

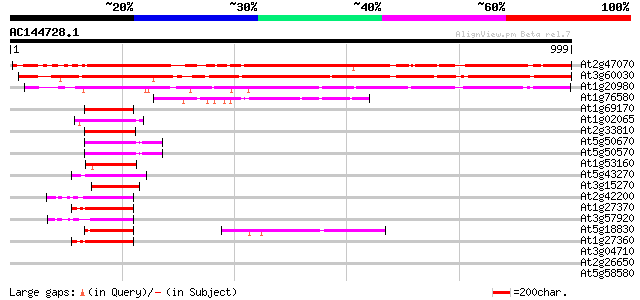
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144728.1 - phase: 0
(999 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g47070 squamosa promoter binding protein-like 1 1000 0.0
At3g60030 squamosa promoter binding protein-like 12 983 0.0
At1g20980 putative SPL1-related protein 426 e-119
At1g76580 unknown protein 130 5e-30
At1g69170 squamosa promoter binding protein-like 6 (spl6) 127 3e-29
At1g02065 squamosa promoter binding protein-like 8 (SPL8) 127 4e-29
At2g33810 squamosa-promoter binding protein-like 3 126 5e-29
At5g50670 putative protein 124 3e-28
At5g50570 putative protein 124 3e-28
At1g53160 squamosa promoter binding protein-like 4 (spl4) 124 3e-28
At5g43270 squamosa promoter binding protein-like 2 (emb|CAB56576.1) 123 6e-28
At3g15270 squamosa promoter binding protein-like 5 118 1e-26
At2g42200 squamosa promoter binding protein-like 9 118 2e-26
At1g27370 squamosa-promoter binding protein-like 10 116 7e-26
At3g57920 squamosa promoter-binding protein homolog 110 3e-24
At5g18830 squamosa promoter binding protein-like 7 109 7e-24
At1g27360 squamosa-promoter binding protein-like 11 107 3e-23
At3g04710 ankyrin-like protein 35 0.21
At2g26650 K+ transporter, AKT1 35 0.21
At5g58580 putative protein 33 0.62
>At2g47070 squamosa promoter binding protein-like 1
Length = 881
Score = 1000 bits (2586), Expect = 0.0
Identities = 566/1002 (56%), Positives = 684/1002 (67%), Gaps = 136/1002 (13%)
Query: 6 GAENYHFYGVGGSSDLSGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPL 65
G E FYG +GKRS EW+LNDW+WDGDLF+A++ + G+QFFPL
Sbjct: 8 GGEAQQFYG--------SVGKRSVEWDLNDWKWDGDLFLATQTTR--------GRQFFPL 51
Query: 66 GSGIPVVGGSSNTSSSCSEEGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVS 125
G+ SSN+SSSCS+EG+ +KKRR + ++ D GAL+LNL G
Sbjct: 52 GN-------SSNSSSSCSDEGN--------DKKRRAVAIQ----GDTNGALTLNLNG--- 89
Query: 126 PVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQ 185
E DG A T + AVCQVE+C ADLS+ KDYHRRHKVCEMHSKA+ A VG +Q
Sbjct: 90 ---ESDGLFP--AKKTKSGAVCQVENCEADLSKVKDYHRRHKVCEMHSKATSATVGGILQ 144
Query: 186 RFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISL 245
RFCQQCSRFH+L+EFDEGKRSCRRRLAGHNKRRRKTN E NG+P+ DD +S+YLLI+L
Sbjct: 145 RFCQQCSRFHLLQEFDEGKRSCRRRLAGHNKRRRKTNPEPGANGNPS-DDHSSNYLLITL 203
Query: 246 LKILSNMHYQPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVS 305
LKILSNMH DQDL++HLL+SL S EQ KNL LL + GG
Sbjct: 204 LKILSNMHNHTGDQDLMSHLLKSLVSHAGEQLGKNLVELLLQ-------GG--------- 247
Query: 306 ALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSET 365
GSQGS I + + Q QE +K + +E
Sbjct: 248 -----GSQGSLN-IGNSALLGIEQAPQE----------------ELKQFSARQDGTATEN 285
Query: 366 RDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYP-WTQQDSHQSSPAQT 424
R S Q KMN+FDLNDIY+DSDD D+ER P TN ATSS+DYP W HQSSP QT
Sbjct: 286 R-SEKQVKMNDFDLNDIYIDSDD--TDVERSPPPTNPATSSLDYPSWI----HQSSPPQT 338
Query: 425 SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPTDIESY 484
S NSDSAS QSPSSSS +AQ RT RIVFKLFGKEPNEFP+VLR QILDWLS SPTD+ESY
Sbjct: 339 SRNSDSASDQSPSSSSEDAQMRTGRIVFKLFGKEPNEFPIVLRGQILDWLSHSPTDMESY 398
Query: 485 IRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQMAFIFN 544
IRPGCIVLTIYLRQAE WEEL DL SL KLLD+SDD W TGW+++RVQ+Q+AF++N
Sbjct: 399 IRPGCIVLTIYLRQAETAWEELSDDLGFSLGKLLDLSDDPLWTTGWIYVRVQNQLAFVYN 458
Query: 545 GQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYLVC 604
GQVV+DTSL +S +YS I +V P+A+ A+++AQF+VKG+NL + TRL+C++EGKYL+
Sbjct: 459 GQVVVDTSLSLKSRDYSHIISVKPLAIAATEKAQFTVKGMNLRQRGTRLLCSVEGKYLIQ 518
Query: 605 EDAHEST----DQYSEELDELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIVAE-ED 659
E H+ST D + + + ++C+ FSC +P+ +GRGF+EIEDQGLSSSFFPF+V E +D
Sbjct: 519 ETTHDSTTREDDDFKDNSEIVECVNFSCDMPILSGRGFMEIEDQGLSSSFFPFLVVEDDD 578
Query: 660 VCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGV 719
VC+EIR+LE LE + TD + QAMDFIHE+GWLLHRS+L + N GV
Sbjct: 579 VCSEIRILETTLEFTGTD----------SAKQAMDFIHEIGWLLHRSKLGES--DPNPGV 626
Query: 720 DLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDETVNKGDHPTLYQALSEMGLLHRAVRR 779
FPL RF WL+EFSMD +WCAV++KLLN+ D V + + LSE+ LLHRAVR+
Sbjct: 627 --FPLIRFQWLIEFSMDREWCAVIRKLLNMFFDGAVGEFSSSS-NATLSELCLLHRAVRK 683
Query: 780 NSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGS 839
NSK +VE+LLRY+P + + LFRPDA GPAGLTPLHIAAGKDGS
Sbjct: 684 NSKPMVEMLLRYIPK----------------QQRNSLFRPDAAGPAGLTPLHIAAGKDGS 727
Query: 840 EDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKINKTQGAA-HV 898
EDVLDALT DP MVGIEAWK RDSTG TPEDYARLRGH++YIHL+Q+KINK HV
Sbjct: 728 EDVLDALTEDPAMVGIEAWKTCRDSTGFTPEDYARLRGHFSYIHLIQRKINKKSTTEDHV 787
Query: 899 VVEIPSNMTE-SNKNPKQNESFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGRSMVYRP 957
VV IP + ++ K PK ++LEI +Q CKLCD K+ T RS+ YRP
Sbjct: 788 VVNIPVSFSDREQKEPKSGPMASALEI-------TQIPCKLCDHKLVYGT-TRRSVAYRP 839
Query: 958 AMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
AMLSMVAIAAVCVCVALLFKS PEVLY+F+PFRWE LD+GTS
Sbjct: 840 AMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLDYGTS 881
>At3g60030 squamosa promoter binding protein-like 12
Length = 927
Score = 983 bits (2542), Expect = 0.0
Identities = 559/1008 (55%), Positives = 704/1008 (69%), Gaps = 115/1008 (11%)
Query: 16 GGSSDLSGMGKRSREWNLNDWRWDGDLFIASRVNQVQAESLRVGQQFFPLGSGIPVVGGS 75
G S + GKRS EW+LNDW+W+GDLF+A+++N GS
Sbjct: 11 GHSLEYGFSGKRSVEWDLNDWKWNGDLFVATQLNH-----------------------GS 47
Query: 76 SNTSSSCSEEGDLE-----KGNKEGEKKRR---VIVLEDDGL-NDKAGALSLNLAGHVSP 126
SN+SS+CS+EG++E + E +KKRR V+ +E+D L +D A L+LNL G+
Sbjct: 48 SNSSSTCSDEGNVEIMERRRIEMEKKKKRRAVTVVAMEEDNLKDDDAHRLTLNLGGNN-- 105
Query: 127 VVERDG-KKSRGAGGTSNRAVC-QVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAM 184
+E +G KK++ GG +RA+C QV++CGADLS+ KDYHRRHKVCE+HSKA+ ALVG M
Sbjct: 106 -IEGNGVKKTKLGGGIPSRAICCQVDNCGADLSKVKDYHRRHKVCEIHSKATTALVGGIM 164
Query: 185 QRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLIS 244
QRFCQQCSRFH+LEEFDEGKRSCRRRLAGHNKRRRK N + + NG+ +DDQTS+Y+LI+
Sbjct: 165 QRFCQQCSRFHVLEEFDEGKRSCRRRLAGHNKRRRKANPDTIGNGTSMSDDQTSNYMLIT 224
Query: 245 LLKILSNMHY----QPTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRN 300
LLKILSN+H Q DQDLL+HLL+SL SQ E +NL LL+ L +S+N
Sbjct: 225 LLKILSNIHSNQSDQTGDQDLLSHLLKSLVSQAGEHIGRNLVGLLQGGGGLQ----ASQN 280
Query: 301 SGMVSALFSNGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTSDHQLISSIKPSISNSPP 360
G +SAL +S+ Q +E + H V + Q + +NS
Sbjct: 281 IGNLSAL-----------------LSLEQAPREDIKHHSVSETPWQEV------YANSAQ 317
Query: 361 AYSETRDSSGQTKMNNFDLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSS 420
S Q K+N+FDLNDIY+DSDD T+ P TN ATSS+DY QDS QSS
Sbjct: 318 ERVAPDRSEKQVKVNDFDLNDIYIDSDDTTDIERSSPPPTNPATSSLDY---HQDSRQSS 374
Query: 421 PAQTSG-NSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFPLVLRAQILDWLSQSPT 479
P QTS NSDSAS QSPSSSSG+AQSRTDRIVFKLFGKEPN+FP+ LR QIL+WL+ +PT
Sbjct: 375 PPQTSRRNSDSASDQSPSSSSGDAQSRTDRIVFKLFGKEPNDFPVALRGQILNWLAHTPT 434
Query: 480 DIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQM 539
D+ESYIRPGCIVLTIYLRQ EA WEELCCDL+ SL +LLD+SDD W GW+++RVQ+Q+
Sbjct: 435 DMESYIRPGCIVLTIYLRQDEASWEELCCDLSFSLRRLLDLSDDPLWTDGWLYLRVQNQL 494
Query: 540 AFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEG 599
AF FNGQVV+DTSLP RS++YS+I TV P+AV +K+AQF+VKG+NL RP TRL+C +EG
Sbjct: 495 AFAFNGQVVLDTSLPLRSHDYSQIITVRPLAV--TKKAQFTVKGINLRRPGTRLLCTVEG 552
Query: 600 KYLVCEDAHESTDQYSE--ELDELQCIQFSCSVPVSNGRGFIEIEDQ-GLSSSFFPFIVA 656
+LV E ++ + E +E+ + FSC +P+++GRGF+EIEDQ GLSSSFFPFIV+
Sbjct: 553 THLVQEATQGGMEERDDLKENNEIDFVNFSCEMPIASGRGFMEIEDQGGLSSSFFPFIVS 612
Query: 657 E-EDVCTEIRVLEPLLESSETDPDIEGTGKIKAKSQAMDFIHEMGWLLHRSQLKYRM-VN 714
E ED+C+EIR LE LE + TD + QAMDFIHE+GWLLHRS+LK R+ +
Sbjct: 613 EDEDICSEIRRLESTLEFTGTD----------SAMQAMDFIHEIGWLLHRSELKSRLAAS 662
Query: 715 LNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDE-TVNKGDHPTLYQALSEMGLL 773
++ DLF L RF +L+EFSMD +WC V+KKLLN+L +E TV+ P+ ALSE+ LL
Sbjct: 663 DHNPEDLFSLIRFKFLIEFSMDREWCCVMKKLLNILFEEGTVD----PSPDAALSELCLL 718
Query: 774 HRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIA 833
HRAVR+NSK +VE+LLR+ P +++ L G LFRPDA GP GLTPLHIA
Sbjct: 719 HRAVRKNSKPMVEMLLRFSPKK-------KNQTLAG------LFRPDAAGPGGLTPLHIA 765
Query: 834 AGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQKKIN-KT 892
AGKDGSEDVLDALT DP M GI+AWKN+RD+TG TPEDYARLRGH++YIHLVQ+K++ K
Sbjct: 766 AGKDGSEDVLDALTEDPGMTGIQAWKNSRDNTGFTPEDYARLRGHFSYIHLVQRKLSRKP 825
Query: 893 QGAAHVVVEIPSNMTESNKNPKQNE-SFTSLEIGKAEVRRSQGNCKLCDTKISCRTAVGR 951
HVVV IP + +K K++ +SLEI + CKLCD K T +
Sbjct: 826 IAKEHVVVNIPESFNIEHKQEKRSPMDSSSLEITQI------NQCKLCDHKRVFVTTHHK 879
Query: 952 SMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRWESLDFGTS 999
S+ YRPAMLSMVAIAAVCVCVALLFKS PEVLY+F+PFRWE L++GTS
Sbjct: 880 SVAYRPAMLSMVAIAAVCVCVALLFKSCPEVLYVFQPFRWELLEYGTS 927
>At1g20980 putative SPL1-related protein
Length = 1035
Score = 426 bits (1095), Expect = e-119
Identities = 331/1087 (30%), Positives = 498/1087 (45%), Gaps = 204/1087 (18%)
Query: 26 KRSREWNLNDWRWDGDLFIASRVN-QVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSE 84
+R EWN W WD F A V+ +VQ
Sbjct: 38 QRRDEWNSKMWDWDSRRFEAKPVDVEVQ-------------------------------- 65
Query: 85 EGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSPVVE--------RDGKKSR 136
E DL N+ GE++ L LNL ++ V E R KK R
Sbjct: 66 EFDLTLRNRSGEER----------------GLDLNLGSGLTAVEETTTTTQNVRPNKKVR 109
Query: 137 GAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHI 196
N +CQV++C DLS KDYHRRHKVCE+HSKA++ALVG MQRFCQQCSRFH+
Sbjct: 110 SGSPGGNYPMCQVDNCTEDLSHAKDYHRRHKVCEVHSKATKALVGKQMQRFCQQCSRFHL 169
Query: 197 LEEFDEGKRSCRRRLAGHNKRRRKTNQ-EAVPNGS--PTNDDQTSSY---------LLIS 244
L EFDEGKRSCRRRLAGHN+RRRKT Q E V +G P N D T++ LL +
Sbjct: 170 LSEFDEGKRSCRRRLAGHNRRRRKTTQPEEVASGVVVPGNHDTTNNTANANMDLMALLTA 229
Query: 245 L------------------------LKILSNMHYQPTDQDLLTHL--LRSLASQNDEQGS 278
L L+IL+ ++ P DL++ L + SLA +N + +
Sbjct: 230 LACAQGKNAVKPPVGSPAVPDREQLLQILNKINALPLPMDLVSKLNNIGSLARKNMDHPT 289
Query: 279 KNLSNLLREQENLLREGGSSRNSGMVSALFSNGSQGSPTVIT----------QHQPVSMN 328
N N + G S +++ L + SP + + ++
Sbjct: 290 VNPQNDMN--------GASPSTMDLLAVLSTTLGSSSPDALAILSQGGFGNKDSEKTKLS 341
Query: 329 QMQQEMVHTHDVRTSDHQLISSIKPSISNSPPAYSETRDSSGQTKMNNFDLNDIYVDSDD 388
+ + + RT + + S SN P S+ DS GQ ++ L +D
Sbjct: 342 SYENGVTTNLEKRTFGFSSVGGERSSSSNQSP--SQDSDSRGQDTRSSLSLQLFTSSPED 399
Query: 389 GTEDL---------------------ERLPVSTNLATSSVDYPWTQQDSHQSSPAQT--- 424
+ PV L + +H++S +T
Sbjct: 400 ESRPTVASSRKYYSSASSNPVEDRSPSSSPVMQELFPLQASPETMRSKNHKNSSPRTGCL 459
Query: 425 -----------------------SGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNE 461
SG + S S SP S + +AQ RT +IVFKL K+P++
Sbjct: 460 PLELFGASNRGAADPNFKGFGQQSGYASSGSDYSPPSLNSDAQDRTGKIVFKLLDKDPSQ 519
Query: 462 FPLVLRAQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVS 521
P LR++I +WLS P+++ESYIRPGC+VL++Y+ + A WE+L L L LL S
Sbjct: 520 LPGTLRSEIYNWLSNIPSEMESYIRPGCVVLSVYVAMSPAAWEQLEQKLLQRLGVLLQNS 579
Query: 522 DDTFWKTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSV 581
FW+ + Q+A NG+V S +R+ N ++ +VSP+AV A + V
Sbjct: 580 PSDFWRNARFIVNTGRQLASHKNGKVRCSKS--WRTWNSPELISVSPVAVVAGEETSLVV 637
Query: 582 KGVNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDELQCIQFSCS--VPVSNGRGFI 639
+G +L + C G Y+ E Q DEL F P GR FI
Sbjct: 638 RGRSLTNDGISIRCTHMGSYMAMEVTRAVCRQ--TIFDELNVNSFKVQNVHPGFLGRCFI 695
Query: 640 EIEDQGLSSSFFPFIVAEEDVCTEI-RVLEPLLESSE--TDPDIEGTGK-IKAKSQAMDF 695
E+E+ G FP I+A +C E+ R+ E S+ T+ + + + ++ + + F
Sbjct: 696 EVEN-GFRGDSFPLIIANASICKELNRLGEEFHPKSQDMTEEQAQSSNRGPTSREEVLCF 754
Query: 696 IHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCAVVKKLLNLLLDET- 754
++E+GWL ++Q L D F L RF +L+ S++ D+CA+++ LL++L++
Sbjct: 755 LNELGWLFQKNQTS----ELREQSD-FSLARFKFLLVCSVERDYCALIRTLLDMLVERNL 809
Query: 755 VNKGDHPTLYQALSEMGLLHRAVRRNSKQLVELLLRYVPDNTSDELGPEDKALVGGKNHS 814
VN + L+E+ LL+RAV+R S ++VELL+ Y+ + L +
Sbjct: 810 VNDELNREALDMLAEIQLLNRAVKRKSTKMVELLIHYLVN-----------PLTLSSSRK 858
Query: 815 YLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYAR 874
++F P+ GP G+TPLH+AA GS+D++D LTNDP +G+ +W RD+TG TP YA
Sbjct: 859 FVFLPNITGPGGITPLHLAACTSGSDDMIDLLTNDPQEIGLSSWNTLRDATGQTPYSYAA 918
Query: 875 LRGHYTYIHLVQKKINKTQGAAHVVVEIPSNMTESNKNPKQNESFTSLEIGKAEVRRSQG 934
+R ++ Y LV +K+ + V + I + + K+ SLE+ K S
Sbjct: 919 IRNNHNYNSLVARKLADKRN-KQVSLNIEHEVVDQTGLSKR----LSLEMNK-----SSS 968
Query: 935 NCKLCDT---KISCRTAVGRSMVYRPAMLSMVAIAAVCVCVALLFKSSPEVLYMFRPFRW 991
+C C T K R + + + P + SM+A+A VCVCV + + P ++ F W
Sbjct: 969 SCASCATVALKYQRRVSGSQRLFPTPIIHSMLAVATVCVCVCVFMHAFP-IVRQGSHFSW 1027
Query: 992 ESLDFGT 998
LD+G+
Sbjct: 1028 GGLDYGS 1034
>At1g76580 unknown protein
Length = 488
Score = 130 bits (326), Expect = 5e-30
Identities = 125/419 (29%), Positives = 187/419 (43%), Gaps = 54/419 (12%)
Query: 256 PTDQDLLTHLLRSLASQNDEQGSKNLSNLLREQENLLREGGSSRNSGMVSAL------FS 309
P+ DLL L SL S E + +E+ R +S + ++L F
Sbjct: 82 PSTMDLLAALSASLGSSAPEAIAFLSQGGFGNKESNDRTKLTSSDHSATTSLEKKTLEFP 141
Query: 310 NGSQGSPTVITQHQPVSMNQMQQEMVHTHDVRTS-DHQLISSI-----KPSISNSPPAYS 363
+ G T T H P + + + D R+S QL +S +P +++S YS
Sbjct: 142 SFGGGERTSSTNHSPSQYSDSRGQ-----DTRSSLSLQLFTSSPEEESRPKVASSTKYYS 196
Query: 364 -------ETRDSSGQTKMNN-FDLN--------DIYVDSDDGTED----LERLPVSTNLA 403
E R S M F L+ + Y D+ LE S A
Sbjct: 197 SASSNPVEDRSPSSSPVMQELFPLHTSPETRRYNNYKDTSTSPRTSCLPLELFGASNRGA 256
Query: 404 TSSVDYPWTQQDSHQSSPAQTSGNSDSASAQSPSSSSGEAQSRTDRIVFKLFGKEPNEFP 463
T++ +Y + HQS G + S S SP S + AQ RT +I FKLF K+P++ P
Sbjct: 257 TANPNYNVLR---HQS------GYASSGSDYSPPSLNSNAQERTGKISFKLFEKDPSQLP 307
Query: 464 LVLRAQILDWLSQSPTDIESYIRPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDD 523
LR +I WLS P+D+ES+IRPGC++L++Y+ + + WE+L +L + L V D
Sbjct: 308 NTLRTEIFRWLSSFPSDMESFIRPGCVILSVYVAMSASAWEQLEENLLQRVRSL--VQDS 365
Query: 524 TFWKTGWVHIRVQHQMAFIFNGQVVIDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKG 583
FW + Q+A +G++ + S +R+ N ++ TVSP+AV A + V+G
Sbjct: 366 EFWSNSRFLVNAGRQLASHKHGRIRLSKS--WRTLNLPELITVSPLAVVAGEETALIVRG 423
Query: 584 VNLMRPATRLMCALEGKYLVCEDAHESTDQYSEELDEL--QCIQFSCSVPVSNGRGFIE 640
NL RL CA G Y E + ++DEL Q + VS GR FIE
Sbjct: 424 RNLTNDGMRLRCAHMGNYASMEVT--GREHRLTKVDELNVSSFQVQSASSVSLGRCFIE 480
>At1g69170 squamosa promoter binding protein-like 6 (spl6)
Length = 405
Score = 127 bits (319), Expect = 3e-29
Identities = 58/88 (65%), Positives = 65/88 (72%)
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
KKSR + S +CQV C DLS KDYH+RH+VCE HSK S +V QRFCQQCS
Sbjct: 110 KKSRASNLCSQNPLCQVYGCSKDLSSSKDYHKRHRVCEAHSKTSVVIVNGLEQRFCQQCS 169
Query: 193 RFHILEEFDEGKRSCRRRLAGHNKRRRK 220
RFH L EFD+GKRSCRRRLAGHN+RRRK
Sbjct: 170 RFHFLSEFDDGKRSCRRRLAGHNERRRK 197
>At1g02065 squamosa promoter binding protein-like 8 (SPL8)
Length = 333
Score = 127 bits (318), Expect = 4e-29
Identities = 67/132 (50%), Positives = 76/132 (56%), Gaps = 14/132 (10%)
Query: 116 LSLNLAGH----------VSPVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRR 165
+ LNL G VS + R G + + CQ E C ADLS K YHRR
Sbjct: 147 IGLNLGGRTYFSAADDDFVSRLYRRSRPGESGMANSLSTPRCQAEGCNADLSHAKHYHRR 206
Query: 166 HKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEA 225
HKVCE HSKAS + QRFCQQCSRFH+L EFD GKRSCR+RLA HN+RRRK +Q A
Sbjct: 207 HKVCEFHSKASTVVAAGLSQRFCQQCSRFHLLSEFDNGKRSCRKRLADHNRRRRKCHQSA 266
Query: 226 VPNGSPTNDDQT 237
S T D T
Sbjct: 267 ----SATQDTGT 274
>At2g33810 squamosa-promoter binding protein-like 3
Length = 131
Score = 126 bits (317), Expect = 5e-29
Identities = 57/92 (61%), Positives = 68/92 (72%), Gaps = 1/92 (1%)
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
KK +G TS+ VCQVE C AD+S+ K YH+RHKVC+ H+KA + QRFCQQCS
Sbjct: 41 KKQKGKA-TSSSGVCQVESCTADMSKAKQYHKRHKVCQFHAKAPHVRISGLHQRFCQQCS 99
Query: 193 RFHILEEFDEGKRSCRRRLAGHNKRRRKTNQE 224
RFH L EFDE KRSCRRRLAGHN+RRRK+ +
Sbjct: 100 RFHALSEFDEAKRSCRRRLAGHNERRRKSTTD 131
>At5g50670 putative protein
Length = 359
Score = 124 bits (311), Expect = 3e-28
Identities = 66/140 (47%), Positives = 84/140 (59%), Gaps = 5/140 (3%)
Query: 133 KKSRGAG-GTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQC 191
K++RG G GT+ +C V+ C +D S ++YH+RHKVC++HSK + QRFCQQC
Sbjct: 86 KRTRGNGVGTNQMPICLVDGCDSDFSNCREYHKRHKVCDVHSKTPVVTINGHKQRFCQQC 145
Query: 192 SRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSN 251
SRFH LEEFDEGKRSCR+RL GHN+RRRK E + G P N + + LL+
Sbjct: 146 SRFHALEEFDEGKRSCRKRLDGHNRRRRKPQPEHI--GRPAN--FFTGFQGSKLLEFSGG 201
Query: 252 MHYQPTDQDLLTHLLRSLAS 271
H PT L SL S
Sbjct: 202 SHVFPTTSVLNPSWGNSLVS 221
>At5g50570 putative protein
Length = 359
Score = 124 bits (311), Expect = 3e-28
Identities = 66/140 (47%), Positives = 84/140 (59%), Gaps = 5/140 (3%)
Query: 133 KKSRGAG-GTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQC 191
K++RG G GT+ +C V+ C +D S ++YH+RHKVC++HSK + QRFCQQC
Sbjct: 86 KRTRGNGVGTNQMPICLVDGCDSDFSNCREYHKRHKVCDVHSKTPVVTINGHKQRFCQQC 145
Query: 192 SRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPNGSPTNDDQTSSYLLISLLKILSN 251
SRFH LEEFDEGKRSCR+RL GHN+RRRK E + G P N + + LL+
Sbjct: 146 SRFHALEEFDEGKRSCRKRLDGHNRRRRKPQPEHI--GRPAN--FFTGFQGSKLLEFSGG 201
Query: 252 MHYQPTDQDLLTHLLRSLAS 271
H PT L SL S
Sbjct: 202 SHVFPTTSVLNPSWGNSLVS 221
>At1g53160 squamosa promoter binding protein-like 4 (spl4)
Length = 174
Score = 124 bits (311), Expect = 3e-28
Identities = 58/94 (61%), Positives = 71/94 (74%), Gaps = 4/94 (4%)
Query: 136 RGAGGTSNRA----VCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQC 191
RG+ G+ NR +CQV+ C AD+ K YHRRHKVCE+H+KAS + QRFCQQC
Sbjct: 39 RGSRGSINRGGSLRLCQVDRCTADMKEAKLYHRRHKVCEVHAKASSVFLSGLNQRFCQQC 98
Query: 192 SRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEA 225
SRFH L+EFDE KRSCRRRLAGHN+RRRK++ E+
Sbjct: 99 SRFHDLQEFDEAKRSCRRRLAGHNERRRKSSGES 132
>At5g43270 squamosa promoter binding protein-like 2 (emb|CAB56576.1)
Length = 419
Score = 123 bits (308), Expect = 6e-28
Identities = 67/135 (49%), Positives = 79/135 (57%), Gaps = 6/135 (4%)
Query: 110 NDKAGALSLNLAGH-VSPVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKV 168
N K L + LA VSPV KKS+ CQVE C DLS KDYHR+H++
Sbjct: 136 NAKGLGLPVTLASSSVSPV-----KKSKSIPQRLQTPHCQVEGCNLDLSSAKDYHRKHRI 190
Query: 169 CEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRKTNQEAVPN 228
CE HSK + +V +RFCQQCSRFH L EFDE KRSCRRRL+ HN RRRK N +
Sbjct: 191 CENHSKFPKVVVSGVERRFCQQCSRFHCLSEFDEKKRSCRRRLSDHNARRRKPNPGRTYD 250
Query: 229 GSPTNDDQTSSYLLI 243
G P D + + LI
Sbjct: 251 GKPQVDFVWNRFALI 265
>At3g15270 squamosa promoter binding protein-like 5
Length = 181
Score = 118 bits (296), Expect = 1e-26
Identities = 53/85 (62%), Positives = 65/85 (76%)
Query: 146 VCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKR 205
+CQV+ C +L+ K Y+RRH+VCE+H+KAS A V QRFCQQCSRFH L EFDE KR
Sbjct: 62 LCQVDRCTVNLTEAKQYYRRHRVCEVHAKASAATVAGVRQRFCQQCSRFHELPEFDEAKR 121
Query: 206 SCRRRLAGHNKRRRKTNQEAVPNGS 230
SCRRRLAGHN+RRRK + ++ GS
Sbjct: 122 SCRRRLAGHNERRRKISGDSFGEGS 146
>At2g42200 squamosa promoter binding protein-like 9
Length = 375
Score = 118 bits (295), Expect = 2e-26
Identities = 74/157 (47%), Positives = 86/157 (54%), Gaps = 23/157 (14%)
Query: 66 GSGIPVVGGSSNTSSSCSEEGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVS 125
G G GGSS SSS S G L G K I ED G G+ S + G
Sbjct: 12 GPGQAESGGSSTESSSFS--GGLMFGQK--------IYFEDGG----GGSGSSSSGG--- 54
Query: 126 PVVERDGKKSRGAGGTSNRAV--CQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNA 183
R ++ RG G + + CQVE CG DL+ K Y+ RH+VC +HSK + V
Sbjct: 55 ----RSNRRVRGGGSGQSGQIPRCQVEGCGMDLTNAKGYYSRHRVCGVHSKTPKVTVAGI 110
Query: 184 MQRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRK 220
QRFCQQCSRFH L EFD KRSCRRRLAGHN+RRRK
Sbjct: 111 EQRFCQQCSRFHQLPEFDLEKRSCRRRLAGHNERRRK 147
>At1g27370 squamosa-promoter binding protein-like 10
Length = 396
Score = 116 bits (290), Expect = 7e-26
Identities = 57/111 (51%), Positives = 73/111 (65%), Gaps = 4/111 (3%)
Query: 110 NDKAGALSLNLAGHVSPVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVC 169
N+ A+S+NL VV R KK++ G + CQ++ C DLS KDYHR+H+VC
Sbjct: 143 NNDLSAVSMNLL--TPSVVAR--KKTKSCGQSMQVPRCQIDGCELDLSSSKDYHRKHRVC 198
Query: 170 EMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRK 220
E HSK + +V +RFCQQCSRFH + EFDE KRSCR+RL+ HN RRRK
Sbjct: 199 ETHSKCPKVVVSGLERRFCQQCSRFHAVSEFDEKKRSCRKRLSHHNARRRK 249
>At3g57920 squamosa promoter-binding protein homolog
Length = 354
Score = 110 bits (276), Expect = 3e-24
Identities = 74/154 (48%), Positives = 86/154 (55%), Gaps = 28/154 (18%)
Query: 67 SGIPVVGGSSNTSSSCSEEGDLEKGNKEGEKKRRVIVLEDDGLNDKAGALSLNLAGHVSP 126
SG GGSS+T SS S G L G K + +DG +G+ S N V
Sbjct: 7 SGQAESGGSSSTESS-SLSGGLRFGQK---------IYFEDG----SGSRSKNRVNTV-- 50
Query: 127 VVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQR 186
+KS S A CQVE C DLS K Y+ RHKVC +HSK+S+ +V QR
Sbjct: 51 ------RKS------STTARCQVEGCRMDLSNVKAYYSRHKVCCIHSKSSKVIVSGLHQR 98
Query: 187 FCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRK 220
FCQQCSRFH L EFD KRSCRRRLA HN+RRRK
Sbjct: 99 FCQQCSRFHQLSEFDLEKRSCRRRLACHNERRRK 132
>At5g18830 squamosa promoter binding protein-like 7
Length = 801
Score = 109 bits (273), Expect = 7e-24
Identities = 83/309 (26%), Positives = 139/309 (44%), Gaps = 23/309 (7%)
Query: 378 DLNDIYVDSDDGTEDLERLPVSTNLATSSVDYPWTQQDSHQSSPAQTS--------GNSD 429
D DI SD E+ L T+ P+T+ + + + T G +D
Sbjct: 239 DGKDITCSSDQRAEEEPSLIFEDRHITTQGSVPFTRSINADNFVSVTGSGEAQPDEGMND 298
Query: 430 SASAQSPSSSSGEAQSRT----DRIVFKLFGKEPNEFPLVLRAQILDWLSQSPTDIESYI 485
+ +SPS+ ++ T RI FKL+ P EFP LR QI WL+ P ++E YI
Sbjct: 299 TKFERSPSNGDNKSAYSTVCPTGRISFKLYDWNPAEFPRRLRHQIFQWLANMPVELEGYI 358
Query: 486 RPGCIVLTIYLRQAEAVWEELCCDLTSSLIKLLDVSDDTFWKTGWVHIRVQHQMAFIFNG 545
RPGC +LT+++ E +W +L D + L + + + G + + + + + + G
Sbjct: 359 RPGCTILTVFIAMPEIMWAKLSKDPVAYLDEFILKPGKMLFGRGSMTVYLNNMIFRLIKG 418
Query: 546 QVV---IDTSLPFRSNNYSKIWTVSPIAVPASKRAQFSVKGVNLMRPATRLMCALEGKYL 602
+D L K+ V P A K + V G NL++P R + + GKYL
Sbjct: 419 GTTLKRVDVKL-----ESPKLQFVYPTCFEAGKPIELVVCGQNLLQPKCRFLVSFSGKYL 473
Query: 603 VCE-DAHESTDQYSEEL--DELQCIQFSCSVPVSNGRGFIEIEDQGLSSSFFPFIVAEED 659
+ DQ + ++ I S P G F+E+E++ S+F P I+ +
Sbjct: 474 PHNYSVVPAPDQDGKRSCNNKFYKINIVNSDPSLFGPAFVEVENESGLSNFIPLIIGDAA 533
Query: 660 VCTEIRVLE 668
VC+E++++E
Sbjct: 534 VCSEMKLIE 542
Score = 99.8 bits (247), Expect = 7e-21
Identities = 47/88 (53%), Positives = 61/88 (68%), Gaps = 1/88 (1%)
Query: 133 KKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVCEMHSKASRALVGNAMQRFCQQCS 192
KK R GG S A CQV DC AD+S K YH+RH+VC + AS ++ +R+CQQC
Sbjct: 125 KKKRVRGG-SGVARCQVPDCEADISELKGYHKRHRVCLRCATASFVVLDGENKRYCQQCG 183
Query: 193 RFHILEEFDEGKRSCRRRLAGHNKRRRK 220
+FH+L +FDEGKRSCRR+L HN RR++
Sbjct: 184 KFHLLPDFDEGKRSCRRKLERHNNRRKR 211
>At1g27360 squamosa-promoter binding protein-like 11
Length = 393
Score = 107 bits (268), Expect = 3e-23
Identities = 55/111 (49%), Positives = 71/111 (63%), Gaps = 5/111 (4%)
Query: 110 NDKAGALSLNLAGHVSPVVERDGKKSRGAGGTSNRAVCQVEDCGADLSRGKDYHRRHKVC 169
N++ A+S+ L ++P V KS+ G + CQ++ C DLS K YHR+HKVC
Sbjct: 143 NNEISAVSMKL---LTPSVVAG--KSKLCGQSMPVPRCQIDGCELDLSSAKGYHRKHKVC 197
Query: 170 EMHSKASRALVGNAMQRFCQQCSRFHILEEFDEGKRSCRRRLAGHNKRRRK 220
E HSK + V +RFCQQCSRFH + EFDE KRSCR+RL+ HN RRRK
Sbjct: 198 EKHSKCPKVSVSGLERRFCQQCSRFHAVSEFDEKKRSCRKRLSHHNARRRK 248
>At3g04710 ankyrin-like protein
Length = 456
Score = 35.0 bits (79), Expect = 0.21
Identities = 40/151 (26%), Positives = 62/151 (40%), Gaps = 15/151 (9%)
Query: 776 AVRRNSKQLVELLLRYVPD---NTSDELGPEDKALVGGKNH--SYLFRPDA---VGPAGL 827
A + K VE+LL + + T D + P A+ G L + A V G
Sbjct: 161 AAGHDQKNAVEVLLEHNANPNAETEDNITPLLSAVAAGSLSCLELLVKAGAKANVFAGGA 220
Query: 828 TPLHIAAGKDGSEDVLDALTNDPCMVGIEAWKNARDSTGSTPEDYARLRGHYTYIHLVQK 887
TPLHIAA E L N C++ A N +D G+ P + A R + + ++
Sbjct: 221 TPLHIAADIGNLE-----LIN--CLLKAGADPNQKDEEGNRPLEVAAARDNRKVVEILFP 273
Query: 888 KINKTQGAAHVVVEIPSNMTESNKNPKQNES 918
K + + V+ ESNK ++N +
Sbjct: 274 LTTKPETVSDWTVDGILAHMESNKEQEENSN 304
>At2g26650 K+ transporter, AKT1
Length = 857
Score = 35.0 bits (79), Expect = 0.21
Identities = 55/210 (26%), Positives = 75/210 (35%), Gaps = 51/210 (24%)
Query: 682 GTGKIKAKSQAMDFIHEMGWLLHRSQLKYRMVNLNSGVDLFPLQRFTWLMEFSMDHDWCA 741
GT I + +A D I E+G L +R QL V L + R T+L +
Sbjct: 425 GTESIVREVKAGDIIGEIGVLCYRPQLF--TVRTKRLCQLLRMNRTTFLNIIQANVGDGT 482
Query: 742 VVKKLLNLLLDETVNKGDHPTLYQALSE-------------MGLLHRAVRRNSKQLVELL 788
++ +N LL D P + L E + L A+R + L +LL
Sbjct: 483 II---MNNLLQHLKEMND-PVMTNVLLEIENMLARGKMDLPLNLCFAAIREDDLLLHQLL 538
Query: 789 LRYVPDNTSDELGPEDKALVGGKNHSYLFRPDAVGPAGLTPLHIAAGKDGSEDVLDALTN 848
R + N SD G TPLHIAA K VL
Sbjct: 539 KRGLDPNESDN-------------------------NGRTPLHIAASKGTLNCVL----- 568
Query: 849 DPCMVGIEAWKNARDSTGSTPEDYARLRGH 878
++ A N RD+ GS P A + GH
Sbjct: 569 --LLLEYHADPNCRDAEGSVPLWEAMVEGH 596
>At5g58580 putative protein
Length = 308
Score = 33.5 bits (75), Expect = 0.62
Identities = 23/61 (37%), Positives = 32/61 (51%), Gaps = 2/61 (3%)
Query: 48 VNQVQAESLRVGQQFFPLGSGIPVVGGSSNTSSSCSE-EGDLEKGNKEGEKKRRVIVLED 106
VN V+ E+ V Q P G V G S S S E DL+ G+ +GE++ R+ V +D
Sbjct: 197 VNAVEEEA-EVRLQMSPAGENESNVSGDRRVSLSLSVMEDDLKTGDDDGEEEVRIEVFDD 255
Query: 107 D 107
D
Sbjct: 256 D 256
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.388
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 23,481,651
Number of Sequences: 26719
Number of extensions: 1061465
Number of successful extensions: 3754
Number of sequences better than 10.0: 56
Number of HSP's better than 10.0 without gapping: 23
Number of HSP's successfully gapped in prelim test: 33
Number of HSP's that attempted gapping in prelim test: 3666
Number of HSP's gapped (non-prelim): 85
length of query: 999
length of database: 11,318,596
effective HSP length: 109
effective length of query: 890
effective length of database: 8,406,225
effective search space: 7481540250
effective search space used: 7481540250
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144728.1


