

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144727.12 - phase: 0
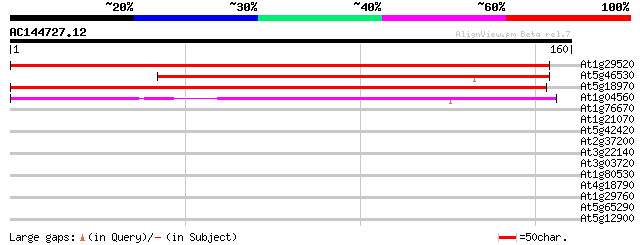
(160 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g29520 plasma membrane associated protein, putative 243 3e-65
At5g46530 putative protein 167 3e-42
At5g18970 plasma membrane associated protein -like 139 7e-34
At1g04560 unknown protein 122 1e-28
At1g76670 unknown protein 33 0.051
At1g21070 unknown protein 33 0.086
At5g42420 unknown protein 31 0.25
At2g37200 unknown protein 29 1.2
At3g22140 hypothetical protein 28 2.1
At3g03720 putative cationic amino acid transporter 27 3.6
At1g80530 nodulin-like protein 27 3.6
At4g18790 ion transporter - like protein 27 4.7
At1g29760 unknown protein 27 4.7
At5g65290 unknown protein 27 6.2
At5g12900 unknown protein 26 8.1
>At1g29520 plasma membrane associated protein, putative
Length = 158
Score = 243 bits (620), Expect = 3e-65
Identities = 117/154 (75%), Positives = 133/154 (85%)
Query: 1 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMG 60
MA EQ+KP+A+ LL LNFCMYVIV+ IGGWAMNRAIDHGFE+GP +LPAHFSPIYFPMG
Sbjct: 1 MAGEQIKPVASGLLVLNFCMYVIVLGIGGWAMNRAIDHGFEVGPNLELPAHFSPIYFPMG 60
Query: 61 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 120
NAATGFFV FAL+AGVVG S ISGL+HIRSWT SLP+AA+ A IAW LTVLAMGF K
Sbjct: 61 NAATGFFVIFALLAGVVGAASTISGLSHIRSWTVGSLPAAATAATIAWTLTVLAMGFAWK 120
Query: 121 EIQLNIRNSRLKTMEAFLIILTATQLFYIAAIHG 154
EI+L RN++L+TMEAFLIIL+ TQL YIAA+HG
Sbjct: 121 EIELQGRNAKLRTMEAFLIILSVTQLIYIAAVHG 154
>At5g46530 putative protein
Length = 138
Score = 167 bits (422), Expect = 3e-42
Identities = 81/118 (68%), Positives = 99/118 (83%), Gaps = 6/118 (5%)
Query: 43 GPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAAS 102
G + LPAHFSPI+FPMGNAATGFF+ FALIAGV G S+ISG++H++SWT+ SLP+A S
Sbjct: 18 GADYSLPAHFSPIHFPMGNAATGFFIMFALIAGVAGAASVISGISHLQSWTTTSLPAAVS 77
Query: 103 VAAIAWALTVLAMGFGCKEIQLNIRNSRL------KTMEAFLIILTATQLFYIAAIHG 154
A IAW+LT+LAMGFGCKEI+L +RN+RL +TMEAFLIIL+ATQL YIAAI+G
Sbjct: 78 AATIAWSLTLLAMGFGCKEIELGMRNARLVSKPLMRTMEAFLIILSATQLLYIAAIYG 135
>At5g18970 plasma membrane associated protein -like
Length = 171
Score = 139 bits (350), Expect = 7e-34
Identities = 70/153 (45%), Positives = 97/153 (62%)
Query: 1 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMG 60
MA+ K A +LL LN +Y ++ I WA+N I+ E LPA PIYFP+G
Sbjct: 1 MASGGSKSAAFMLLMLNLGLYFVITIIASWAVNHGIERTRESASTLSLPAKIFPIYFPVG 60
Query: 61 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 120
N ATGFFV F LIAGVVG+ + ++G+ ++ W S +L SAA+ + I+W+LT+LAMG CK
Sbjct: 61 NMATGFFVIFTLIAGVVGMATSLTGIINVLQWDSPNLHSAAASSLISWSLTLLAMGLACK 120
Query: 121 EIQLNIRNSRLKTMEAFLIILTATQLFYIAAIH 153
EI + + L+T+E II++ATQL AIH
Sbjct: 121 EINIGWTEANLRTLEVLTIIVSATQLLCTGAIH 153
>At1g04560 unknown protein
Length = 186
Score = 122 bits (305), Expect = 1e-28
Identities = 68/157 (43%), Positives = 94/157 (59%), Gaps = 14/157 (8%)
Query: 1 MANEQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMG 60
MA + IA LL LN MY+IV+ W +N+ I+ G P F G
Sbjct: 1 MATTVGRNIAAPLLFLNLVMYLIVLGFASWCLNKYIN-GQTNHPSFG------------G 47
Query: 61 NAATGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGFGCK 120
N AT FF+TF+++A VVG+ S ++G NHIR W ++SL +A + + +AWA+T LAMG CK
Sbjct: 48 NGATPFFLTFSILAAVVGVASKLAGANHIRFWRNDSLAAAGASSIVAWAITALAMGLACK 107
Query: 121 EIQL-NIRNSRLKTMEAFLIILTATQLFYIAAIHGAA 156
+I + R RLK +EAF+IILT TQL Y+ IH +
Sbjct: 108 QINIGGWRGWRLKMIEAFIIILTFTQLLYLMLIHAGS 144
>At1g76670 unknown protein
Length = 347
Score = 33.5 bits (75), Expect = 0.051
Identities = 25/96 (26%), Positives = 43/96 (44%), Gaps = 4/96 (4%)
Query: 24 VISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLI 83
V +G WAMN G + + + S F TGF F + G+V + +
Sbjct: 12 VSDVGAWAMNVISSVGIIMANKQLMSS--SGFGFGFATTLTGFHFAFTALVGMVSNATGL 69
Query: 84 SGLNHIRSWTS--ESLPSAASVAAIAWALTVLAMGF 117
S H+ W S+ + S+AA+ ++L + ++GF
Sbjct: 70 SASKHVPLWELLWFSIVANISIAAMNFSLMLNSVGF 105
>At1g21070 unknown protein
Length = 348
Score = 32.7 bits (73), Expect = 0.086
Identities = 25/96 (26%), Positives = 42/96 (43%), Gaps = 4/96 (4%)
Query: 24 VISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLI 83
V +G WAMN G + + + S F TGF + G+V + +
Sbjct: 13 VSDVGAWAMNVTSSVGIIMANKQLMSS--SGFGFSFATTLTGFHFALTALVGMVSNATGL 70
Query: 84 SGLNHIRSWTS--ESLPSAASVAAIAWALTVLAMGF 117
S H+ W SL + S+AA+ ++L + ++GF
Sbjct: 71 SASKHVPLWELLWFSLVANISIAAMNFSLMLNSVGF 106
>At5g42420 unknown protein
Length = 350
Score = 31.2 bits (69), Expect = 0.25
Identities = 24/96 (25%), Positives = 41/96 (42%), Gaps = 4/96 (4%)
Query: 24 VISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLI 83
V +G WAMN G + + + S F TGF + G+V +
Sbjct: 13 VSDMGAWAMNVISSVGIIMANKQLMSS--SGFAFSFATTLTGFHFALTALVGMVSNATGF 70
Query: 84 SGLNHIRSWTS--ESLPSAASVAAIAWALTVLAMGF 117
S H+ W S+ + S+AA+ ++L + ++GF
Sbjct: 71 SASKHVPMWELIWFSIVANVSIAAMNFSLMLNSVGF 106
>At2g37200 unknown protein
Length = 179
Score = 28.9 bits (63), Expect = 1.2
Identities = 24/95 (25%), Positives = 41/95 (42%), Gaps = 2/95 (2%)
Query: 15 ALNFCMYVIVISIGG-WAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALI 73
A FC V+ +S+ W+++ I + + L H F +G+ T +TFA
Sbjct: 69 ATAFCCLVLAVSLQSLWSLSLFIIDAYALLVRRSLRNHSVVQCFTIGDGVTST-LTFAAA 127
Query: 74 AGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAW 108
+ GI LI+ L +A ++A I+W
Sbjct: 128 SASAGITVLINDLGQCNVNHCTRFETATAMAFISW 162
>At3g22140 hypothetical protein
Length = 1237
Score = 28.1 bits (61), Expect = 2.1
Identities = 20/61 (32%), Positives = 30/61 (48%), Gaps = 2/61 (3%)
Query: 25 ISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLIS 84
+ IGG A GFEIG G ++ F+ +G +TG F T + G V +G + +
Sbjct: 895 VVIGGVAAGGVATGGFEIG-GVEIGG-FAVGGVAIGGVSTGGFATGGVAVGGVVVGGVAT 952
Query: 85 G 85
G
Sbjct: 953 G 953
Score = 26.6 bits (57), Expect = 6.2
Identities = 20/61 (32%), Positives = 28/61 (45%), Gaps = 2/61 (3%)
Query: 25 ISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLIS 84
+ IGG A GFEIG G + F+ +G ATG F T + G +G + +
Sbjct: 525 VVIGGVAAGGVATGGFEIG-GVKIGG-FAVGGVAIGGVATGGFATGGVAVGGAVVGGVAT 582
Query: 85 G 85
G
Sbjct: 583 G 583
Score = 26.6 bits (57), Expect = 6.2
Identities = 20/60 (33%), Positives = 24/60 (39%), Gaps = 12/60 (20%)
Query: 26 SIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLISG 85
+IG +AM G IG F MG ATG VT G V IG + +G
Sbjct: 115 AIGDFAMGGVAIGGVTIGG------------FTMGGVATGGVVTGGFATGGVSIGDVATG 162
Score = 26.6 bits (57), Expect = 6.2
Identities = 18/61 (29%), Positives = 25/61 (40%), Gaps = 12/61 (19%)
Query: 25 ISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLIS 84
++IGG A+ + G G F MG ATG F T + G V +G +
Sbjct: 300 VTIGGIAVGGFVTGGVATGG------------FVMGGIATGGFTTGGIATGGVAVGGVAV 347
Query: 85 G 85
G
Sbjct: 348 G 348
Score = 26.6 bits (57), Expect = 6.2
Identities = 20/61 (32%), Positives = 29/61 (46%), Gaps = 2/61 (3%)
Query: 25 ISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGSLIS 84
+ IGG A FEIG G ++ F+ +G ATG F T + G V +G + +
Sbjct: 775 VVIGGVAAGGVATGDFEIG-GVEIGG-FAVGGVAIGGVATGGFATGGVAVGGVVVGGVTT 832
Query: 85 G 85
G
Sbjct: 833 G 833
Score = 26.2 bits (56), Expect = 8.1
Identities = 19/68 (27%), Positives = 29/68 (41%), Gaps = 3/68 (4%)
Query: 25 ISIGGWAMNRAIDHGFEIG---PGFDLPAHFSPIYFPMGNAATGFFVTFALIAGVVGIGS 81
+++GG + GF IG G + F MG ATG F + G V +G
Sbjct: 940 VAVGGVVVGGVATGGFSIGGVATGGVAVGGVAIGDFAMGGVATGGFAGGGVFIGDVAVGG 999
Query: 82 LISGLNHI 89
+ +G + I
Sbjct: 1000 VATGDSEI 1007
>At3g03720 putative cationic amino acid transporter
Length = 614
Score = 27.3 bits (59), Expect = 3.6
Identities = 22/103 (21%), Positives = 45/103 (43%), Gaps = 12/103 (11%)
Query: 4 EQMKPIATLLLALNFCMYVIVISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNAA 63
++ + ++ ++N C V +I +GG+ A G+ G+DLP+ + P F +
Sbjct: 192 KESSTVQAIVTSVNVCTLVFIIVVGGYL---ACKTGW---VGYDLPSGYFP--FGLNGIL 243
Query: 64 TGFFVTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAI 106
G V F +G ++ S +++ LP +A +
Sbjct: 244 AGSAVVF---FSYIGFDTVTSTAEEVKN-PQRDLPLGIGIALL 282
>At1g80530 nodulin-like protein
Length = 561
Score = 27.3 bits (59), Expect = 3.6
Identities = 12/25 (48%), Positives = 19/25 (76%)
Query: 66 FFVTFALIAGVVGIGSLISGLNHIR 90
F VTF ++AGV G+G+L+S + +R
Sbjct: 514 FRVTFLVLAGVCGLGTLLSIILTVR 538
>At4g18790 ion transporter - like protein
Length = 530
Score = 26.9 bits (58), Expect = 4.7
Identities = 27/86 (31%), Positives = 33/86 (37%), Gaps = 10/86 (11%)
Query: 24 VISIGGWAMNRAIDHGFEIGPGFDLPAHFSPIYFPMGNA----------ATGFFVTFALI 73
VI + N I F+IGP + A ++ M N GF V F +
Sbjct: 433 VIPLLTMVSNEHIMGVFKIGPSLEKLAWTVAVFVMMINGYLLLDFFMAEVEGFLVGFLVF 492
Query: 74 AGVVGIGSLISGLNHIRSWTSESLPS 99
GVVG S I L RS S S S
Sbjct: 493 GGVVGYISFIIYLVSYRSSQSSSWSS 518
>At1g29760 unknown protein
Length = 526
Score = 26.9 bits (58), Expect = 4.7
Identities = 15/48 (31%), Positives = 23/48 (47%), Gaps = 2/48 (4%)
Query: 18 FCMYVIVISIGGWAMNRAIDHGFEIGP--GFDLPAHFSPIYFPMGNAA 63
F + V + IGG+ +NR D FE+ FD + Y P+ + A
Sbjct: 273 FGLLVSSLMIGGYVINRIADKPFEVKETLNFDYTKNSPEAYVPISSCA 320
>At5g65290 unknown protein
Length = 733
Score = 26.6 bits (57), Expect = 6.2
Identities = 17/56 (30%), Positives = 27/56 (47%), Gaps = 1/56 (1%)
Query: 68 VTFALIAGVVGIGSLISGLNHIRSWTSESLPSAASVAAIAWALT-VLAMGFGCKEI 122
+ F L+ G +G+ LI + R+WT L A + + +T +GFG EI
Sbjct: 115 LVFYLVLGFIGLLGLILLIMMHRNWTGSILGYAMACSNTFGLVTGAFLLGFGLSEI 170
>At5g12900 unknown protein
Length = 562
Score = 26.2 bits (56), Expect = 8.1
Identities = 12/36 (33%), Positives = 18/36 (49%)
Query: 82 LISGLNHIRSWTSESLPSAASVAAIAWALTVLAMGF 117
+I+ LN R W+ + L + A W L +LA F
Sbjct: 461 IITTLNWTRPWSEQMLQAFFVAAKCVWLLHLLAFSF 496
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.140 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 3,137,119
Number of Sequences: 26719
Number of extensions: 117921
Number of successful extensions: 381
Number of sequences better than 10.0: 15
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 358
Number of HSP's gapped (non-prelim): 25
length of query: 160
length of database: 11,318,596
effective HSP length: 91
effective length of query: 69
effective length of database: 8,887,167
effective search space: 613214523
effective search space used: 613214523
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.8 bits)
S2: 56 (26.2 bits)
Medicago: description of AC144727.12


