

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144726.1 + phase: 0
(240 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
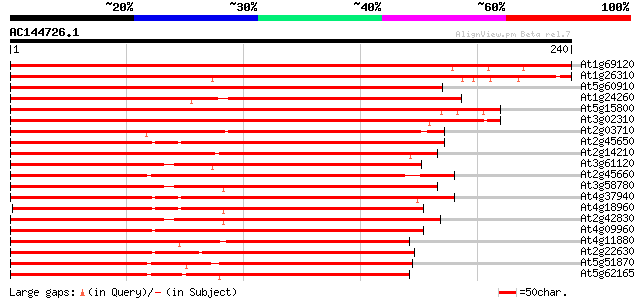
Score E
Sequences producing significant alignments: (bits) Value
At1g69120 unknown protein 354 2e-98
At1g26310 cauliflower (CAL) 307 3e-84
At5g60910 floral homeotic protein AGL8 270 5e-73
At1g24260 floral homeotic protein, AGL9 189 1e-48
At5g15800 transcription factor (AGL2) 184 4e-47
At3g02310 floral homeotic protein AGL4 182 2e-46
At2g03710 MADS-box protein (AGL3) 170 7e-43
At2g45650 MADS-box protein (AGL6) 169 9e-43
At2g14210 pseudogene 160 6e-40
At3g61120 MADS-box protein AGL13 158 3e-39
At2g45660 MADS-box protein (AGL20) 151 3e-37
At3g58780 shatterproof 1 (SHP1)/ agamous -like 1 (AGL1) 151 3e-37
At4g37940 MADS-box protein AGL17 -like protein 149 1e-36
At4g18960 floral homeotic protein agamous 147 5e-36
At2g42830 MADS box transcription factor (AGL5) 146 1e-35
At4g09960 MADS-box protein AGL11 144 3e-35
At4g11880 MADS-box protein AGL14 141 4e-34
At2g22630 MADS-box protein AGL17 (AGL17) 140 6e-34
At5g51870 MADS box transcription factor-like 139 1e-33
At5g62165 unknown protein 136 1e-32
>At1g69120 unknown protein
Length = 256
Score = 354 bits (909), Expect = 2e-98
Identities = 182/254 (71%), Positives = 210/254 (82%), Gaps = 14/254 (5%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVAL+VFSHKGKLFEY+TD
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALVVFSHKGKLFEYSTD 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
SCMEKILERYERYSYAERQL+A +S+ NW++EY RLKAKI+LL+RN RHY+GEDL +M
Sbjct: 61 SCMEKILERYERYSYAERQLIAPESDVNTNWSMEYNRLKAKIELLERNQRHYLGEDLQAM 120
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAA 180
S KELQ+LEQQLDTALK IRTR+NQ+MYESI+ELQKKEK IQEQN+ML+K+IKE+EKI
Sbjct: 121 SPKELQNLEQQLDTALKHIRTRKNQLMYESINELQKKEKAIQEQNSMLSKQIKEREKILR 180
Query: 181 QQQAQWEH------------PNHHGVNPNYLLQQQ-LPTLNMGGNYREEAP-EMGRNELD 226
QQ QW+ P H + Y+L Q P LNMGG Y+E+ P M RN+L+
Sbjct: 181 AQQEQWDQQNQGHNMPPPLPPQQHQIQHPYMLSHQPSPFLNMGGLYQEDDPMAMRRNDLE 240
Query: 227 LTLEPLYTCHLGCF 240
LTLEP+Y C+LGCF
Sbjct: 241 LTLEPVYNCNLGCF 254
>At1g26310 cauliflower (CAL)
Length = 255
Score = 307 bits (787), Expect = 3e-84
Identities = 166/254 (65%), Positives = 203/254 (79%), Gaps = 15/254 (5%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTFSKRR GLLKKA EISVLCDAEV+LIVFSHKGKLFEY+++
Sbjct: 1 MGRGRVELKRIENKINRQVTFSKRRTGLLKKAQEISVLCDAEVSLIVFSHKGKLFEYSSE 60
Query: 61 SCMEKILERYERYSYAERQLVANDS--ESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLG 118
SCMEK+LERYERYSYAERQL+A DS +Q NW++EY+RLKAKI+LL+RN RHY+GE+L
Sbjct: 61 SCMEKVLERYERYSYAERQLIAPDSHVNAQTNWSMEYSRLKAKIELLERNQRHYLGEELE 120
Query: 119 SMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKI 178
MSLK+LQ+LEQQL+TALK IR+R+NQ+M ES++ LQ+KEK IQE+N+ML K+IKE+E I
Sbjct: 121 PMSLKDLQNLEQQLETALKHIRSRKNQLMNESLNHLQRKEKEIQEENSMLTKQIKERENI 180
Query: 179 AAQQQAQWEHPNHH---------GVNPN-YLLQQQL-PTLNMGGNYREE-APEMGRNELD 226
+Q Q E N +P+ Y++ Q P LNMGG Y+EE M RN LD
Sbjct: 181 LRTKQTQCEQLNRSVDDVPQPQPFQHPHLYMIAHQTSPFLNMGGLYQEEDQTAMRRNNLD 240
Query: 227 LTLEPLYTCHLGCF 240
LTLEP+Y +LGC+
Sbjct: 241 LTLEPIYN-YLGCY 253
>At5g60910 floral homeotic protein AGL8
Length = 242
Score = 270 bits (690), Expect = 5e-73
Identities = 131/185 (70%), Positives = 159/185 (85%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRVQLKRIENKINRQVTFSKRR+GLLKKAHEISVLCDAEVALIVFS KGKLFEY+TD
Sbjct: 1 MGRGRVQLKRIENKINRQVTFSKRRSGLLKKAHEISVLCDAEVALIVFSSKGKLFEYSTD 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
SCME+ILERY+RY Y+++QLV D NW +E+ +LKA++++L++N R++MGEDL S+
Sbjct: 61 SCMERILERYDRYLYSDKQLVGRDVSQSENWVLEHAKLKARVEVLEKNKRNFMGEDLDSL 120
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAA 180
SLKELQSLE QLD A+K IR+R+NQ M+ESIS LQKK+K +Q+ NN L KKIKE+EK
Sbjct: 121 SLKELQSLEHQLDAAIKSIRSRKNQAMFESISALQKKDKALQDHNNSLLKKIKEREKKTG 180
Query: 181 QQQAQ 185
QQ+ Q
Sbjct: 181 QQEGQ 185
>At1g24260 floral homeotic protein, AGL9
Length = 251
Score = 189 bits (480), Expect = 1e-48
Identities = 100/200 (50%), Positives = 140/200 (70%), Gaps = 11/200 (5%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALI+FS++GKL+E+ +
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 SCMEKILERYERYSYA-------ERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYM 113
S M + LERY++ +Y R+ +A + SQ EY +LK + D LQR R+ +
Sbjct: 61 SSMLRTLERYQKCNYGAPEPNVPSREALAVELSSQQ----EYLKLKERYDALQRTQRNLL 116
Query: 114 GEDLGSMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIK 173
GEDLG +S KEL+SLE+QLD++LK IR R Q M + +++LQ KE+++ E N L ++
Sbjct: 117 GEDLGPLSTKELESLERQLDSSLKQIRALRTQFMLDQLNDLQSKERMLTETNKTLRLRLA 176
Query: 174 EKEKIAAQQQAQWEHPNHHG 193
+ ++ Q E +H+G
Sbjct: 177 DGYQMPLQLNPNQEEVDHYG 196
>At5g15800 transcription factor (AGL2)
Length = 251
Score = 184 bits (467), Expect = 4e-47
Identities = 103/221 (46%), Positives = 144/221 (64%), Gaps = 11/221 (4%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEVALI+FS++GKL+E+ +
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVALIIFSNRGKLYEFCSS 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S M K L+RY++ SY ++ ++ N EY +LK + + LQR R+ +GEDLG +
Sbjct: 61 SNMLKTLDRYQKCSYGSIEVNNKPAKELENSYREYLKLKGRYENLQRQQRNLLGEDLGPL 120
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAA 180
+ KEL+ LE+QLD +LK +R+ + Q M + +S+LQ KE+++ E N LA K+ + + +
Sbjct: 121 NSKELEQLERQLDGSLKQVRSIKTQYMLDQLSDLQNKEQMLLETNRALAMKLDDMIGVRS 180
Query: 181 QQQ---AQWEHPN------HHGVNPNYLLQ--QQLPTLNMG 210
WE HH L Q + PTL MG
Sbjct: 181 HHMGGGGGWEGGEQNVTYAHHQAQSQGLYQPLECNPTLQMG 221
>At3g02310 floral homeotic protein AGL4
Length = 250
Score = 182 bits (461), Expect = 2e-46
Identities = 101/221 (45%), Positives = 143/221 (64%), Gaps = 12/221 (5%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAEV+LIVFS++GKL+E+ +
Sbjct: 1 MGRGRVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEVSLIVFSNRGKLYEFCST 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S M K LERY++ SY ++ ++ N EY +LK + + LQR R+ +GEDLG +
Sbjct: 61 SNMLKTLERYQKCSYGSIEVNNKPAKELENSYREYLKLKGRYENLQRQQRNLLGEDLGPL 120
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKI-- 178
+ KEL+ LE+QLD +LK +R + Q M + +S+LQ KE ++ + N L+ K+++ +
Sbjct: 121 NSKELEQLERQLDGSLKQVRCIKTQYMLDQLSDLQGKEHILLDANRALSMKLEDMIGVRH 180
Query: 179 ---------AAQQQAQWEHPNHHGVNPNYLLQQQLPTLNMG 210
QQ + HP H L+ PTL +G
Sbjct: 181 HHIGGGWEGGDQQNIAYGHPQAHSQGLYQSLECD-PTLQIG 220
>At2g03710 MADS-box protein (AGL3)
Length = 257
Score = 170 bits (430), Expect = 7e-43
Identities = 84/187 (44%), Positives = 139/187 (73%), Gaps = 4/187 (2%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEY-AT 59
MGRG+V+LKRIENKINRQVTF+KRR GLLKKA+E+SVLCDAE+AL++FS++GKL+E+ ++
Sbjct: 1 MGRGKVELKRIENKINRQVTFAKRRNGLLKKAYELSVLCDAEIALLIFSNRGKLYEFCSS 60
Query: 60 DSCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGS 119
S M + +++Y ++SYA + + Q + +Y +LK+++++LQ + RH +GE+L
Sbjct: 61 PSGMARTVDKYRKHSYATMDPNQSAKDLQDKYQ-DYLKLKSRVEILQHSQRHLLGEELSE 119
Query: 120 MSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIA 179
M + EL+ LE+Q+D +L+ IR+ + + M + +S+L+ KE+++ E N L +K+++ + A
Sbjct: 120 MDVNELEHLERQVDASLRQIRSTKARSMLDQLSDLKTKEEMLLETNRDLRRKLEDSD--A 177
Query: 180 AQQQAQW 186
A Q+ W
Sbjct: 178 ALTQSFW 184
>At2g45650 MADS-box protein (AGL6)
Length = 252
Score = 169 bits (429), Expect = 9e-43
Identities = 94/186 (50%), Positives = 125/186 (66%), Gaps = 2/186 (1%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRGRV++KRIENKINRQVTFSKRR GLLKKA+E+SVLCDAEVALI+FS +GKL+E+ +
Sbjct: 1 MGRGRVEMKRIENKINRQVTFSKRRNGLLKKAYELSVLCDAEVALIIFSSRGKLYEFGSV 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
+E +ERY R Y E+ +W E T+LK+K + L R R+ +GEDLG M
Sbjct: 61 G-IESTIERYNR-CYNCSLSNNKPEETTQSWCQEVTKLKSKYESLVRTNRNLLGEDLGEM 118
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAA 180
+KELQ+LE+QL+ AL R R+ QVM E + +L+KKE+ + + N L K + +
Sbjct: 119 GVKELQALERQLEAALTATRQRKTQVMMEEMEDLRKKERQLGDINKQLKIKFETEGHAFK 178
Query: 181 QQQAQW 186
Q W
Sbjct: 179 TFQDLW 184
>At2g14210 pseudogene
Length = 234
Score = 160 bits (405), Expect = 6e-40
Identities = 84/185 (45%), Positives = 128/185 (68%), Gaps = 3/185 (1%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++ ++RI+N +RQVTFSKRR+GLLKKA E+S+LCDAEV +I+FS GKL++YA++
Sbjct: 1 MGRGKIVIRRIDNSTSRQVTFSKRRSGLLKKAKELSILCDAEVGVIIFSSTGKLYDYASN 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S M+ I+ERY R + QL+ + SE + W E L+ ++ LQ +R +GE+L M
Sbjct: 61 SSMKTIIERYNRVKEEQHQLLNHASEIK-FWQREVASLQQQLQYLQECHRKLVGEELSGM 119
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAK--KIKEKEKI 178
+ +LQ+LE QL T+LK +R +++Q+M I EL +K ++IQ++N+ L I KE I
Sbjct: 120 NANDLQNLEDQLVTSLKGVRLKKDQLMTNEIRELNRKGQIIQKENHELQNIVDIMRKENI 179
Query: 179 AAQQQ 183
Q++
Sbjct: 180 KLQKK 184
>At3g61120 MADS-box protein AGL13
Length = 244
Score = 158 bits (399), Expect = 3e-39
Identities = 87/177 (49%), Positives = 123/177 (69%), Gaps = 5/177 (2%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG+V++KRIENKI RQVTFSKR++GLLKKA+E+SVLCDAEV+LI+FS GKL+E++
Sbjct: 1 MGRGKVEVKRIENKITRQVTFSKRKSGLLKKAYELSVLCDAEVSLIIFSTGGKLYEFSNV 60
Query: 61 SCMEKILERYERYSYAERQLVANDS-ESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGS 119
I ERY + L+ ND+ E E T+LK K + L R +R+ +GEDL
Sbjct: 61 GVGRTI----ERYYRCKDNLLDNDTLEDTQGLRQEVTKLKCKYESLLRTHRNLVGEDLEG 116
Query: 120 MSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKE 176
MS+KELQ+LE+QL+ AL R ++ QVM E + EL++KE+ + + NN L + ++ +
Sbjct: 117 MSIKELQTLERQLEGALSATRKQKTQVMMEQMEELRRKERELGDINNKLKLETEDHD 173
>At2g45660 MADS-box protein (AGL20)
Length = 214
Score = 151 bits (382), Expect = 3e-37
Identities = 83/190 (43%), Positives = 122/190 (63%), Gaps = 7/190 (3%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M RG+ Q+KRIEN +RQVTFSKRR GLLKKA E+SVLCDAEV+LI+FS KGKL+E+A+
Sbjct: 1 MVRGKTQMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVSLIIFSPKGKLYEFAS- 59
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S M+ ++RY R++ E+ + E + KI+ L+ + R +GE +G+
Sbjct: 60 SNMQDTIDRYLRHTKDRVSTKPVSEENMQHLKYEAANMMKKIEQLEASKRKLLGEGIGTC 119
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEKIAA 180
S++ELQ +EQQL+ ++K IR R+ QV E I +L++KEK + +N L+ EK +
Sbjct: 120 SIEELQQIEQQLEKSVKCIRARKTQVFKEQIEQLKQKEKALAAENEKLS------EKWGS 173
Query: 181 QQQAQWEHPN 190
+ W + N
Sbjct: 174 HESEVWSNKN 183
>At3g58780 shatterproof 1 (SHP1)/ agamous -like 1 (AGL1)
Length = 248
Score = 151 bits (381), Expect = 3e-37
Identities = 81/186 (43%), Positives = 123/186 (65%), Gaps = 7/186 (3%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
+GRG++++KRIEN NRQVTF KRR GLLKKA+E+SVLCDAEVAL++FS +G+L+EYA +
Sbjct: 16 LGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEYANN 75
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGN---WTIEYTRLKAKIDLLQRNYRHYMGEDL 117
S I ERY A V S ++ N + E ++L+ +I +Q + RH +GE L
Sbjct: 76 SVRGTI----ERYKKACSDAVNPPSVTEANTQYYQQEASKLRRQIRDIQNSNRHIVGESL 131
Query: 118 GSMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEK 177
GS++ KEL++LE +L+ + +R+++N+++ I +QK+E +Q N L KI E +
Sbjct: 132 GSLNFKELKNLEGRLEKGISRVRSKKNELLVAEIEYMQKREMELQHNNMYLRAKIAEGAR 191
Query: 178 IAAQQQ 183
+ QQ
Sbjct: 192 LNPDQQ 197
>At4g37940 MADS-box protein AGL17 -like protein
Length = 228
Score = 149 bits (377), Expect = 1e-36
Identities = 77/192 (40%), Positives = 128/192 (66%), Gaps = 4/192 (2%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++ ++RI++ +RQVTFSKRR GL+KKA E+++LCDAEV LI+FS GKL+++A+
Sbjct: 1 MGRGKIVIQRIDDSTSRQVTFSKRRKGLIKKAKELAILCDAEVGLIIFSSTGKLYDFASS 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S M+ +++RY + S E+Q + N + W E L+ ++ LQ N+R MGE L +
Sbjct: 61 S-MKSVIDRYNK-SKIEQQQLLNPASEVKFWQREAAVLRQELHALQENHRQMMGEQLNGL 118
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIK--EKEKI 178
S+ EL SLE Q++ +L+ IR R+ Q++ + I EL +K +I ++N L++K++ +E +
Sbjct: 119 SVNELNSLENQIEISLRGIRMRKEQLLTQEIQELSQKRNLIHQENLDLSRKVQRIHQENV 178
Query: 179 AAQQQAQWEHPN 190
++A + N
Sbjct: 179 ELYKKAYMANTN 190
>At4g18960 floral homeotic protein agamous
Length = 284
Score = 147 bits (371), Expect = 5e-36
Identities = 78/177 (44%), Positives = 124/177 (69%), Gaps = 3/177 (1%)
Query: 2 GRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATDS 61
GRG++++KRIEN NRQVTF KRR GLLKKA+E+SVLCDAEVALIVFS +G+L+EY+ +S
Sbjct: 50 GRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSSRGRLYEYSNNS 109
Query: 62 CMEKILERYERYSYAERQLVANDSESQGN-WTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
++ +ERY++ + ++ + +E + E +L+ +I +Q + R MGE +GSM
Sbjct: 110 -VKGTIERYKK-AISDNSNTGSVAEINAQYYQQESAKLRQQIISIQNSNRQLMGETIGSM 167
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEK 177
S KEL++LE +L+ ++ IR+++N++++ I +QK+E + N +L KI E E+
Sbjct: 168 SPKELRNLEGRLERSITRIRSKKNELLFSEIDYMQKREVDLHNDNQILRAKIAENER 224
>At2g42830 MADS box transcription factor (AGL5)
Length = 246
Score = 146 bits (368), Expect = 1e-35
Identities = 78/187 (41%), Positives = 123/187 (65%), Gaps = 7/187 (3%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
+GRG++++KRIEN NRQVTF KRR GLLKKA+E+SVLCDAEVAL++FS +G+L+EYA +
Sbjct: 16 IGRGKIEIKRIENTTNRQVTFCKRRNGLLKKAYELSVLCDAEVALVIFSTRGRLYEYANN 75
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGN---WTIEYTRLKAKIDLLQRNYRHYMGEDL 117
S I ERY A V + ++ N + E ++L+ +I +Q RH +GE L
Sbjct: 76 SVRGTI----ERYKKACSDAVNPPTITEANTQYYQQEASKLRRQIRDIQNLNRHILGESL 131
Query: 118 GSMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEK 177
GS++ KEL++LE +L+ + +R+++++++ I +QK+E +Q N L KI E+
Sbjct: 132 GSLNFKELKNLESRLEKGISRVRSKKHEMLVAEIEYMQKREIELQNDNMYLRSKITERTG 191
Query: 178 IAAQQQA 184
+ Q+ +
Sbjct: 192 LQQQESS 198
>At4g09960 MADS-box protein AGL11
Length = 230
Score = 144 bits (364), Expect = 3e-35
Identities = 74/177 (41%), Positives = 117/177 (65%), Gaps = 1/177 (0%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++++KRIEN NRQVTF KRR GLLKKA+E+SVLCDAEVALIVFS +G+L+EYA +
Sbjct: 1 MGRGKIEIKRIENSTNRQVTFCKRRNGLLKKAYELSVLCDAEVALIVFSTRGRLYEYANN 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
+ + +ERY++ + + E +L+ +I +Q + R+ MG+ L S+
Sbjct: 61 N-IRSTIERYKKACSDSTNTSTVQEINAAYYQQESAKLRQQIQTIQNSNRNLMGDSLSSL 119
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIKEKEK 177
S+KEL+ +E +L+ A+ IR+++++++ I QK+E + +N L K+ E E+
Sbjct: 120 SVKELKQVENRLEKAISRIRSKKHELLLVEIENAQKREIELDNENIYLRTKVAEVER 176
>At4g11880 MADS-box protein AGL14
Length = 221
Score = 141 bits (355), Expect = 4e-34
Identities = 81/172 (47%), Positives = 114/172 (66%), Gaps = 3/172 (1%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M RG+ ++KRIEN +RQVTFSKRR GLLKKA E+SVLCDAEVALI+FS +GKL+E+++
Sbjct: 1 MVRGKTEMKRIENATSRQVTFSKRRNGLLKKAFELSVLCDAEVALIIFSPRGKLYEFSSS 60
Query: 61 SCMEKILERYE-RYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGS 119
S + K +ERY+ R ND+ Q E L KI+ L+ + R MGE L +
Sbjct: 61 SSIPKTVERYQKRIQDLGSNHKRNDNSQQSK--DETYGLARKIEHLEISTRKMMGEGLDA 118
Query: 120 MSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKK 171
S++ELQ LE QLD +L IR ++ Q++ E +L++KE+ + +N ML +K
Sbjct: 119 SSIEELQQLENQLDRSLMKIRAKKYQLLREETEKLKEKERNLIAENKMLMEK 170
>At2g22630 MADS-box protein AGL17 (AGL17)
Length = 227
Score = 140 bits (353), Expect = 6e-34
Identities = 69/173 (39%), Positives = 117/173 (66%), Gaps = 2/173 (1%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
MGRG++ +++I++ +RQVTFSKRR GL+KKA E+++LCDAEV LI+FS+ KL+++A+
Sbjct: 1 MGRGKIVIQKIDDSTSRQVTFSKRRKGLIKKAKELAILCDAEVCLIIFSNTDKLYDFASS 60
Query: 61 SCMEKILERYERYSYAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGEDLGSM 120
S ++ +ER+ E++L+ N + W E L+ ++ LQ NYR G +L +
Sbjct: 61 S-VKSTIERFNTAKMEEQELM-NPASEVKFWQREAETLRQELHSLQENYRQLTGVELNGL 118
Query: 121 SLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKIK 173
S+KELQ++E QL+ +L+ IR +R Q++ I EL +K ++ +N L++K++
Sbjct: 119 SVKELQNIESQLEMSLRGIRMKREQILTNEIKELTRKRNLVHHENLELSRKVQ 171
>At5g51870 MADS box transcription factor-like
Length = 207
Score = 139 bits (351), Expect = 1e-33
Identities = 74/176 (42%), Positives = 122/176 (69%), Gaps = 8/176 (4%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M RG++++K+IEN +RQVTFSKRR+GL KKAHE+SVLCDA+VA IVFS G+L EY++
Sbjct: 1 MVRGKIEIKKIENVTSRQVTFSKRRSGLFKKAHELSVLCDAQVAAIVFSQSGRLHEYSS- 59
Query: 61 SCMEKILERYERYS----YAERQLVANDSESQGNWTIEYTRLKAKIDLLQRNYRHYMGED 116
S MEKI++RY ++S AER V + +E R+ KIDLL+ ++R +G+
Sbjct: 60 SQMEKIIDRYGKFSNAFYVAERPQVERYLQ---ELKMEIDRMVKKIDLLEVHHRKLLGQG 116
Query: 117 LGSMSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKKI 172
L S S+ ELQ ++ Q++ +L+++R+R+ ++ + + +L++KE+ + + L +++
Sbjct: 117 LDSCSVTELQEIDTQIEKSLRIVRSRKAELYADQLKKLKEKERELLNERKRLLEEV 172
>At5g62165 unknown protein
Length = 210
Score = 136 bits (342), Expect = 1e-32
Identities = 73/172 (42%), Positives = 120/172 (69%), Gaps = 3/172 (1%)
Query: 1 MGRGRVQLKRIENKINRQVTFSKRRAGLLKKAHEISVLCDAEVALIVFSHKGKLFEYATD 60
M RG++++K+IEN +RQVTFSKRR GLLKKA+E+SVLCDA+++LI+FS +G+L+E+++
Sbjct: 1 MVRGKIEMKKIENATSRQVTFSKRRNGLLKKAYELSVLCDAQLSLIIFSQRGRLYEFSS- 59
Query: 61 SCMEKILERYERYSYAERQLVANDSESQ-GNWTIEYTRLKAKIDLLQRNYRHYMGEDLGS 119
S M+K +ERY +Y+ + + +DS+ E + + KI+LL+ + R +G+ + S
Sbjct: 60 SDMQKTIERYRKYT-KDHETSNHDSQIHLQQLKQEASHMITKIELLEFHKRKLLGQGIAS 118
Query: 120 MSLKELQSLEQQLDTALKLIRTRRNQVMYESISELQKKEKVIQEQNNMLAKK 171
SL+ELQ ++ QL +L +R R+ Q+ E + +L+ KEK + E+N L +K
Sbjct: 119 CSLEELQEIDSQLQRSLGKVRERKAQLFKEQLEKLKAKEKQLLEENVKLHQK 170
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.316 0.132 0.373
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,079,355
Number of Sequences: 26719
Number of extensions: 203376
Number of successful extensions: 1229
Number of sequences better than 10.0: 194
Number of HSP's better than 10.0 without gapping: 119
Number of HSP's successfully gapped in prelim test: 76
Number of HSP's that attempted gapping in prelim test: 1008
Number of HSP's gapped (non-prelim): 252
length of query: 240
length of database: 11,318,596
effective HSP length: 96
effective length of query: 144
effective length of database: 8,753,572
effective search space: 1260514368
effective search space used: 1260514368
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.6 bits)
S2: 58 (26.9 bits)
Medicago: description of AC144726.1


