +VT ++ L + + + +SRRL + P WM R
Sbjct: 186 TIKKALKTAVQLTHNGLAMVTEMSNYLGQMQIP---EMNSRRLLSQEF---PSWMDARAR 239
Query: 240 KLL-ADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQI 298
+LL A MS V P+ VVA+DGSG++KT+ +A+N PK +V+++K G+Y EY+Q+
Sbjct: 240 RLLNAPMS---EVKPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHIKEGIYKEYVQV 296
Query: 299 DKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKH 358
+++ +++ GDGP KT+I+G K++ DG+ T +TAT + V + FIAK +AFENTAGA KH
Sbjct: 297 NRSMTHLVFIGDGPDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNIAFENTAGAIKH 356
Query: 359 QAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKI 418
QAVA+RV D+S F++C GYQDTLYAH+HRQFYR+C ISGT+DF+FG A+ V QN +
Sbjct: 357 QAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFGDAAAVFQNCTL 416
Query: 419 VVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSR 478
+VRKP NQ I A G TG VLQ C I+ EP + + +++L RPWK YSR
Sbjct: 417 LVRKPLLNQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKTYLGRPWKEYSR 476
Query: 479 AIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADA 537
I M I D + P+G+ PW G L+T F++E NTGPG+ + RV W G LS +
Sbjct: 477 TIIMNTFIPDFVPPEGWQPWLGEFGLNTLFYSEVQNTGPGAAITKRVTWPGIKKLSDEEI 536
Query: 538 TKYTAAQWIEGGVWLPATGIPFDLGFTKG 566
K+T AQ+I+G W+P G+P+ LG G
Sbjct: 537 LKFTPAQYIQGDAWIPGKGVPYILGLFSG 565
>At3g62170 PECTINESTERASE-like protein
Length = 588
Score = 399 bits (1025), Expect = e-111
Identities = 228/590 (38%), Positives = 333/590 (55%), Gaps = 38/590 (6%)
Query: 4 KIFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFCAD 63
K+ VS SL+LVVGVA+GV+ V G A+ ++ G + SH KAV +CQ++ D CA
Sbjct: 5 KVVVSVASLLLVVGVAIGVITFVNKGGG-ANGDSNGPINSHQKAVQTICQSTTDQGSCAK 63
Query: 64 TLGSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKN-NQSTKMALDDCKDLLEF 122
TL V + DP+ +KA + + +++ K+ N T N N ++K LD CK +L +
Sbjct: 64 TLDPVKSDDPSKLVKAFLMATKDAITKSSNFTASTEGGMGTNMNATSKAVLDYCKRVLMY 123
Query: 123 AIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQT 182
A+++L+ +I+ + + LK WL VF YQ CLD + K++ +
Sbjct: 124 ALEDLE--TIVEEMGEDLQQSGTKLDQLKQWLTGVFNYQTDCLDDIEEVELKKIMGE--- 178
Query: 183 GSLDHVGKLTALALD----VVTAITKVLAALDLDLNVKP------SSRRLFEV-DEDGNP 231
+ + LT+ A+D VVTA+ ++ +D N+ ++RRL E D G P
Sbjct: 179 -GISNSKVLTSNAIDIFHSVVTAMAQMGVKVDDMKNITMGAGAGGAARRLLEDNDSKGLP 237
Query: 232 EWMSGADRKLLADMSTGM----------------SVTPNAVVAKDGSGKFKTVLDAINSY 275
+W SG DRKL+A G + VVAKDGSG+FKT+ +A+ +
Sbjct: 238 KWFSGKDRKLMAKAGRGAPAGGDDGIGEGGGGGGKIKATHVVAKDGSGQFKTISEAVMAC 297
Query: 276 PKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKN--FVDGVKTIQTA 333
P + GR +I++KAG+Y+E ++I K K NI ++GDG T+TIIT ++ G T +
Sbjct: 298 PDKNPGRCIIHIKAGIYNEQVRIPKKKNNIFMFGDGATQTIITFDRSVKLSPGTTTSLSG 357
Query: 334 TFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFY 393
T +EGF+AK + F+NTAG HQAVALRV GD++ F+C GYQDTLY + RQFY
Sbjct: 358 TVQVESEGFMAKWIGFKNTAGPLGHQAVALRVNGDRAVIFNCRFDGYQDTLYVNNGRQFY 417
Query: 394 RNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQ-KNMPTGVVLQNCEI 452
RN +SGTVDFIFG ++TVIQNS I+VRK Q N + ADG + M G+VL NC I
Sbjct: 418 RNIVVSGTVDFIFGKSATVIQNSLILVRKGSPGQSNYVTADGNEKGAAMKIGIVLHNCRI 477
Query: 453 MPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEY 512
+P+ L+ D+L ++S+L RPWK ++ + + IGDLI+P+G+ W G Q T + E+
Sbjct: 478 IPDKELEADKLTIKSYLGRPWKKFATTVIIGTEIGDLIKPEGWTEWQGEQNHKTAKYIEF 537
Query: 513 ANTGPGSNVQARVKWGKGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
N GPG+ R W K S A+ YT A W+ W+ +P LG
Sbjct: 538 NNRGPGAATTQRPPWVKVAKSAAEVETYTVANWVGPANWIQEANVPVQLG 587
>At5g49180 pectin methylesterase
Length = 571
Score = 389 bits (998), Expect = e-108
Identities = 223/565 (39%), Positives = 323/565 (56%), Gaps = 14/565 (2%)
Query: 2 ERKIFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFC 61
++K ++GV L+V + + V N ++ ++ + T AV AVC +D + C
Sbjct: 9 KKKCIIAGVITALLVLMVVAVGITTSRNTSHSEKIVPVQIKTATTAVEAVCAPTDYKETC 68
Query: 62 ADTL--GSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDL 119
++L S +++ P D IK +I S+ + N++ TK AL+ C+ L
Sbjct: 69 VNSLMKASPDSTQPLDLIKLGFNVTIRSIEDSIKKASVELTAKAANDKDTKGALELCEKL 128
Query: 120 LEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQ 179
+ A D+L+ + D S+ + D DL+ WL AYQQ+C+D F+ K Q
Sbjct: 129 MNDATDDLK-KCLDNFDGFSIPQIEDFVEDLRVWLSGSIAYQQTCMDTFEETNSKLSQDM 187
Query: 180 LQTGSLDHVGKLTALALDVVTAITKVLAALDLDL---NVKPSSRRLFEVDEDGNPEWMSG 236
+ +LT+ L ++T I+ +L ++ ++ +R+L EDG P W+
Sbjct: 188 QKIFKTSR--ELTSNGLAMITNISNLLGEFNVTGVTGDLGKYARKLLSA-EDGIPSWVGP 244
Query: 237 ADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYI 296
R+L+A T V N VVA DGSG++KT+ +A+N+ PK +Q +VIY+K GVY+E +
Sbjct: 245 NTRRLMA---TKGGVKANVVVAHDGSGQYKTINEALNAVPKANQKPFVIYIKQGVYNEKV 301
Query: 297 QIDKTKKNILIYGDGPTKTIITGKKNFVDG-VKTIQTATFSTVAEGFIAKAMAFENTAGA 355
+ K ++ GDGPTKT ITG N+ G VKT TAT + + F AK + FENTAG
Sbjct: 302 DVTKKMTHVTFIGDGPTKTKITGSLNYYIGKVKTYLTATVAINGDNFTAKNIGFENTAGP 361
Query: 356 NKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQN 415
HQAVALRV D + F++C I GYQDTLY H+HRQF+R+C +SGTVDFIFG V+QN
Sbjct: 362 EGHQAVALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRDCTVSGTVDFIFGDGIVVLQN 421
Query: 416 SKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKA 475
IVVRKP +Q +I A G K TG+VLQNC I EPA P + +++L RPWK
Sbjct: 422 CNIVVRKPMKSQSCMITAQGRSDKRESTGLVLQNCHITGEPAYIPVKSINKAYLGRPWKE 481
Query: 476 YSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSK 534
+SR I M TI D+I P G+LPW G L+T ++AEY N GPGSN RVKW G LS
Sbjct: 482 FSRTIIMGTTIDDVIDPAGWLPWNGDFALNTLYYAEYENNGPGSNQAQRVKWPGIKKLSP 541
Query: 535 ADATKYTAAQWIEGGVWLPATGIPF 559
A ++T A+++ G +W+P +P+
Sbjct: 542 KQALRFTPARFLRGNLWIPPNRVPY 566
>At2g47040 putative pectinesterase
Length = 595
Score = 389 bits (998), Expect = e-108
Identities = 225/605 (37%), Positives = 328/605 (54%), Gaps = 60/605 (9%)
Query: 4 KIFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFCAD 63
K+ VS S++L+VGVA+GVVA + N DAN L+ KAV +C+ + D C
Sbjct: 4 KVVVSVASILLIVGVAIGVVAYINKNG---DAN----LSPQMKAVRGICEATSDKASCVK 56
Query: 64 TLGSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVE-NEKNNQSTKMALDDCKDLLEF 122
TL V + DPN IKA + + +++ ++ N T K + + K LD CK + +
Sbjct: 57 TLEPVKSDDPNKLIKAFMLATRDAITQSSNFTGKTEGNLGSGISPNNKAVLDYCKKVFMY 116
Query: 123 AIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDG-EKQVQSQLQ 181
A+++L S+I+ ++ + + LK WL V+ YQ CLD + D K + +
Sbjct: 117 ALEDL--STIVEEMGEDLNQIGSKIDQLKQWLTGVYNYQTDCLDDIEEDDLRKTIGEGIA 174
Query: 182 TGSLDHVGKLTALALDVVTAITKVLAALDLDLN--------------------------- 214
+ + LT+ A+D+ + +A L+L +
Sbjct: 175 SSKI-----LTSNAIDIFHTVVSAMAKLNLKVEDFKNMTGGIFAPSDKGAAPVNKGTPPV 229
Query: 215 ---------VKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMS-----VTPNAVVAKD 260
P+ R L ++DE G P W+SGADRKL+ G + + VVAKD
Sbjct: 230 ADDSPVADPDGPARRLLEDIDETGIPTWVSGADRKLMTKAGRGSNDGGARIRATFVVAKD 289
Query: 261 GSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGK 320
GSG+FKTV A+N+ P+ + GR +I++KAG+Y E + I K K NI ++GDG KT+I+
Sbjct: 290 GSGQFKTVQQAVNACPEKNPGRCIIHIKAGIYREQVIIPKKKNNIFMFGDGARKTVISYN 349
Query: 321 KN--FVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIR 378
++ G T + T +EGF+AK + F+NTAG HQAVA+RV GD++ F+C
Sbjct: 350 RSVKLSPGTTTSLSGTVQVESEGFMAKWIGFKNTAGPMGHQAVAIRVNGDRAVIFNCRFD 409
Query: 379 GYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQ 438
GYQDTLY + RQFYRN +SGTVDFIFG ++TVIQNS IVVRK Q N + ADG +
Sbjct: 410 GYQDTLYVNNGRQFYRNIVVSGTVDFIFGKSATVIQNSLIVVRKGNKGQFNTVTADGNEK 469
Query: 439 -KNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLP 497
M G+VLQNC I+P+ L +RL V S+L RPWK +S + + + IGD+I+P+G+
Sbjct: 470 GLAMKIGIVLQNCRIVPDKKLAAERLIVESYLGRPWKKFSTTVIINSEIGDVIRPEGWKI 529
Query: 498 WAGTQFLDTCFFAEYANTGPGSNVQARVKWGKGVLSKADATKYTAAQWIEGGVWLPATGI 557
W G F +C + EY N GPG+ RV W K S A+ +T A W+ W+ +
Sbjct: 530 WDGESFHKSCRYVEYNNRGPGAITNRRVNWVKIARSAAEVNDFTVANWLGPINWIQEANV 589
Query: 558 PFDLG 562
P LG
Sbjct: 590 PVTLG 594
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 382 bits (980), Expect = e-106
Identities = 224/576 (38%), Positives = 327/576 (55%), Gaps = 30/576 (5%)
Query: 12 LILVVGVALGVVALVR-TNNGPADANNGGELTSHTKAVT-AVCQNSDDHKFCAD---TLG 66
++L VAL VA V + G + AN L+ + AV + C ++ + C T G
Sbjct: 21 VLLSAAVALLFVAAVAGISAGASKANEKRTLSPSSHAVLRSSCSSTRYPELCISAVVTAG 80
Query: 67 SVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAIDE 126
V + D I+A V +I +V + KL + + K AL DC + ++ +DE
Sbjct: 81 GVELTSQKDVIEASVNLTITAVEHNYFTVKKLIKKRKGLTPREKTALHDCLETIDETLDE 140
Query: 127 LQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFD-TDGEKQVQSQLQTGSL 185
L + + + + A DLK + + Q++CLDGF D +KQV+ L G +
Sbjct: 141 LHETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSHDDADKQVRKALLKGQI 200
Query: 186 DHVGKLTALALDVVTAITKV-LAALDLDLNVKPSSRRLFE----------------VDED 228
HV + + AL ++ +T +A + + ++R+L E +D +
Sbjct: 201 -HVEHMCSNALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQETTVAVDIAGAGELDSE 259
Query: 229 GNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVK 288
G P W+S DR+LL G V +A VA DGSG FKTV A+ + P+N RYVI++K
Sbjct: 260 GWPTWLSAGDRRLLQ----GSGVKADATVAADGSGTFKTVAAAVAAAPENSNKRYVIHIK 315
Query: 289 AGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMA 348
AGVY E +++ K KKNI+ GDG T+TIITG +N VDG T +AT + V E F+A+ +
Sbjct: 316 AGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAAVGERFLARDIT 375
Query: 349 FENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGY 408
F+NTAG +KHQAVALRV D SAF++C + YQDTLY H++RQF+ C I+GTVDFIFG
Sbjct: 376 FQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCLIAGTVDFIFGN 435
Query: 409 ASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSF 468
A+ V+Q+ I R+P + Q+N++ A G N TG+V+Q C I LQ + ++
Sbjct: 436 AAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLQSVKGSFPTY 495
Query: 469 LARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG 528
L RPWK YS+ + M++ I D+I+P+G+ W GT L+T + EY+NTG G+ RVKW
Sbjct: 496 LGRPWKEYSQTVIMQSAISDVIRPEGWSEWTGTFALNTLTYREYSNTGAGAGTANRVKWR 555
Query: 529 --KGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
K + + A+A KYTA Q+I GG WL +TG PF LG
Sbjct: 556 GFKVITAAAEAQKYTAGQFIGGGGWLSSTGFPFSLG 591
>At2g26450 putative pectinesterase
Length = 496
Score = 375 bits (964), Expect = e-104
Identities = 202/484 (41%), Positives = 303/484 (61%), Gaps = 18/484 (3%)
Query: 81 VKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAIDELQASSILAADNSSV 140
+K++IE+V + ++ + + + NQ K A++ CK L+E A +E AS + + + V
Sbjct: 25 LKSAIEAVNEDLDLVLEKVLSLKTENQDDKDAIEQCKLLVEDAKEETVAS-LNKINVTEV 83
Query: 141 HNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVT 200
++ DL++WL AV +YQ++CLDGF+ E ++S+++T S++ LT+ +L ++
Sbjct: 84 NSFEKVVPDLESWLSAVMSYQETCLDGFE---EGNLKSEVKT-SVNSSQVLTSNSLALIK 139
Query: 201 AITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKD 260
T+ L+ + +K R L D P W+S DR++L + ++ PNA VAKD
Sbjct: 140 TFTENLSPV-----MKVVERHLL----DDIPSWVSNDDRRMLRAVDV-KALKPNATVAKD 189
Query: 261 GSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGK 320
GSG F T+ DA+ + P+ ++GRY+IYVK G+YDEY+ +DK K N+ + GDG KTI+TG
Sbjct: 190 GSGDFTTINDALRAMPEKYEGRYIIYVKQGIYDEYVTVDKKKANLTMVGDGSQKTIVTGN 249
Query: 321 KNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGY 380
K+ ++T TATF EGF+A++M F NTAG HQAVA+RVQ D+S F +C GY
Sbjct: 250 KSHAKKIRTFLTATFVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQSDRSIFLNCRFEGY 309
Query: 381 QDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKN 440
QDTLYA+ HRQ+YR+C I GT+DFIFG A+ + QN I +RK Q+N + A G V K
Sbjct: 310 QDTLYAYTHRQYYRSCVIVGTIDFIFGDAAAIFQNCNIFIRKGLPGQKNTVTAQGRVDKF 369
Query: 441 MPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAG 500
TG V+ NC+I L+P + + +S+L RPWK YSR I ME+ I ++I P G+L W
Sbjct: 370 QTTGFVVHNCKIAANEDLKPVKEEYKSYLGRPWKNYSRTIIMESKIENVIDPVGWLRWQE 429
Query: 501 TQF-LDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIP 558
T F +DT ++AEY N G + +RVKW G V++K +A YT +++G W+ A+G P
Sbjct: 430 TDFAIDTLYYAEYNNKGSSGDTTSRVKWPGFKVINKEEALNYTVGPFLQGD-WISASGSP 488
Query: 559 FDLG 562
LG
Sbjct: 489 VKLG 492
>At4g33230 pectinesterase - like protein
Length = 609
Score = 373 bits (957), Expect = e-103
Identities = 223/596 (37%), Positives = 334/596 (55%), Gaps = 48/596 (8%)
Query: 3 RKIFVSGVSLILVVGVAL--GVVALVR-----------TNNGPADANNGGELTSHT---- 45
RK + GV +LVV A+ G A V TNN D+ E S
Sbjct: 22 RKRIILGVVSVLVVAAAIIGGAFAYVTYENKTQEQGKTTNNKSKDSPTKSESPSPKPPSS 81
Query: 46 -----------KAVTAVCQNSDDHKFCADTLGSVNTSD-PNDYIKAVVKTSIESVIKAFN 93
K + +C ++ C +TL + D P ++++K++I +V +
Sbjct: 82 AAQTVKAGQVDKIIQTLCNSTLYKPTCQNTLKNETKKDTPQTDPRSLLKSAIVAVNDDLD 141
Query: 94 MTDKLAVENEKNNQSTKMALDDCKDLLEFAIDELQASSILAADNSSVHNVNDRAADLKNW 153
K + + N+ K A+ CK L++ A +EL +S+ ++S V+N DL +W
Sbjct: 142 QVFKRVLSLKTENKDDKDAIAQCKLLVDEAKEEL-GTSMKRINDSEVNNFAKIVPDLDSW 200
Query: 154 LGAVFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDL 213
L AV +YQ++C+DGF+ E +++++++ + + LT+ +L ++ ++ L+++
Sbjct: 201 LSAVMSYQETCVDGFE---EGKLKTEIRK-NFNSSQVLTSNSLAMIKSLDGYLSSVP--- 253
Query: 214 NVKPSSRRLFEV-----DEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTV 268
K +R L E + D W+S +R++L + ++ PNA VAKDGSG F T+
Sbjct: 254 --KVKTRLLLEARSSAKETDHITSWLSNKERRMLKAVDV-KALKPNATVAKDGSGNFTTI 310
Query: 269 LDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVK 328
A+ + P +QGRY IY+K G+YDE + IDK K N+ + GDG KTI+TG K+ ++
Sbjct: 311 NAALKAMPAKYQGRYTIYIKHGIYDESVIIDKKKPNVTMVGDGSQKTIVTGNKSHAKKIR 370
Query: 329 TIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHA 388
T TATF EGF+A++M F NTAG HQAVA+RVQ D+S F +C GYQDTLYA+
Sbjct: 371 TFLTATFVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQSDRSVFLNCRFEGYQDTLYAYT 430
Query: 389 HRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQ 448
HRQ+YR+C I GTVDFIFG A+ + QN I +RK Q+N + A G V K TG V+
Sbjct: 431 HRQYYRSCVIIGTVDFIFGDAAAIFQNCDIFIRKGLPGQKNTVTAQGRVDKFQTTGFVIH 490
Query: 449 NCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQF-LDTC 507
NC + P L+P + + +S+L RPWK +SR + ME+TI D+I P G+L W T F +DT
Sbjct: 491 NCTVAPNEDLKPVKAQFKSYLGRPWKPHSRTVVMESTIEDVIDPVGWLRWQETDFAIDTL 550
Query: 508 FFAEYANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
+AEY N GP ARVKW G VL+K +A K+T +++ G W+ A G P LG
Sbjct: 551 SYAEYKNDGPSGATAARVKWPGFRVLNKEEAMKFTVGPFLQ-GEWIQAIGSPVKLG 605
>At3g06830 pectin methylesterase like protein
Length = 568
Score = 370 bits (951), Expect = e-103
Identities = 220/561 (39%), Positives = 314/561 (55%), Gaps = 21/561 (3%)
Query: 7 VSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFCADTL- 65
VSG +I+VV VA+ V + + P D N + TKAV AVC +D C ++L
Sbjct: 17 VSGFLVIMVVSVAV-----VTSKHSPKDDEN--HIRKTTKAVQAVCAPTDFKDTCVNSLM 69
Query: 66 -GSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAI 124
S ++ DP D IK K +I+S+ ++ N K A + C+ L+ AI
Sbjct: 70 GASPDSDDPVDLIKLGFKVTIKSINESLEKASGDIKAKADKNPEAKGAFELCEKLMIDAI 129
Query: 125 DELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQTGS 184
D+L+ SV + DL+ WL A+QQ+C+D F +Q L+
Sbjct: 130 DDLKKCM---DHGFSVDQIEVFVEDLRVWLSGSIAFQQTCMDSFGEIKSNLMQDMLKIFK 186
Query: 185 LDHVGKLTALALDVVTAITKVLAALDLDL---NVKPSSRRLFEVDEDGNPEWMSGADRKL 241
+L++ +L +VT I+ ++ +L + +R+L ED P W+ R+L
Sbjct: 187 TSR--ELSSNSLAMVTRISTLIPNSNLTGLTGALAKYARKLLST-EDSIPTWVGPEARRL 243
Query: 242 LADMSTGMS-VTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDK 300
+A G V NAVVA+DG+G+FKT+ DA+N+ PK ++ ++I++K G+Y E + + K
Sbjct: 244 MAAQGGGPGPVKANAVVAQDGTGQFKTITDALNAVPKGNKVPFIIHIKEGIYKEKVTVTK 303
Query: 301 TKKNILIYGDGPTKTIITGKKNFVDG-VKTIQTATFSTVAEGFIAKAMAFENTAGANKHQ 359
++ GDGP KT+ITG NF G VKT TAT + + F AK + ENTAG Q
Sbjct: 304 KMPHVTFIGDGPNKTLITGSLNFGIGKVKTFLTATITIEGDHFTAKNIGIENTAGPEGGQ 363
Query: 360 AVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIV 419
AVALRV D + F C I G+QDTLY H+HRQFYR+C +SGTVDFIFG A ++QN KIV
Sbjct: 364 AVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTVSGTVDFIFGDAKCILQNCKIV 423
Query: 420 VRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRA 479
VRKP Q ++ A G TG+VL C I +PA P + +++L RPWK +SR
Sbjct: 424 VRKPNKGQTCMVTAQGRSNVRESTGLVLHGCHITGDPAYIPMKSVNKAYLGRPWKEFSRT 483
Query: 480 IFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKADAT 538
I M+ TI D+I P G+LPW+G L T ++AE+ NTGPGSN RVKW G L+ DA
Sbjct: 484 IIMKTTIDDVIDPAGWLPWSGDFALKTLYYAEHMNTGPGSNQAQRVKWPGIKKLTPQDAL 543
Query: 539 KYTAAQWIEGGVWLPATGIPF 559
YT +++ G W+P T +P+
Sbjct: 544 LYTGDRFLRGDTWIPQTQVPY 564
>At3g10710 putative pectinesterase
Length = 561
Score = 367 bits (943), Expect = e-102
Identities = 213/567 (37%), Positives = 326/567 (56%), Gaps = 37/567 (6%)
Query: 3 RKIFVSGVSLILVVGVALGVV-ALVRTNNGP--ADANNGGELTSHTKAVTAVCQNSDDHK 59
+ I + VSL+++ G+ +G V + P + NN G+ S + +V AVC + +
Sbjct: 24 KNIAIIAVSLVILAGIVIGAVFGTMAHKKSPETVETNNNGD--SISVSVKAVCDVTLHKE 81
Query: 60 FCADTLGSV---NTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDC 116
C +TLGS ++ +P + + VK +I V KA N ++ +EKNN + ++ C
Sbjct: 82 KCFETLGSAPNASSLNPEELFRYAVKITIAEVSKAINAFSS-SLGDEKNN----ITMNAC 136
Query: 117 KDLLEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQV 176
+LL+ ID L +++ ++ N V V + DL+ WL + YQ++C++ D
Sbjct: 137 AELLDLTIDNLN-NTLTSSSNGDV-TVPELVDDLRTWLSSAGTYQRTCVETLAPDMRPFG 194
Query: 177 QSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWMSG 236
+S L+ + +LT+ AL ++T + K+ + L RR D ++ +G
Sbjct: 195 ESHLKNST-----ELTSNALAIITWLGKIADSFKL--------RRRLLTTADVEVDFHAG 241
Query: 237 ADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYI 296
R+LL ST + + VVAKDGSGK++T+ A+ P+ + R +IYVK GVY E +
Sbjct: 242 --RRLL--QSTDLRKVADIVVAKDGSGKYRTIKRALQDVPEKSEKRTIIYVKKGVYFENV 297
Query: 297 QIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGAN 356
+++K N+++ GDG +K+I++G+ N +DG T +TATF+ +GF+A+ M F NTAG +
Sbjct: 298 KVEKKMWNVIVVGDGESKSIVSGRLNVIDGTPTFKTATFAVFGKGFMARDMGFINTAGPS 357
Query: 357 KHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNS 416
KHQAVAL V D +AF+ C + YQDTLY HA RQFYR C I GTVDFIFG +++V+Q+
Sbjct: 358 KHQAVALMVSADLTAFYRCTMNAYQDTLYVHAQRQFYRECTIIGTVDFIFGNSASVLQSC 417
Query: 417 KIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAY 476
+I+ R+P QQN I A G NM TG+ + C I P D V +FL RPWK +
Sbjct: 418 RILPRRPMKGQQNTITAQGRTDPNMNTGISIHRCNISP----LGDLTDVMTFLGRPWKNF 473
Query: 477 SRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVLSKA 535
S + M++ + I G+LPW G DT F+ EY NTGPG++ + RVKW G LS
Sbjct: 474 STTVIMDSYLHGFIDRKGWLPWTGDSAPDTIFYGEYKNTGPGASTKNRVKWKGLRFLSTK 533
Query: 536 DATKYTAAQWIEGGVWLPATGIPFDLG 562
+A ++T +I+GG WLPAT +PF G
Sbjct: 534 EANRFTVKPFIDGGRWLPATKVPFRSG 560
>At1g53830 unknown protein
Length = 587
Score = 355 bits (910), Expect = 5e-98
Identities = 210/575 (36%), Positives = 320/575 (55%), Gaps = 23/575 (4%)
Query: 3 RKIFVSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFCA 62
+K+ +S ++ L++ ++ +A TN TSH + +VC ++ + C
Sbjct: 18 KKLILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHA-ILKSVCSSTLYPELCF 76
Query: 63 DTL---GSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDL 119
+ G + + I+A + + ++V + KL + + AL DC +
Sbjct: 77 SAVAATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREVTALHDCLET 136
Query: 120 LEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFD-TDGEKQVQS 178
++ +DEL + ++ A DLK + + Q +CLDGF D +++V+
Sbjct: 137 IDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFSYDDADRKVRK 196
Query: 179 QLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSS------RRLFEV----DED 228
L G + HV + + AL ++ +T+ A + +L K S+ R+L EV D D
Sbjct: 197 ALLKGQV-HVEHMCSNALAMIKNMTETDIA-NFELRDKSSTFTNNNNRKLKEVTGDLDSD 254
Query: 229 GNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVK 288
G P+W+S DR+LL G ++ +A VA DGSG F TV A+ + P+ R+VI++K
Sbjct: 255 GWPKWLSVGDRRLLQ----GSTIKADATVADDGSGDFTTVAAAVAAAPEKSNKRFVIHIK 310
Query: 289 AGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMA 348
AGVY E +++ K K NI+ GDG KTIITG +N VDG T +AT + V E F+A+ +
Sbjct: 311 AGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGERFLARDIT 370
Query: 349 FENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGY 408
F+NTAG +KHQAVALRV D SAF+ C + YQDTLY H++RQF+ C I+GTVDFIFG
Sbjct: 371 FQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGTVDFIFGN 430
Query: 409 ASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSF 468
A+ V+Q+ I R+P + Q+N++ A G N TG+V+QNC I L + ++
Sbjct: 431 AAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAVKGTFPTY 490
Query: 469 LARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG 528
L RPWK YSR + M++ I D+I+P+G+ W+G+ LDT + EY N G G+ RVKW
Sbjct: 491 LGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAGTANRVKWK 550
Query: 529 --KGVLSKADATKYTAAQWIEGGVWLPATGIPFDL 561
K + S +A +TA Q+I GG WL +TG PF L
Sbjct: 551 GYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFSL 585
>At3g05620 putative pectinesterase
Length = 543
Score = 354 bits (909), Expect = 6e-98
Identities = 207/531 (38%), Positives = 294/531 (54%), Gaps = 43/531 (8%)
Query: 48 VTAVCQNSDDHKFCADTLG-----SVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVEN 102
V CQ D H+ C + S + +P+ ++A VK + + A + + +
Sbjct: 37 VAKACQFIDAHELCVSNIWTHVKESGHGLNPHSVLRAAVKEAHDKAKLAMERIPTVMMLS 96
Query: 103 EKNNQSTKMALDDCKDLLEFAIDELQASSI----------LAADNSSVHNVNDRAADLKN 152
++ + ++A++DCK+L+ F++ EL S + + D+ S H+ +LK
Sbjct: 97 IRSRE--QVAIEDCKELVGFSVTELAWSMLEMNKLHGGGGIDLDDGS-HDAAAAGGNLKT 153
Query: 153 WLGAVFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLD 212
WL A + Q +CL+GF+ G ++ +L GSL V +L + LD+ T
Sbjct: 154 WLSAAMSNQDTCLEGFE--GTERKYEELIKGSLRQVTQLVSNVLDMYT-----------Q 200
Query: 213 LNVKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAI 272
LN P E +PEW++ D L+ + + PN VVA DG GK++T+ +AI
Sbjct: 201 LNALPFKASRNE-SVIASPEWLTETDESLMMRHDPSV-MHPNTVVAIDGKGKYRTINEAI 258
Query: 273 NSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQT 332
N P + RYVIYVK GVY E I + K K NI++ GDG +TIITG +NF+ G+ T +T
Sbjct: 259 NEAPNHSTKRYVIYVKKGVYKENIDLKKKKTNIMLVGDGIGQTIITGDRNFMQGLTTFRT 318
Query: 333 ATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQF 392
AT + GFIAK + F NTAG QAVALRV D+SAF+ C++ GYQDTLYAH+ RQF
Sbjct: 319 ATVAVSGRGFIAKDITFRNTAGPQNRQAVALRVDSDQSAFYRCSVEGYQDTLYAHSLRQF 378
Query: 393 YRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEI 452
YR+CEI GT+DFIFG + V+QN KI R P Q+ I A G N TG V+QN +
Sbjct: 379 YRDCEIYGTIDFIFGNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPNQNTGFVIQNSYV 438
Query: 453 MPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEY 512
+ A QP ++L RPWK YSR ++M + L+QP G+L W G LDT ++ EY
Sbjct: 439 L---ATQP------TYLGRPWKLYSRTVYMNTYMSQLVQPRGWLEWFGNFALDTLWYGEY 489
Query: 513 ANTGPGSNVQARVKW-GKGVLSKADATKYTAAQWIEGGVWLPATGIPFDLG 562
N GPG RVKW G ++ K A +T +I+G WLPATG+ F G
Sbjct: 490 NNIGPGWRSSGRVKWPGYHIMDKRTALSFTVGSFIDGRRWLPATGVTFTAG 540
>At5g04960 pectinesterase
Length = 564
Score = 352 bits (903), Expect = 3e-97
Identities = 214/576 (37%), Positives = 319/576 (55%), Gaps = 47/576 (8%)
Query: 2 ERKIFVSGVSLILVVGVALGVVA--LVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHK 59
+++I + +S I++V + +G V R N+ N GE S +V A+C + +
Sbjct: 22 KKRIAIIAISSIVLVCIVVGAVVGTTARDNSKKPPTENNGEPIS--VSVKALCDVTLHKE 79
Query: 60 FCADTLGSV---NTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDC 116
C +TLGS + S P + K VK +I + K + E + +T A+ C
Sbjct: 80 KCFETLGSAPNASRSSPEELFKYAVKVTITELSKVLDGFSN----GEHMDNATSAAMGAC 135
Query: 117 KDLLEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQV 176
+L+ A+D+L + SS+ N +D L+ WL +V YQ++C+D + K
Sbjct: 136 VELIGLAVDQLNETM-----TSSLKNFDD----LRTWLSSVGTYQETCMDAL-VEANKPS 185
Query: 177 QSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPE---- 232
+ L + ++T+ AL ++T + K+ VK RRL E GN +
Sbjct: 186 LTTFGENHLKNSTEMTSNALAIITWLGKIADT------VKFRRRRLLET---GNAKVVVA 236
Query: 233 ---WMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKA 289
M G R+LL S + VVAKDGSGK++T+ +A+ + ++ +IYVK
Sbjct: 237 DLPMMEG--RRLLE--SGDLKKKATIVVAKDGSGKYRTIGEALAEVEEKNEKPTIIYVKK 292
Query: 290 GVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAF 349
GVY E ++++KTK N+++ GDG +KTI++ NF+DG T +TATF+ +GF+A+ M F
Sbjct: 293 GVYLENVRVEKTKWNVVMVGDGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMARDMGF 352
Query: 350 ENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYA 409
NTAG KHQAVAL V D S F+ C + +QDT+YAHA RQFYR+C I GTVDFIFG A
Sbjct: 353 INTAGPAKHQAVALMVSADLSVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFIFGNA 412
Query: 410 STVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFL 469
+ V Q +I+ R+P QQN I A G N TG+ + NC I P L +++FL
Sbjct: 413 AVVFQKCEILPRRPMKGQQNTITAQGRKDPNQNTGISIHNCTIKPLDNL----TDIQTFL 468
Query: 470 ARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG- 528
RPWK +S + M++ + I P G+LPW G DT F+AEY N+GPG++ + RVKW
Sbjct: 469 GRPWKDFSTTVIMKSFMDKFINPKGWLPWTGDTAPDTIFYAEYLNSGPGASTKNRVKWQG 528
Query: 529 -KGVLSKADATKYTAAQWIEGGVWLPATGIPFDLGF 563
K L+K +A K+T +I+G WLPAT +PF+ F
Sbjct: 529 LKTSLTKKEANKFTVKPFIDGNNWLPATKVPFNSDF 564
>At3g43270 pectinesterase like protein
Length = 527
Score = 343 bits (881), Expect = 1e-94
Identities = 197/493 (39%), Positives = 281/493 (56%), Gaps = 15/493 (3%)
Query: 73 PNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAIDELQ-ASS 131
P ++ +A KT ++++ KA + K + K+ S A+ DC DLL+ A +EL S
Sbjct: 44 PLEFAEAA-KTVVDAITKAVAIVSKFDKKAGKSRVSN--AIVDCVDLLDSAAEELSWIIS 100
Query: 132 ILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQTGSLDHVGKL 191
+ N ++ D +DL+ W+ A + Q +CLDGF+ G + ++ G L VG
Sbjct: 101 ASQSPNGKDNSTGDVGSDLRTWISAALSNQDTCLDGFE--GTNGIIKKIVAGGLSKVGTT 158
Query: 192 TALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWMSGADRKLLADMSTGMSV 251
L +V + + + F P W+ DRKLL T
Sbjct: 159 VRNLLTMVHSPPSKPKPKPIKAQTMTKAHSGFSKF----PSWVKPGDRKLL---QTDNIT 211
Query: 252 TPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTKKNILIYGDG 311
+AVVA DG+G F T+ DA+ + P RYVI+VK GVY E ++I K K NI++ GDG
Sbjct: 212 VADAVVAADGTGNFTTISDAVLAAPDYSTKRYVIHVKRGVYVENVEIKKKKWNIMMVGDG 271
Query: 312 PTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVALRVQGDKSA 371
T+ITG ++F+DG T ++ATF+ GFIA+ + F+NTAG KHQAVA+R D
Sbjct: 272 IDATVITGNRSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQAVAIRSDTDLGV 331
Query: 372 FFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRKPEANQQNII 431
F+ CA+RGYQDTLYAH+ RQF+R C I+GTVDFIFG A+ V Q+ +I ++ NQ+N I
Sbjct: 332 FYRCAMRGYQDTLYAHSMRQFFRECIITGTVDFIFGDATAVFQSCQIKAKQGLPNQKNSI 391
Query: 432 VADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFMENTIGDLIQ 491
A G N PTG +Q I + L + ++L RPWK YSR +FM+N + D I
Sbjct: 392 TAQGRKDPNEPTGFTIQFSNIAADTDLLLNLNTTATYLGRPWKLYSRTVFMQNYMSDAIN 451
Query: 492 PDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVL-SKADATKYTAAQWIEGG 549
P G+L W G LDT ++ EY N+GPG+++ RVKW G VL + A+A +T +Q I+G
Sbjct: 452 PVGWLEWNGNFALDTLYYGEYMNSGPGASLDRRVKWPGYHVLNTSAEANNFTVSQLIQGN 511
Query: 550 VWLPATGIPFDLG 562
+WLP+TGI F G
Sbjct: 512 LWLPSTGITFIAG 524
>At1g53840 unknown protein
Length = 586
Score = 343 bits (879), Expect = 2e-94
Identities = 203/571 (35%), Positives = 317/571 (54%), Gaps = 32/571 (5%)
Query: 3 RKIFVSGVSLILVVGVALGVVA--LVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKF 60
+++ + +S+++++ V + V +V N + + ELT T ++ A+C + +
Sbjct: 27 KRLLLLSISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPST-SLKAICSVTRFPES 85
Query: 61 CADTLG---SVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCK 117
C ++ S NT+DP K +K I+ + ++ +KL+ E E ++ K AL C
Sbjct: 86 CISSISKLPSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETE--DERIKSALRVCG 143
Query: 118 DLLEFAIDELQASSILAADNSSVHNV--NDRAADLKNWLGAVFAYQQSCLDGFD------ 169
DL+E A+D L ++ A D+ + + DLK WL A ++C D D
Sbjct: 144 DLIEDALDRLN-DTVSAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCFDSLDELKQNK 202
Query: 170 TDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDG 229
T+ +Q ++ + T+ +L +V+ I L+ L + ++ + RRL
Sbjct: 203 TEYANSTITQNLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIHRR---RRLMSHHHQQ 259
Query: 230 NPEWMSGADRKLLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKA 289
+ ++ A R+LL + G+ P+ VA DG+G TV +A+ PK +VIYVK+
Sbjct: 260 SVDFEKWARRRLL--QTAGLK--PDVTVAGDGTGDVLTVNEAVAKVPKKSLKMFVIYVKS 315
Query: 290 GVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAF 349
G Y E + +DK+K N++IYGDG KTII+G KNFVDG T +TATF+ +GFI K +
Sbjct: 316 GTYVENVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFAIQGKGFIMKDIGI 375
Query: 350 ENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYA 409
NTAGA KHQAVA R D S ++ C+ G+QDTLY H++RQFYR+C+++GT+DFIFG A
Sbjct: 376 INTAGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDVTGTIDFIFGSA 435
Query: 410 STVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFL 469
+ V Q KI+ R+P +NQ N I A G N +G+ +Q C I + ++L
Sbjct: 436 AVVFQGCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTISAN-----GNVIAPTYL 490
Query: 470 ARPWKAYSRAIFMENTIGDLIQPDGFLPW-AGTQFLDTCFFAEYANTGPGSNVQARVKWG 528
RPWK +S + ME IG +++P G++ W +G + + EY NTGPGS+V RVKW
Sbjct: 491 GRPWKEFSTTVIMETVIGAVVRPSGWMSWVSGVDPPASIVYGEYKNTGPGSDVTQRVKWA 550
Query: 529 --KGVLSKADATKYTAAQWIEGGVWLPATGI 557
K V+S A+A K+T A + G W+PATG+
Sbjct: 551 GYKPVMSDAEAAKFTVATLLHGADWIPATGV 581
>At1g11580 unknown protein
Length = 557
Score = 341 bits (875), Expect = 6e-94
Identities = 198/559 (35%), Positives = 296/559 (52%), Gaps = 38/559 (6%)
Query: 12 LILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFCADTLGSVNTS 71
L+L LG VA + NN + T + +C + D C L T
Sbjct: 22 LVLSFVAILGSVAFFTAQLISVNTNNNDDSLLTT---SQICHGAHDQDSCQALLSEFTTL 78
Query: 72 DPN-----DYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAIDE 126
+ D + +K S+ + M + + + N K DC+++++ + D
Sbjct: 79 SLSKLNRLDLLHVFLKNSVWRLESTMTMVSEARIRS--NGVRDKAGFADCEEMMDVSKDR 136
Query: 127 LQAS-SILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFD---TDGEKQVQSQLQT 182
+ +S L N ++ + ++ + WL +V +CL+ + ++ V+ QL+
Sbjct: 137 MMSSMEELRGGNYNLESYSN----VHTWLSSVLTNYMTCLESISDVSVNSKQIVKPQLED 192
Query: 183 GSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGNPEWMSGADRKLL 242
+++ AL + ++V P+ L + + P W++ DRKLL
Sbjct: 193 ------------------LVSRARVALAIFVSVLPARDDLKMIISNRFPSWLTALDRKLL 234
Query: 243 ADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEYIQIDKTK 302
+ VT N VVAKDG+GKFKTV +A+ + P+N RYVIYVK GVY E I I K K
Sbjct: 235 ESSPKTLKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKGVYKETIDIGKKK 294
Query: 303 KNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQAVA 362
KN+++ GDG TIITG N +DG T ++AT + +GF+A+ + F+NTAG KHQAVA
Sbjct: 295 KNLMLVGDGKDATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQNTAGPAKHQAVA 354
Query: 363 LRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVVRK 422
LRV D++ C I YQDTLY H RQFYR+ I+GTVDFIFG ++ V QN IV R
Sbjct: 355 LRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSAVVFQNCDIVARN 414
Query: 423 PEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAIFM 482
P A Q+N++ A G +N T + +Q C+I L P + V++FL RPWK YSR + M
Sbjct: 415 PGAGQKNMLTAQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLGRPWKLYSRTVIM 474
Query: 483 ENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG--KGVLSKADATKY 540
++ I + I P G+ PW G L T ++ EYANTGPG++ RV W K + +A ++
Sbjct: 475 QSFIDNHIDPAGWFPWDGEFALSTLYYGEYANTGPGADTSKRVNWKGFKVIKDSKEAEQF 534
Query: 541 TAAQWIEGGVWLPATGIPF 559
T A+ I+GG+WL TG+ F
Sbjct: 535 TVAKLIQGGLWLKPTGVTF 553
>At5g53370 pectinesterase
Length = 587
Score = 339 bits (869), Expect = 3e-93
Identities = 205/567 (36%), Positives = 310/567 (54%), Gaps = 34/567 (5%)
Query: 13 ILVVGVA-LGVVALVRT-NNGPADANNGGELTSH-TKAVTAVCQNSDDHKFCADTL---- 65
+LVVGV G+ A +R ++G + +LT T+A++ C S C DTL
Sbjct: 41 VLVVGVVCFGIFAGIRAVDSGKTEP----KLTRKPTQAISRTCSKSLYPNLCIDTLLDFP 96
Query: 66 GSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEFAID 125
GS+ T+D N+ I +++ KA + + + A D C +LL+ ++D
Sbjct: 97 GSL-TADENELIHISFNATLQKFSKALYTSSTITYTQMPPR--VRSAYDSCLELLDDSVD 153
Query: 126 ELQA--SSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDT-DGEKQVQSQLQT 182
L SS++ H+ D+ WL + +C DGFD +G+
Sbjct: 154 ALTRALSSVVVVSGDESHS------DVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQVI 207
Query: 183 GSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDEDGN-PEWMSGADRKL 241
G++ + ++ + L + K L+ + + N R+L +E P W+ DR+L
Sbjct: 208 GAVKDLSEMVSNCLAIFAGKVKDLSGVPVVNN-----RKLLGTEETEELPNWLKREDREL 262
Query: 242 LADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEY-IQIDK 300
L ++ ++ + V+KDGSG FKT+ +AI P++ R+VIYVKAG Y+E +++ +
Sbjct: 263 LGTPTS--AIQADITVSKDGSGTFKTIAEAIKKAPEHSSRRFVIYVKAGRYEEENLKVGR 320
Query: 301 TKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQA 360
K N++ GDG KT+ITG K+ D + T TATF+ GFI + M FEN AG KHQA
Sbjct: 321 KKTNLMFIGDGKGKTVITGGKSIADDLTTFHTATFAATGAGFIVRDMTFENYAGPAKHQA 380
Query: 361 VALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIVV 420
VALRV GD + + C I GYQD LY H++RQF+R CEI GTVDFIFG A+ ++Q+ I
Sbjct: 381 VALRVGGDHAVVYRCNIIGYQDALYVHSNRQFFRECEIYGTVDFIFGNAAVILQSCNIYA 440
Query: 421 RKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRAI 480
RKP A Q+ I A N TG+ + C+++ P L+ + ++L RPWK YSR +
Sbjct: 441 RKPMAQQKITITAQNRKDPNQNTGISIHACKLLATPDLEASKGSYPTYLGRPWKLYSRVV 500
Query: 481 FMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKW-GKGVL-SKADAT 538
+M + +GD I P G+L W G LD+ ++ EY N G GS + RVKW G V+ S +A+
Sbjct: 501 YMMSDMGDHIDPRGWLEWNGPFALDSLYYGEYMNKGLGSGIGQRVKWPGYHVITSTVEAS 560
Query: 539 KYTAAQWIEGGVWLPATGIPFDLGFTK 565
K+T AQ+I G WLP+TG+ F G ++
Sbjct: 561 KFTVAQFISGSSWLPSTGVSFFSGLSQ 587
>At3g49220 pectinesterase - like protein
Length = 598
Score = 321 bits (822), Expect = 8e-88
Identities = 194/567 (34%), Positives = 303/567 (53%), Gaps = 29/567 (5%)
Query: 7 VSGVSLILVVGVALGVVALVRTNNGPADANNGGELTSHTKAVTAVCQNSDDHKFCADTL- 65
V +SLIL + GV + ++ N G ++A++ C+ + + C D+L
Sbjct: 51 VLAISLILAAAIFAGVRSRLKLNQSVP-----GLARKPSQAISKACELTRFPELCVDSLM 105
Query: 66 ---GSVNTSDPNDYIKAVVKTSIESVIKAFNMTDKLAVENEKNNQSTKMALDDCKDLLEF 122
GS+ S D I V ++ A + L+ + + A D C +LL+
Sbjct: 106 DFPGSLAASSSKDLIHVTVNMTLHHFSHALYSSASLSFVDMPPR--ARSAYDSCVELLDD 163
Query: 123 AIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQQSCLDGFDTDGEKQVQSQLQT 182
++D L + SSV + + + D+ WL A +C +GFD + V+ + T
Sbjct: 164 SVDALSRAL------SSVVSSSAKPQDVTTWLSAALTNHDTCTEGFDGVDDGGVKDHM-T 216
Query: 183 GSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRRLFEVDE--DGNPEWMSGADRK 240
+L ++ +L + L + +A D V +RRL V+E + P WM +R+
Sbjct: 217 AALQNLSELVSNCLAIFSASHDG----DDFAGVPIQNRRLLGVEEREEKFPRWMRPKERE 272
Query: 241 LLADMSTGMSVTPNAVVAKDGSGKFKTVLDAINSYPKNHQGRYVIYVKAGVYDEY-IQID 299
+L +M + + +V+KDG+G KT+ +AI P+N R +IYVKAG Y+E +++
Sbjct: 273 IL-EMPVSQ-IQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYVKAGRYEENNLKVG 330
Query: 300 KTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVAEGFIAKAMAFENTAGANKHQ 359
+ K N++ GDG KT+I+G K+ D + T TA+F+ GFIA+ + FEN AG KHQ
Sbjct: 331 RKKINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARDITFENWAGPAKHQ 390
Query: 360 AVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEISGTVDFIFGYASTVIQNSKIV 419
AVALR+ D + + C I GYQDTLY H++RQF+R C+I GTVDFIFG A+ V+QN I
Sbjct: 391 AVALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGNAAVVLQNCSIY 450
Query: 420 VRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQPDRLKVRSFLARPWKAYSRA 479
RKP Q+N I A N TG+ + ++ LQ +++L RPWK +SR
Sbjct: 451 ARKPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTYLGRPWKLFSRT 510
Query: 480 IFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGSNVQARVKWG--KGVLSKADA 537
++M + IG + G+L W T LDT ++ EY N+GPGS + RV W + + S A+A
Sbjct: 511 VYMMSYIGGHVHTRGWLEWNTTFALDTLYYGEYLNSGPGSGLGQRVSWPGYRVINSTAEA 570
Query: 538 TKYTAAQWIEGGVWLPATGIPFDLGFT 564
++T A++I G WLP+TG+ F G +
Sbjct: 571 NRFTVAEFIYGSSWLPSTGVSFLAGLS 597
>At2g45220 pectinesterase like protein
Length = 511
Score = 317 bits (811), Expect = 1e-86
Identities = 194/525 (36%), Positives = 285/525 (53%), Gaps = 50/525 (9%)
Query: 46 KAVTAVCQNSDDHKFCADTLGSVNTSDP----NDYIKAVVKTSIESVIKAFNMTDKLAVE 101
K V A C + + K C L + ++P ++++K +K ++ I A T +
Sbjct: 28 KDVKAWCSQTPNPKPCEYFLTHNSNNEPIKSESEFLKISMKLVLDRAILA--KTHAFTLG 85
Query: 102 NEKNNQSTKMALDDCKDLLEFAIDELQASSILAADNSSVHNVNDRAADLKNWLGAVFAYQ 161
+ + K A +DC L + + ++ + + NV D + WL
Sbjct: 86 PKCRDTREKAAWEDCIKLYDLTVSKI--------NETMDPNVKCSKLDAQTWLSTALTNL 137
Query: 162 QSCLDGFDTDGEKQVQSQLQTGSLDHVGKLTALALDVVTAITKVLAALDLDLNVKPSSRR 221
+C GF L+ G D V L ++ +V + LA + N P
Sbjct: 138 DTCRAGF-----------LELGVTDIV--LPLMSNNVSNLLCNTLAINKVPFNYTPP--- 181
Query: 222 LFEVDEDGNPEWMSGADRKLLADMSTGMSVTP--NAVVAKDGSGKFKTVLDAINSYPKNH 279
++DG P W+ DRKLL S TP NAVVAKDGSG FKT+ +AI++ +
Sbjct: 182 ----EKDGFPSWVKPGDRKLL------QSSTPKDNAVVAKDGSGNFKTIKEAIDA--ASG 229
Query: 280 QGRYVIYVKAGVYDEYIQIDKTKKNILIYGDGPTKTIITGKKNFVDGVKTIQTATFSTVA 339
GR+VIYVK GVY E ++I KKN+++ GDG KTIITG K+ G T +AT + V
Sbjct: 230 SGRFVIYVKQGVYSENLEI--RKKNVMLRGDGIGKTIITGSKSVGGGTTTFNSATVAAVG 287
Query: 340 EGFIAKAMAFENTAGANKHQAVALRVQGDKSAFFDCAIRGYQDTLYAHAHRQFYRNCEIS 399
+GFIA+ + F NTAGA+ QAVALR D S F+ C+ YQDTLY H++RQFYR+C++
Sbjct: 288 DGFIARGITFRNTAGASNEQAVALRSGSDLSVFYQCSFEAYQDTLYVHSNRQFYRDCDVY 347
Query: 400 GTVDFIFGYASTVIQNSKIVVRKPEANQQNIIVADGTVQKNMPTGVVLQNCEIMPEPALQ 459
GTVDFIFG A+ V+QN I R+P ++ N I A G N TG+++ N + L+
Sbjct: 348 GTVDFIFGNAAAVLQNCNIFARRPR-SKTNTITAQGRSDPNQNTGIIIHNSRVTAASDLR 406
Query: 460 PDRLKVRSFLARPWKAYSRAIFMENTIGDLIQPDGFLPWAGTQFLDTCFFAEYANTGPGS 519
P +++L RPW+ YSR +FM+ ++ LI P G+L W G L T F+AE+ NTGPG+
Sbjct: 407 PVLGSTKTYLGRPWRQYSRTVFMKTSLDSLIDPRGWLEWDGNFALKTLFYAEFQNTGPGA 466
Query: 520 NVQARVKW-GKGVL-SKADATKYTAAQWIEGGVWLPATGIPFDLG 562
+ RV W G VL S ++A+K+T ++ GG W+P++ +PF G
Sbjct: 467 STSGRVTWPGFRVLGSASEASKFTVGTFLAGGSWIPSS-VPFTSG 510
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.387
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,199,759
Number of Sequences: 26719
Number of extensions: 505680
Number of successful extensions: 1609
Number of sequences better than 10.0: 91
Number of HSP's better than 10.0 without gapping: 72
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1301
Number of HSP's gapped (non-prelim): 106
length of query: 566
length of database: 11,318,596
effective HSP length: 105
effective length of query: 461
effective length of database: 8,513,101
effective search space: 3924539561
effective search space used: 3924539561
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144728.4


