

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.10 + phase: 0 /pseudo
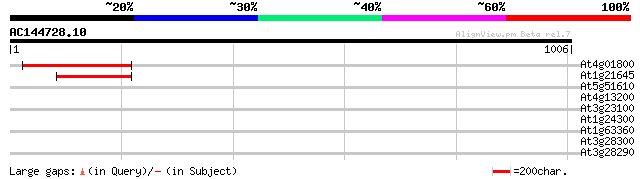
(1006 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g01800 SecA-type chloroplast protein transport factor like pr... 265 1e-70
At1g21645 putative SecA-type chloroplast protein transport factor 169 7e-42
At5g51610 50S ribosomal protein L11-like 33 0.81
At4g13200 unknown protein 32 1.8
At3g23100 putative double strand break repair protein (XRCC4) 31 3.1
At1g24300 unknown protein 31 4.0
At1g63360 disease resistance protein, putative 30 5.3
At3g28300 At14a-2 protein 30 6.9
At3g28290 At14a-1 protein 30 6.9
>At4g01800 SecA-type chloroplast protein transport factor like
protein
Length = 1021
Score = 265 bits (677), Expect = 1e-70
Identities = 143/196 (72%), Positives = 160/196 (80%), Gaps = 2/196 (1%)
Query: 24 RKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSVAV-ASLGGLLGGIFKGNDTGEAT 82
R+ L SSF +F N+ + + S R S +V ASLGGLL GIFKG+D GE+T
Sbjct: 33 RQNRLLSSSSFWGTKFG-NTVKLGVSGCSSCSRKRSTSVNASLGGLLSGIFKGSDNGEST 91
Query: 83 RKQYAATVNVINGLEANISKLSDSELRDKTFELRERAQKRESLDSLLPEAFAVVREASKR 142
R+QYA+ V +N LE IS LSDSELR++T L++RAQK ES+DSLLPEAFAVVREASKR
Sbjct: 92 RQQYASIVASVNRLETEISALSDSELRERTDALKQRAQKGESMDSLLPEAFAVVREASKR 151
Query: 143 VLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILPAYLNALVGKGVHVVTVNDYLAR 202
VLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILPAYLNAL GKGVHVVTVNDYLAR
Sbjct: 152 VLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILPAYLNALSGKGVHVVTVNDYLAR 211
Query: 203 RDCEWVGQVPRFLGMK 218
RDCEWVGQVPRFLG+K
Sbjct: 212 RDCEWVGQVPRFLGLK 227
>At1g21645 putative SecA-type chloroplast protein transport factor
Length = 882
Score = 169 bits (428), Expect = 7e-42
Identities = 85/134 (63%), Positives = 102/134 (75%)
Query: 84 KQYAATVNVINGLEANISKLSDSELRDKTFELRERAQKRESLDSLLPEAFAVVREASKRV 143
+ Y V +N LE I LSD +L+ KT E RER + ESL + EAFAVVREA+KR
Sbjct: 35 RDYYRLVESVNSLEPQIQSLSDEQLKAKTAEFRERLARGESLADMQAEAFAVVREAAKRT 94
Query: 144 LGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAILPAYLNALVGKGVHVVTVNDYLARR 203
+G+R FDVQ+IGG VLH G IAEM+TGEGKTLV+ L AYLNAL G+GVHVVTVNDYLA+R
Sbjct: 95 IGMRHFDVQIIGGGVLHDGSIAEMKTGEGKTLVSTLAAYLNALTGEGVHVVTVNDYLAQR 154
Query: 204 DCEWVGQVPRFLGM 217
D EW+G+V RFLG+
Sbjct: 155 DAEWMGRVHRFLGL 168
>At5g51610 50S ribosomal protein L11-like
Length = 182
Score = 33.1 bits (74), Expect = 0.81
Identities = 27/69 (39%), Positives = 36/69 (52%), Gaps = 3/69 (4%)
Query: 1 MAASSLCSSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSAS--VSKTRRSRSRRSG 58
MA+SSL S S TS H L H + NL + LS +F N S +S T RS + +G
Sbjct: 1 MASSSL-SHLCSCTSSLHHPLSHSISANLTSKTNLSFQFLANRQSPLLSSTPRSLTVIAG 59
Query: 59 SVAVASLGG 67
+ A + L G
Sbjct: 60 ATAPSKLRG 68
>At4g13200 unknown protein
Length = 185
Score = 32.0 bits (71), Expect = 1.8
Identities = 32/120 (26%), Positives = 49/120 (40%), Gaps = 10/120 (8%)
Query: 8 SSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSVAVASLGG 67
SSF+ + S++Q H TL S+F EFH V RRS S RS + G
Sbjct: 10 SSFSLSNSYNQTS-PHSFTLRNSRSNF---EFHRLRLDVESRRRSTSLRSNCSTKGTDSG 65
Query: 68 ------LLGGIFKGNDTGEATRKQYAATVNVINGLEANISKLSDSELRDKTFELRERAQK 121
+L F G E ++ +TV + ++++ E+ ERA+K
Sbjct: 66 ENENKSVLDAFFLGKALAEVINERIESTVGEVLSTIGKFQAEQQKQVQEIQEEVLERAKK 125
>At3g23100 putative double strand break repair protein (XRCC4)
Length = 264
Score = 31.2 bits (69), Expect = 3.1
Identities = 28/108 (25%), Positives = 44/108 (39%), Gaps = 5/108 (4%)
Query: 35 LSREFHLNSASVSKTRRSRSRRSGSVAVASLGGLLGGIFKGNDTGEATRKQYAATVNVIN 94
+ L+ V+KTR RS + + G L ++ E YA ++V+N
Sbjct: 147 MEANIRLSEEVVNKTRSFEKMRSEAERCLAQGEKLC-----DEKTEFESATYAKFLSVLN 201
Query: 95 GLEANISKLSDSELRDKTFELRERAQKRESLDSLLPEAFAVVREASKR 142
+A + L D E + E E K ES +S + EASK+
Sbjct: 202 AKKAKLRALRDKEDSVRVVEEEESTDKAESFESGRSDDEKSEEEASKK 249
>At1g24300 unknown protein
Length = 1417
Score = 30.8 bits (68), Expect = 4.0
Identities = 40/154 (25%), Positives = 64/154 (40%), Gaps = 21/154 (13%)
Query: 1 MAASSLCSSFTSTTSHSQHRLHHRKTLNLPGSSFLSREFHLNSASVSKTRRSRSRRSGSV 60
MA S L + +S H H N P S+F +R F S + RS R+
Sbjct: 1127 MAPSGLTPGIQTLGGYSDHGGSH----NAP-STFGARAF-----SDEQINRSSGDRNNMG 1176
Query: 61 AVASLGGLLGGIFKGNDTGEATRKQYAATVNVI------NGLEA--NISKLSDSELRDKT 112
++ LL GI G G +T+ + A N+ N ++ N+ ++ R +
Sbjct: 1177 SLHRNNSLLSGIIDG---GRSTQNETQAFSNMYAMNKDANDIKTWNNVPPKNEGMGRMMS 1233
Query: 113 FELRERAQKRESLDSLLPEAFAVVREASKRVLGL 146
FE ++R K+ LDSL+ VV + L +
Sbjct: 1234 FEAQDRMGKQAVLDSLVHGELPVVTPGQQSSLNI 1267
>At1g63360 disease resistance protein, putative
Length = 884
Score = 30.4 bits (67), Expect = 5.3
Identities = 21/54 (38%), Positives = 29/54 (52%), Gaps = 2/54 (3%)
Query: 17 SQHRL-HHRKTLNLPGSSFLSREFHLNSASVSKTR-RSRSRRSGSVAVASLGGL 68
S HRL H + L + GSS S HL S SVS + R +S S++ +GG+
Sbjct: 682 SSHRLLSHSRLLEIYGSSVSSLNRHLESLSVSTDKLREFQIKSCSISEIKMGGI 735
>At3g28300 At14a-2 protein
Length = 385
Score = 30.0 bits (66), Expect = 6.9
Identities = 40/147 (27%), Positives = 59/147 (39%), Gaps = 21/147 (14%)
Query: 74 KGNDTGEATRKQYAATVNVINGLEANISKLSDSELRDKTFELRERAQK------RESLDS 127
K ND G+ T K+Y T+ + EA + D + F+L + Q+ RE+ +
Sbjct: 140 KENDVGKKT-KRYENTLRELKKFEA-MGNPFDGDKFTTLFKLMHKEQESLLERVRETKEK 197
Query: 128 LLPEAFAVVREASKR--------VLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAIL 179
L E + E S R VL + F +G MVL + G G + +L
Sbjct: 198 LDEELKNIEMEISSRKKWSIISNVLFIGAFVAVAVGSMVLVCTGV-----GAGVGVAGLL 252
Query: 180 PAYLNALVGKGVHVVTVNDYLARRDCE 206
L A+ GVH + N AR E
Sbjct: 253 SLPLIAIGWVGVHTILENKIQAREKQE 279
>At3g28290 At14a-1 protein
Length = 385
Score = 30.0 bits (66), Expect = 6.9
Identities = 40/147 (27%), Positives = 59/147 (39%), Gaps = 21/147 (14%)
Query: 74 KGNDTGEATRKQYAATVNVINGLEANISKLSDSELRDKTFELRERAQK------RESLDS 127
K ND G+ T K+Y T+ + EA + D + F+L + Q+ RE+ +
Sbjct: 140 KENDVGKKT-KRYENTLRELKKFEA-MGNPFDGDKFTTLFKLMHKEQESLLERVRETKEK 197
Query: 128 LLPEAFAVVREASKR--------VLGLRPFDVQLIGGMVLHKGEIAEMRTGEGKTLVAIL 179
L E + E S R VL + F +G MVL + G G + +L
Sbjct: 198 LDEELKNIEMEISSRKKWSIISNVLFIGAFVAVAVGSMVLVCTGV-----GAGVGVAGLL 252
Query: 180 PAYLNALVGKGVHVVTVNDYLARRDCE 206
L A+ GVH + N AR E
Sbjct: 253 SLPLIAIGWVGVHTILENKIQAREKQE 279
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.353 0.155 0.518
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,929,251
Number of Sequences: 26719
Number of extensions: 644720
Number of successful extensions: 3322
Number of sequences better than 10.0: 9
Number of HSP's better than 10.0 without gapping: 2
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 3319
Number of HSP's gapped (non-prelim): 9
length of query: 1006
length of database: 11,318,596
effective HSP length: 109
effective length of query: 897
effective length of database: 8,406,225
effective search space: 7540383825
effective search space used: 7540383825
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.5 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144728.10


