

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.7 - phase: 0 /pseudo
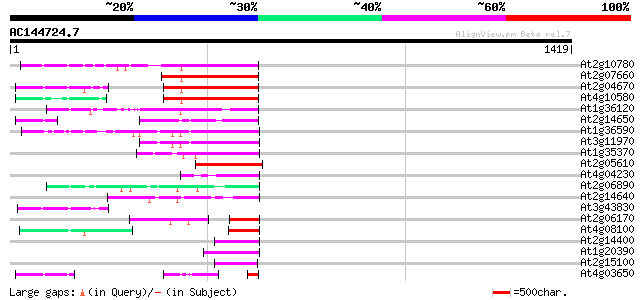
(1419 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 298 1e-80
At2g07660 putative retroelement pol polyprotein 227 4e-59
At2g04670 putative retroelement pol polyprotein 227 4e-59
At4g10580 putative reverse-transcriptase -like protein 226 7e-59
At1g36120 putative reverse transcriptase gb|AAD22339.1 210 4e-54
At2g14650 putative retroelement pol polyprotein 189 7e-48
At1g36590 hypothetical protein 177 3e-44
At3g11970 hypothetical protein 177 5e-44
At1g35370 hypothetical protein 157 5e-38
At2g05610 putative retroelement pol polyprotein 148 2e-35
At4g04230 putative transposon protein 122 1e-27
At2g06890 putative retroelement integrase 122 1e-27
At2g14640 putative retroelement pol polyprotein 121 3e-27
At3g43830 putative protein 103 7e-22
At2g06170 putative Ty3-gypsy-like retroelement pol polyprotein 85 3e-16
At4g08100 putative polyprotein 84 8e-16
At2g14400 putative retroelement pol polyprotein 79 1e-14
At1g20390 hypothetical protein 79 2e-14
At2g15100 putative retroelement pol polyprotein 77 5e-14
At4g03650 putative reverse transcriptase 76 2e-13
>At2g10780 pseudogene
Length = 1611
Score = 298 bits (764), Expect = 1e-80
Identities = 210/650 (32%), Positives = 315/650 (48%), Gaps = 91/650 (14%)
Query: 28 YNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWWTNAKGRLEIVGEV 87
+ G DP A W ++R F + +D + + H L +A WW + R EV
Sbjct: 182 FMGSTDPIVADEWRSRLKRNFKSTRCPEDYQRDIAVHFLEGDAHNWWLTVEKRRG--DEV 239
Query: 88 VTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKLSRFCPYIIAEDAM 147
++A F+ EF +KYFP + R E A+L+L G+ +V EY +F +L R+ + E+
Sbjct: 240 RSFADFEDEFNKKYFPPEAWDRLECAYLDLVQGNRTVREYDEEFNRLRRYVGRELEEEQ- 298
Query: 148 VSKCVKFESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDDAGRVKANYYKAQSEKRGKGHG 207
++ +F GLR +I + V+ LV + ++ ++ Y + + + +
Sbjct: 299 -AQLRRFIRGLRIEIRNHCLVRTFNSVSELVERAAMIEEG--IEEERYLNREKAPIRNNQ 355
Query: 208 VGKPYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDI-SCHKCGKAGHKAAE 266
KP DK++K + +K +C CG +++ C K I +C +CG H
Sbjct: 356 STKP--ADKKRKFDKVDNTKSDAKTGECVTCGK--NHSGTCWKAIGACGRCGSKDHAIQS 411
Query: 267 CKDVA---------RDVTCYNCGEKGHISTKCTK---------------------PKK-- 294
C + TC+ CG+ GH+ +C K PK+
Sbjct: 412 CPKMEPGQSKVLGEETRTCFYCGKTGHLKRECPKLTAEKQAGQRDNRGGNGLPPPPKRQA 471
Query: 295 AAGKVFALNAEEVEQPDNL--IRGMCFINSTHLIGIIDIGATHSFISVSCVKRLKLVVTP 352
A +V+ L+ EE N I G G++ + V+ + +VV
Sbjct: 472 VASRVYELS-EEANDAGNFRAITGGFRKEPNTDYGMVRAAGGQAMYPTGLVRGISVVVN- 529
Query: 353 LLRGMVIDTPARGSVTTSFMCAKCPVNFGNVDFELDLVCLPLKHMDVIFGMDWLLFFGVS 412
V+ DL+ +PLK DVI GMDWL +
Sbjct: 530 -----------------------------GVNMPADLIIVPLKKHDVILGMDWLGKYKAH 560
Query: 413 INCLTKSVTFSKPVEKLDRK---------FLTAEQVKKSLDGEAC-VFMMFASLKE-NSE 461
I+C + F + L + ++A Q ++ L G+ C ++ + KE +
Sbjct: 561 IDCHRGRIQFERDEGMLKFQGIRTTSGSLVISAIQAERML-GKGCEAYLATITTKEVGAS 619
Query: 462 KGVGDLPIVQEFPE---DITELPLEREVEFAIDLVPGMSPIWITPYPMSASELGELKKQL 518
+ D+PIV EF + ++ +P +R F I+L PG +PI PY M+ +E+ +LKKQL
Sbjct: 620 AELKDIPIVNEFSDVFAAVSGVPPDRSDPFTIELEPGTTPISKAPYRMAPAEMAKLKKQL 679
Query: 519 EELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQL 578
EELL+K FIRPS SPWGAPVL VKKKDGS RLC+DYR LNKVT+KN+YPL RID+LMDQL
Sbjct: 680 EELLDKGFIRPSSSPWGAPVLFVKKKDGSFRLCIDYRGLNKVTVKNKYPLPRIDELMDQL 739
Query: 579 VGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVP 628
GA+ FSKID SGYH ++ D+ KT FRTRY H+E+ VM FG+ N P
Sbjct: 740 GGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGLTNAP 789
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 227 bits (578), Expect = 4e-59
Identities = 126/260 (48%), Positives = 167/260 (63%), Gaps = 15/260 (5%)
Query: 383 VDFELDLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFSKPVEKLDRK---------F 433
V+ DL+ +PLK DVI GMDWL + I+C + + F + L +
Sbjct: 26 VNMPADLIIVPLKKHDVILGMDWLGKYKGHIDCHRERIQFERDEGMLKFQGIRTTSGSLV 85
Query: 434 LTAEQVKKSLDGEAC-VFMMFASLKE-NSEKGVGDLPIVQEFPE---DITELPLEREVEF 488
++A Q ++ L G+ C ++ + KE + + D+ IV EF + ++ +P +R F
Sbjct: 86 ISAIQAERML-GKGCEAYLATITTKEVGASAELKDILIVNEFSDVFAAVSGVPPDRSDPF 144
Query: 489 AIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSV 548
I+L PG +PI PY M+ +E+ ELKKQLEELL K FIRPS SPWGAPVL VKKKDGS
Sbjct: 145 TIELEPGTTPISKAPYRMAPAEMAELKKQLEELLAKGFIRPSSSPWGAPVLFVKKKDGSF 204
Query: 549 RLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVF 608
RLC+DYR LNKVT+KN+YPL RID+LMDQL GA+ FSKID SGYH ++ D+ KT F
Sbjct: 205 RLCIDYRGLNKVTVKNKYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAF 264
Query: 609 RTRYGHYEYFVMQFGVYNVP 628
RTRYGH+E+ VM FG+ N P
Sbjct: 265 RTRYGHFEFVVMPFGLTNAP 284
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 227 bits (578), Expect = 4e-59
Identities = 122/255 (47%), Positives = 164/255 (63%), Gaps = 15/255 (5%)
Query: 388 DLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFSKPVEKLDRK---------FLTAEQ 438
DL+ P++ DVI GMDWL + V ++C V+F +P +L + ++A Q
Sbjct: 394 DLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPTSGSLVISAVQ 453
Query: 439 VKKSLDGEACVFMMFASLKEN-SEKGVGDLPIVQEFPEDITE----LPLEREVEFAIDLV 493
KK ++ +++ S+ E+ + V D+ ++QEF ED+ + LP R F I+L
Sbjct: 454 AKKMIEKGCEAYLVTISMPESLGQVAVSDIRVIQEF-EDVFQSLQGLPPSRSDPFTIELE 512
Query: 494 PGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVD 553
PG +P+ PY M+ +E+ ELKKQLE+LL K FIRPS SPWGAPVL VKKKDGS RLC+D
Sbjct: 513 PGTAPLSKAPYRMAPAEMTELKKQLEDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCID 572
Query: 554 YRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYG 613
YR LN VT+KN+YPL RID+L+DQL GA FSKID SGYH + D+ KT FRTRYG
Sbjct: 573 YRGLNWVTVKNKYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYG 632
Query: 614 HYEYFVMQFGVYNVP 628
H+E+ VM F + N P
Sbjct: 633 HFEFVVMPFALTNAP 647
Score = 80.1 bits (196), Expect = 8e-15
Identities = 66/243 (27%), Positives = 101/243 (41%), Gaps = 19/243 (7%)
Query: 15 RRLDRFLRNNPPTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWW 74
R +++ R ++ PE A +W ++R F + + +V L H L +A WW
Sbjct: 125 RMMEQLQRIGTWYFSSGTSPEEADSWRSRVQRNFGSSRCPAEYRVDLAVHFLEGDAHLWW 184
Query: 75 TNAKGRLEIVGEVVTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKL 134
+ R ++WA F AEF KYFP++ R E FL L G SV EY +F +L
Sbjct: 185 RSVTARRRQAD--MSWADFVAEFNAKYFPQEALDRMEARFLELTQGEWSVREYDREFNRL 242
Query: 135 SRFCPYIIAEDAMVSKCVKFESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDD------AG 188
+ + +D ++ +F GLRPD+ V + LV + ++ G
Sbjct: 243 LAYAGRGMEDDQ--AQMRRFLRGLRPDLRVQCRVSQYATKAALVETAAEVEEDLQRQVVG 300
Query: 189 RVKANYYKAQSEKRGKGHGVGKPYNKDKRK---KREVGGGSKPSLADIKCYRCGTLGHYA 245
A K ++ G GKP KRK G G + + CG+L H
Sbjct: 301 VSPAVQTKNTQQQVTPSKG-GKPAQGQKRKWDHPSRAGQGGRAGY-----FSCGSLDHTV 354
Query: 246 NDC 248
DC
Sbjct: 355 ADC 357
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 226 bits (576), Expect = 7e-59
Identities = 119/254 (46%), Positives = 164/254 (63%), Gaps = 13/254 (5%)
Query: 388 DLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFSKPVEKLDRK---------FLTAEQ 438
DL+ P++ DVI GMDWL ++ V ++ V F +P +L + ++A Q
Sbjct: 368 DLIISPVELYDVILGMDWLDYYRVHLDWHRGRVFFERPEGRLVYQGVRPISGSLVISAVQ 427
Query: 439 VKKSLDGEACVFMMFASLKEN-SEKGVGDLPIVQEFPE---DITELPLEREVEFAIDLVP 494
+K ++ +++ S+ E+ + V D+ +VQEF + + LP + F I+L P
Sbjct: 428 AEKMIEKGCEAYLVTISMPESVGQVAVSDIRVVQEFQDVFQSLQGLPPSQSDPFTIELEP 487
Query: 495 GMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDY 554
G +P+ PY M+ +E+ ELKKQL++LL K FIRPS SPWGAPVL VKKKDGS RLC+DY
Sbjct: 488 GTAPLSKAPYRMAPAEMAELKKQLKDLLGKGFIRPSTSPWGAPVLFVKKKDGSFRLCIDY 547
Query: 555 RQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGH 614
R+LN+VT+KNRYPL RID+L+DQL GA FSKID SGYH + D+ KT FRTRYGH
Sbjct: 548 RELNRVTVKNRYPLPRIDELLDQLRGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGH 607
Query: 615 YEYFVMQFGVYNVP 628
+E+ VM FG+ N P
Sbjct: 608 FEFVVMPFGLTNAP 621
Score = 52.4 bits (124), Expect = 2e-06
Identities = 53/237 (22%), Positives = 90/237 (37%), Gaps = 44/237 (18%)
Query: 15 RRLDRFLRNNPPTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWW 74
R +++ R + ++G PE A +W + R F + + +V L H L +A WW
Sbjct: 139 RMMEQLQRIDTGYFSGGTSPEEADSWRSRVGRNFGSSRCPAEYRVDLAVHFLEGDAHLWW 198
Query: 75 TNAKGRLEIVGEVVTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKL 134
+ R ++WA F AEF KYFP+ E L
Sbjct: 199 RSVTARRRQTD--MSWADFVAEFKAKYFPQ---------------------------EAL 229
Query: 135 SRFCPYIIAEDAMVSKCVKFESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDDAGR----- 189
+ + +D ++ +F GLRPD+ V + LV + ++ +
Sbjct: 230 DPYAGQGMEDDQ--AQMRRFLRGLRPDLRVRCRVSQYATKAALVETAAEVEEDFQRQVVG 287
Query: 190 VKANYYKAQSEKRGKGHGVGKPYNKDKRK---KREVGGGSKPSLADIKCYRCGTLGH 243
V +++++ GKP KRK G G + +C+ CG+L H
Sbjct: 288 VSPVVQPKKTQQQVTPSKGGKPAQGQKRKWDHPSRAGQGGR-----ARCFSCGSLDH 339
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 210 bits (535), Expect = 4e-54
Identities = 179/556 (32%), Positives = 249/556 (44%), Gaps = 85/556 (15%)
Query: 93 FKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKLSRFCPYIIAEDAMVSKCV 152
F AEF KYFP++ R E FL L G +SV EY +F++L + + +D +
Sbjct: 157 FVAEFNAKYFPQEALDRMEARFLELTKGELSVREYGREFKRLLVYAGRGMEDDQAQMR-- 214
Query: 153 KFESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDDAGRVKANYYKAQSEK------RGKGH 206
+F GLRPD+ V + L + D +V A Q +K KG
Sbjct: 215 RFLRGLRPDLRVRCRVSQYATKAALTAAEVEEDLQRQVVAVSPSVQPKKIQQQVAPSKG- 273
Query: 207 GVGKPYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAE 266
GKP KRK +R G G A D S G A A
Sbjct: 274 --GKPAQVQKRKWDHP-------------FRAGQGGR-AGSAADDSSTSTAGGAAQSAGA 317
Query: 267 CKDVARDVTCYNCGEKGHISTKCTKPKKAAGKVFALNAEEVEQPDNLIRGMCFINSTHLI 326
+ A E +ST C+ G + A+ LI G C H++
Sbjct: 318 ARSAA---------ECAAVSTYCS------GTTWLYYAD-------LISGRC---RGHVL 352
Query: 327 GIIDIGATHSFISVSCVKRLKLVVTPLLRGMVIDTPARGSVTTSFMCAKCPVNFGNVDFE 386
D GA+H FI+ R + P + + +T +
Sbjct: 353 --FDSGASHCFITPESASRDNIRGEPGEQFGAVKIAGGQFLTVLGRAKGVNIQIEEESMP 410
Query: 387 LDLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFSKPVEKL--------DRKFLTAE- 437
DL+ P+ D I GMDWL + V ++C V+F +P +L R + +E
Sbjct: 411 ADLIINPVVLYDAILGMDWLDHYRVHLDCHRGRVSFERPEGRLVYQGVRPTSRSLVISEV 470
Query: 438 QVKKSLDGEACVFMMFASLKEN-SEKGVGDLPIVQEFPEDITE----LPLEREVEFAIDL 492
QV+K ++ +++ S+ E+ + V D+ +VQEF ED+ + LP R F I+L
Sbjct: 471 QVEKMIEKGCEAYLVTISMSESVGQVAVSDIWVVQEF-EDVFQSLQGLPPSRSDPFTIEL 529
Query: 493 VPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCV 552
G +P+ TPY M +E+ ELKKQLE+LL K FIRPS S WGAP
Sbjct: 530 ELGTAPLSKTPYRMVPAEIAELKKQLEDLLGKGFIRPSTSRWGAP--------------- 574
Query: 553 DYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRY 612
LN+VT+KN+YPL RID+L+DQL GA FSKID GYH + D+ KT FRTRY
Sbjct: 575 ---GLNRVTLKNKYPLPRIDELLDQLRGATCFSKIDLTPGYHQFPIAEADVRKTAFRTRY 631
Query: 613 GHYEYFVMQFGVYNVP 628
GH+E+ VM FG+ N P
Sbjct: 632 GHFEFVVMPFGLTNAP 647
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 189 bits (481), Expect = 7e-48
Identities = 116/301 (38%), Positives = 157/301 (51%), Gaps = 20/301 (6%)
Query: 328 IIDIGATHSFISVSCVKRLKLVVTPLLRGMVIDTPARGSVTTSFMCAKCPVNFGNVDFEL 387
+ D GA+H FI+ R + P + + V +
Sbjct: 316 LFDSGASHCFITPESASRGNIRGDPGEQLGAVKVAGGQFVAVLGRTKGVDIQIAGESMPA 375
Query: 388 DLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFSKPVEKLDRKFLTAEQVKKSLDGEA 447
DL+ P++ DVI GMDWL + V ++C V+F +P L Q + G
Sbjct: 376 DLIISPVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGSL------VYQGVRPTSGSL 429
Query: 448 CVFMMFASLKENSEKGVGDLPIVQEFPEDITELPLEREVEFAIDLVPGMSPIWITPYPMS 507
+ + A ++ EKG ++ + + LP R F I+L PG +P+ PY M+
Sbjct: 430 VISAVQA--EKMIEKGCEAYLEFEDVFQSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMA 487
Query: 508 ASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYP 567
+E+ ELKKQLE+LL GAPVL VKKKDGS RLC+DYR LN VT+KN+YP
Sbjct: 488 PAEMAELKKQLEDLL------------GAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYP 535
Query: 568 LLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNV 627
L RID+L+DQL GA FSKID SGYH + D+ KT FRTRYGH+E+ VM FG+ N
Sbjct: 536 LPRIDELLDQLRGATCFSKIDLTSGYHLIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNA 595
Query: 628 P 628
P
Sbjct: 596 P 596
Score = 59.7 bits (143), Expect = 1e-08
Identities = 33/106 (31%), Positives = 50/106 (47%), Gaps = 2/106 (1%)
Query: 15 RRLDRFLRNNPPTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWW 74
R +++ R ++G PE A +W +ER F + + ++ L H L +A WW
Sbjct: 143 RMMEQLQRIGTGYFSGGTSPEVADSWRSRVERNFGSSRCPAEYRIDLAVHFLEGDAHLWW 202
Query: 75 TNAKGRLEIVGEVVTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*G 120
+ R ++WA F AEF KYFP++ R E FL L G
Sbjct: 203 RSVTARRRQAD--ISWADFVAEFNAKYFPQEALDRMEAHFLELTQG 246
>At1g36590 hypothetical protein
Length = 1499
Score = 177 bits (450), Expect = 3e-44
Identities = 175/662 (26%), Positives = 283/662 (42%), Gaps = 106/662 (16%)
Query: 31 RFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYW---WTNAKGRLEIVGEV 87
RFD + W+ +E F T +D KV++ A W + + LE++ +
Sbjct: 116 RFDGTRLKEWLFKVEEFFGVDSTPEDMKVKMAAIHFDSHASTWHQSFIQSGVGLEVLYD- 174
Query: 88 VTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKLSRFCPYIIAEDAM 147
W + + L++ F +D +A L + + +Y KFE + ++E+ +
Sbjct: 175 --WKGY-VKLLKERFEDDCDD--PMAELKHLQETDGIIDYHQKFELIKTRVN--LSEEYL 227
Query: 148 VSKCVKFESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDDAGRVKANYYKAQSEKRGKGHG 207
VS + +GLR D ++ + + + + + ++ A +K S R G
Sbjct: 228 VSV---YLAGLRTDTQMHVRMFQPQTVRHCLFLGKTYEKA-HLKKPANTTWSTNRSAPTG 283
Query: 208 VGKPYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCKKDISCHKCGKAGHKAAEC 267
Y K+ K + HY N + K + E
Sbjct: 284 GYNKYQKEGESKTD---------------------HYGNKGNFKPVSQQPKKMSQQ--EM 320
Query: 268 KDVARDVTCYNCGEKGH-----ISTKCTKPKKAAGKVFALNAEEVEQPD---------NL 313
D CY C EK + K + + F EE+ D N
Sbjct: 321 SDRRSKGLCYFCDEKYTPEHYLVHKKTQLFRMDVDEEFEDAREELVNDDDEHMPQISVNA 380
Query: 314 IRGMCFINSTHLIG---------IIDIGATHSFISVSCVKRL--KLVVTPLLRGMVID-- 360
+ G+ + + G +ID G+TH+F+ + +L K+ L R V D
Sbjct: 381 VSGIAGYKTMRVKGTYDKKIIFILIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGR 440
Query: 361 -TPARGSVTTSFMCAKCPVNFGNVDFELDLVCLPLKHMDVIFGMDWLLFFG--------- 410
G VT F+ D++ +PL+ +D++ G+ WL G
Sbjct: 441 KLRVEGKVTDFSW------KLQTTTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKL 494
Query: 411 ----------VSINCLTKSVTFSKPVEKLDR--------KFLTAEQVKKSLDGEACVFMM 452
V ++ LT +KL + L ++V +S +GE C
Sbjct: 495 EMRFKFNNQKVLLHGLTSGSVREVKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINA 554
Query: 453 FASLKENSEKGVGDLPIVQEFPE---DITELPLEREVE-FAIDLVPGMSPIWITPYPMSA 508
S E E+ V + ++ E+P+ + T LP RE I L+ G +P+ PY S
Sbjct: 555 LTS--ELGEESVVE-EVLNEYPDIFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSI 611
Query: 509 SELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPL 568
+ E+ K +E+LL ++ S SP+ +PV+LVKKKDG+ RLCVDYR+LN +T+K+ +P+
Sbjct: 612 HQKNEIDKLVEDLLTNGTVQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPI 671
Query: 569 LRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVP 628
I+DLMD+L GA +FSKID R+GYH R+ +DI KT F+T GH+EY VM FG+ N P
Sbjct: 672 PLIEDLMDELGGAVIFSKIDLRAGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAP 731
Query: 629 GT 630
T
Sbjct: 732 AT 733
>At3g11970 hypothetical protein
Length = 1499
Score = 177 bits (448), Expect = 5e-44
Identities = 117/339 (34%), Positives = 176/339 (51%), Gaps = 45/339 (13%)
Query: 328 IIDIGATHSFISVSCVKRL--KLVVTPLLRGMVID---TPARGSVTTSFMCAKCPVNFGN 382
+ID G+TH+F+ + +L K+ L R V D G VT
Sbjct: 404 LIDSGSTHNFLDPNTAAKLGCKVDTAGLTRVSVADGRKLRVEGKVTDFSW------KLQT 457
Query: 383 VDFELDLVCLPLKHMDVIFGMDWLLFFG-------------------VSINCLTKSVTFS 423
F+ D++ +PL+ +D++ G+ WL G V ++ LT
Sbjct: 458 TTFQSDILLIPLQGIDMVLGVQWLETLGRISWEFKKLEMRFKFNNQKVLLHGLTSGSVRE 517
Query: 424 KPVEKLDR--------KFLTAEQVKKSLDGEACVFMMFASLKENSEKGVGDLPIVQEFPE 475
+KL + L ++V +S +GE C S E E+ V + ++ E+P+
Sbjct: 518 VKAQKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTS--ELGEESVVE-EVLNEYPD 574
Query: 476 ---DITELPLEREVE-FAIDLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSV 531
+ T LP RE I L+ G +P+ PY S + E+ K +E+LL ++ S
Sbjct: 575 IFIEPTALPPFREKHNHKIKLLEGSNPVNQRPYRYSIHQKNEIDKLVEDLLTNGTVQASS 634
Query: 532 SPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRS 591
SP+ +PV+LVKKKDG+ RLCVDYR+LN +T+K+ +P+ I+DLMD+L GA +FSKID R+
Sbjct: 635 SPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLRA 694
Query: 592 GYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGT 630
GYH R+ +DI KT F+T GH+EY VM FG+ N P T
Sbjct: 695 GYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPAT 733
>At1g35370 hypothetical protein
Length = 1447
Score = 157 bits (396), Expect = 5e-38
Identities = 107/349 (30%), Positives = 170/349 (48%), Gaps = 49/349 (14%)
Query: 320 INSTHLIGIIDIGATHSFISVSCVKRLKLVVTPLLRGMVIDTPARGSVTTSFMCAKCPVN 379
++ L +ID G+TH+FI + +L V V R +
Sbjct: 375 VDKRDLFILIDSGSTHNFIDSTVAAKLGCHVESAGLTKVAVADGR-KLNVDGQIKGFTWK 433
Query: 380 FGNVDFELDLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFSKPVEKLDRKFLTAEQ- 438
+ F+ D++ +PL+ +D++ G+ WL G S +KL+ +F Q
Sbjct: 434 LQSTTFQSDILLIPLQGVDMVLGVQWLETLG----------RISWEFKKLEMQFFYKNQR 483
Query: 439 -------------------VKKSLDGEACVFMMFASLKENSEKGVGDLP----------- 468
K D + + + E+ +G +
Sbjct: 484 VWLHGIITGSVRDIKAHKLQKTQADQIQLAMVCVREVVSDEEQEIGSISALTSDVVEESV 543
Query: 469 ---IVQEFPE---DITELPLEREV-EFAIDLVPGMSPIWITPYPMSASELGELKKQLEEL 521
IV+EFP+ + T+LP RE + I L+ G +P+ PY + E+ K ++++
Sbjct: 544 VQNIVEEFPDVFAEPTDLPPFREKHDHKIKLLEGANPVNQRPYRYVVHQKDEIDKIVQDM 603
Query: 522 LEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGA 581
++ I+ S SP+ +PV+LVKKKDG+ RLCVDY +LN +T+K+R+ + I+DLMD+L G+
Sbjct: 604 IKSGTIQVSSSPFASPVVLVKKKDGTWRLCVDYTELNGMTVKDRFLIPLIEDLMDELGGS 663
Query: 582 EVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGT 630
VFSKID R+GYH R+ +DI KT F+T GH+EY VM FG+ N P T
Sbjct: 664 VVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFEYLVMLFGLTNAPAT 712
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 148 bits (373), Expect = 2e-35
Identities = 77/175 (44%), Positives = 115/175 (65%), Gaps = 4/175 (2%)
Query: 469 IVQEFPE---DITELPLEREV-EFAIDLVPGMSPIWITPYPMSASELGELKKQLEELLEK 524
+V EFP+ + T+LP R + I+L+ +P+ PY + E+ K ++++L
Sbjct: 10 VVTEFPDIFVEPTDLPPFRAPHDHKIELLEDSNPVNQRPYRYVVHQKNEIDKIVDDMLAS 69
Query: 525 QFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVF 584
I+ S SP+ +PV+LVKKKDG+ RLCVDYR+LN +T+K+R+P+ I+DLMD+L G+ V+
Sbjct: 70 GTIQASSSPYASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMDELGGSNVY 129
Query: 585 SKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGTSWNI*IEYF 639
SKID R+GYH R+ DI KT F+T GHYEY VM FG+ N P + ++ +F
Sbjct: 130 SKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMNSFF 184
>At4g04230 putative transposon protein
Length = 315
Score = 122 bits (306), Expect = 1e-27
Identities = 76/202 (37%), Positives = 109/202 (53%), Gaps = 37/202 (18%)
Query: 433 FLTAEQVKKSLDGEACVFMMFASLKENSEKGVGDLPIVQEFPEDITELPLEREVEFAI-- 490
+L+++ V K++ + V +M KE G+GD PE L EV+ +
Sbjct: 65 YLSSKDVCKTMSAKGTVLLMV--FKECLSTGIGD-------PE------LSAEVQVVLHW 109
Query: 491 --DLVPGMSPIWITPYPMSASELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSV 548
DL P P + P + K +IR S+SP PVLLV KKDG+
Sbjct: 110 YKDLFPEEIPPGLPP------------------IHKGYIRESLSPCAVPVLLVPKKDGTW 151
Query: 549 RLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVF 608
R+C+D R +N +TIK R+P+ R+ D++D+L GA +FSK+D RSGYH R++ D KT F
Sbjct: 152 RMCLDCRAINNITIKYRHPIPRLYDMLDELSGAIIFSKVDLRSGYHQVRMREGDEWKTAF 211
Query: 609 RTRYGHYEYFVMQFGVYNVPGT 630
+T+ G YE VM FG+ N P T
Sbjct: 212 KTKQGLYECLVMPFGLTNAPST 233
>At2g06890 putative retroelement integrase
Length = 1215
Score = 122 bits (306), Expect = 1e-27
Identities = 148/601 (24%), Positives = 234/601 (38%), Gaps = 115/601 (19%)
Query: 94 KAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKLSRFCPYIIAEDAMVSKCVK 153
KA +++ P + NL G+ SV EY + E L +A +S+
Sbjct: 2 KAIMRKRFVPSHYHRELHLKLRNLTQGNRSVEEYYKEMETLMLRADISEDREATLSR--- 58
Query: 154 FESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDDAGRVKANYYKAQSEKRGKGHG-VGKPY 212
F L DI + Q + ++HK F+ + K+ S + G G + KP
Sbjct: 59 FLGDLNRDIQDRLETQYYVQIEEMLHKAILFEQQVKRKS------SSRSSYGSGTIAKPT 112
Query: 213 NKDKRKKREVGGGSKPSL---ADIKCYRCGT----LGHYANDCKKDISCHKCGKAGHKAA 265
+ R++R +KP + A+ K Y + +D+ C+KC GH A
Sbjct: 113 YQ--REERTSSYHNKPIVSPRAESKPYAAVQDHKGKAEISTSRVRDVRCYKCQGKGHYAN 170
Query: 266 ECKDVARDVTCYNCGE--------KGHISTKCTKPKKAAGKVFALN--------AEEVEQ 309
EC + R + + GE S K + A G++ +E EQ
Sbjct: 171 ECPN-KRVMILLDNGEIEPEEEIPDSPSSLKENEELPAQGELLVARRTLSVQTKTDEQEQ 229
Query: 310 PDNLIRGMCFINSTHLIGIIDIGATHSFISVSCVKRLKLVVTPLLRGMVIDTPARGSVTT 369
NL C ++ IID G+ + S + VK+L L ++ + V
Sbjct: 230 RKNLFHTRCHVHGKVCSLIIDGGSCTNVASETMVKKLGL--------KWLNDSGKMRVKN 281
Query: 370 SFMCAKCPVNFGNVDFELDLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFS------ 423
+ P+ G + E+ LP++ ++ G W V + T +F
Sbjct: 282 QVVV---PIVIGKYEDEILCDVLPMEAGHILLGRPWQSDRKVMHDGFTNRHSFEFKGGKT 338
Query: 424 ---------------------KPVEKLDRKFLTAEQVKKSLDGEACVFMMFASLKENSEK 462
+ V K F + +VK + + M+ KE
Sbjct: 339 ILVSMTPHEVYQDQIHLKQKKEQVVKQPNFFAKSGEVKSAYSSKQP--MLLFVFKEALTS 396
Query: 463 GVGDLPIVQE------------FPEDITE-LPLEREVEFAIDLVPGMSPIWITPYPMSAS 509
P++ FPED + LP R +E ID VPG S Y +
Sbjct: 397 LTNFAPVLPSEMTSLLQDYKDVFPEDNPKGLPPIRGIEHQIDFVPGASLPNRPAYRTNPV 456
Query: 510 ELGELKKQLEELLEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLL 569
E EL++Q KDGS R+C D R +N VT+K +P+
Sbjct: 457 ETKELQRQ--------------------------KDGSWRMCFDCRAINNVTVKYCHPIP 490
Query: 570 RIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPG 629
R+DD++D+L G+ +FSKID +SGYH R+ D KT F+T++G YE+ VM FG+ + P
Sbjct: 491 RLDDMLDELHGSSIFSKIDLKSGYHQIRMNEGDEWKTAFKTKHGLYEWLVMPFGLTHAPS 550
Query: 630 T 630
T
Sbjct: 551 T 551
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 121 bits (304), Expect = 3e-27
Identities = 113/409 (27%), Positives = 171/409 (41%), Gaps = 69/409 (16%)
Query: 248 CKKDISCHKCGKAGHKAAECKDVARDVTCYNCGEKGHISTKCTKPKKAAGKVFALNAEEV 307
C K S + + K E K V V+ GE + + + L A E
Sbjct: 252 CAKRQSNRRQRDSVRKNYERKGVWAYVSAVMKGENDDEAVEEESTEAEGKPEITLYALEG 311
Query: 308 EQPDNLIRGMCFINSTHLIGIIDIGATHSFISVSCVKRLKLVVTP-------LLRGMVID 360
+ IR I+ LI +I+ G+TH+FI V+ + L T ++ GM +
Sbjct: 312 VDTTSTIRVRATIHRNRLIALINSGSTHNFIGEKAVRGMNLKATTTKPFTVRVVNGMPLV 371
Query: 361 TPARGSVTTSFMCAKCPVNFGNVDFELDLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSV 420
+R PV G V F + L LPL +D+ G+ WL G ++ C K
Sbjct: 372 CRSRYEAI--------PVVMGGVVFPVTLYALPLMGLDLAMGVQWLSTLGPTL-CNWKEQ 422
Query: 421 TFS-------------KP--VEKLDRKFLTAEQVKKSLDGEACVFMMFASLKENSEKGVG 465
T KP + ++ K +T KK+ G + A + G
Sbjct: 423 TLQFHWAGDEVRLMGIKPTGLRGVEHKTIT----KKARMGHTIFAITMAHNSSDPNTPDG 478
Query: 466 DLP-IVQEFP---EDITELPLEREVEFAIDLVPGMSPIWITPYPMSASELGELKKQLEEL 521
++ +++EF E T+LP ER + I L G P+ + PY + Q +E+
Sbjct: 479 EIKKLIEEFAGLFETPTQLPSERPIVHRIALKKGTDPVNVRPYRYAFY-------QKDEM 531
Query: 522 LEKQFIRPSVSPWGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGA 581
IRPS SP+ +PVLL R+P+ +DD++D+L GA
Sbjct: 532 SRAGIIRPSSSPFLSPVLL-----------------------ERFPIPTVDDMLDELNGA 568
Query: 582 EVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGT 630
F+K+D +GY R+ + DI K F+T GHYEY VM FG+ N P T
Sbjct: 569 VYFTKLDLTAGYQQVRMHSPDIPKIAFQTHNGHYEYLVMPFGLCNAPST 617
>At3g43830 putative protein
Length = 329
Score = 103 bits (257), Expect = 7e-22
Identities = 81/235 (34%), Positives = 110/235 (46%), Gaps = 24/235 (10%)
Query: 19 RFLRN-NPPTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWWTNA 77
RF+R+ + T+NG DP A W +E F +M + L + L EA+ WW
Sbjct: 106 RFMRDLDIETFNGGTDPVKAYNWRNMLECKFKSMRCPVEFWRDLASCYLRGEAQEWWERV 165
Query: 78 KGRLEIVGEVVTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKLSRF 137
K R E VG V W+ FK EF R+Y PE+ E+ FL L+ G+ +V +Y +F L RF
Sbjct: 166 KQR-EQVGCVDQWSFFKEEFTRRYLPEETIDDLEMKFLRLQQGTKTVRKYEKEFHSLERF 224
Query: 138 CPYIIAEDAMVSKCVKFESGLRPDIYQYMCVQEIRDFDT---LVHKCRKFDDAGRVKANY 194
E ++ KF SGLR DI + C +RDFD LV K + +A Y
Sbjct: 225 ERRKRGEHELIH---KFISGLRVDI-RLCC--HVRDFDNMIDLVEKAASLEIGLEEEARY 278
Query: 195 YKAQSEKRGKGHGVGKPYNKDKRKKREVGGGSKPSLADIKCYRCGTLGHYANDCK 249
+ EK G Y + G SK ++ CYRCG GH A DC+
Sbjct: 279 KRIAQEKEAMG-----SYGQT--------GHSKRRCQEVTCYRCGVAGHIARDCR 320
>At2g06170 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 587
Score = 85.1 bits (209), Expect = 3e-16
Identities = 38/76 (50%), Positives = 57/76 (75%)
Query: 555 RQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGH 614
+ +NK+TIK R+P+ ++DD++D+L G++VFSKID RSGYH R++ D KT F+++ G
Sbjct: 401 KAINKITIKYRFPIPQLDDMLDELSGSKVFSKIDLRSGYHQIRIRPGDEWKTDFKSKDGL 460
Query: 615 YEYFVMQFGVYNVPGT 630
YE+ VM FG+ N P T
Sbjct: 461 YEWQVMPFGMSNAPST 476
Score = 44.3 bits (103), Expect = 5e-04
Identities = 57/235 (24%), Positives = 97/235 (41%), Gaps = 34/235 (14%)
Query: 302 LNAEEVEQPDNLIRGMCFINSTHLIGIIDIGATHSFISVSCVKRLKLVVTPLLRGMVIDT 361
L+++E Q NL R C IN I+DIG++ + +S V+ LKL T + +
Sbjct: 213 LSSKEEGQRRNLFRTRCSINDKVCNLIVDIGSSENLVSQKLVEYLKLPTTLHQKPYSLGW 272
Query: 362 PARGSVTTSFMCAKCPVNFGNVDFELDLVC--LPLKHMDVIFGMDW-------------- 405
++GS + + P++ G ++ +++C L + +I G W
Sbjct: 273 VSKGSQFCVSLSCRVPISIGK-HYKEEVLCDVLNMDVCHIILGRSWQYDNDITYRGKDNV 331
Query: 406 LLFFGVSINCLTKSVT-FSKPVEKLDRKFLTAEQVKKSLDGE----ACVF------MMFA 454
L+F + V+ F + + K + FL Q +K LD C+ +M A
Sbjct: 332 LMFTWNGHKIVMAPVSHFDQNLVKKNSNFLVVTQSEKELDEAIKEIECICPVVIKGLMSA 391
Query: 455 SLKEN--SEKGVGDLPIVQEFP----EDITELPLEREVEFAIDLVPGMSPIWITP 503
K N +K + + I FP +D+ + +V IDL G I I P
Sbjct: 392 EKKTNRFRQKAINKITIKYRFPIPQLDDMLDELSGSKVFSKIDLRSGYHQIRIRP 446
>At4g08100 putative polyprotein
Length = 1054
Score = 83.6 bits (205), Expect = 8e-16
Identities = 37/78 (47%), Positives = 53/78 (67%)
Query: 553 DYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRY 612
D R +N +T+K R+P+ R+DD +D+L G+ +FSKID +SGYH R+K D KT +T+
Sbjct: 527 DCRAINNITVKYRHPIPRLDDTLDKLHGSSIFSKIDLKSGYHQTRMKEGDEWKTAIKTKQ 586
Query: 613 GHYEYFVMQFGVYNVPGT 630
YE+ VM FG+ N P T
Sbjct: 587 RLYEWLVMPFGLTNAPNT 604
Score = 67.0 bits (162), Expect = 7e-11
Identities = 72/307 (23%), Positives = 119/307 (38%), Gaps = 25/307 (8%)
Query: 26 PTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWWTN--AKGRLEI 83
P ++G +P+ W Q +E +F KV++ A WW R
Sbjct: 97 PAFHGTDNPDTYLEWEQKIELVFLCQECLQSNKVKIAATKFYNYALSWWDQLVTSRRRTR 156
Query: 84 VGEVVTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKLSRFCPYIIA 143
+ TW + K +++ P NL GS +V EY + E L
Sbjct: 157 DYPIKTWNQLKFVMRKRFVPSYYHRELHQRLRNLVQGSKTVEEYFLEMETLMLRADLQED 216
Query: 144 EDAMVSKCVKFESGLRPDIYQYMCVQEIRDFDTLVHKCRKFDD--------AGRVKANYY 195
+A++S +F GL +I + Q + + ++HK F+ + K NY
Sbjct: 217 GEAVMS---RFMGGLNREIQDRLETQHYVELEEMLHKAVMFEQQIKRKNARSSHTKTNYS 273
Query: 196 KA----QSEKRGKGHGVGKPYNKDKRKKREVGGGSKPSLA---DIKCYRCGTLGHYANDC 248
Q E++ KP+ K K + G K + DI+C++ L HYA+ C
Sbjct: 274 SGKPSYQKEEKFGYQNDSKPFVKPKPVDLDPKGKGKEVITRARDIRCFKSQGLRHYASKC 333
Query: 249 -KKDISCHKCG---KAGHKAAECKDVARDVTCYNCGEKGHIS-TKCTKPKKAAGKVFALN 303
K I H+ ++ ++AE + DV GE S + TK ++ A V L
Sbjct: 334 SNKRIMVHRDNGEVESKDESAEEDSMEEDVESPARGEFARRSLSVLTKAEEQANMVEKLG 393
Query: 304 AEEVEQP 310
E ++ P
Sbjct: 394 LEVLKHP 400
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 79.3 bits (194), Expect = 1e-14
Identities = 46/115 (40%), Positives = 67/115 (58%), Gaps = 1/115 (0%)
Query: 517 QLEELLEKQFIRPSVSP-WGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLM 575
++++LL+ IR P W A ++VKKK+G R+C+D+ LNK K+ +PL ID L+
Sbjct: 483 EVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLV 542
Query: 576 DQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYNVPGT 630
+ G E+ S +D SGY+ + ED KT+F T G Y Y VM FG+ N T
Sbjct: 543 ESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPFGLRNAGAT 597
>At1g20390 hypothetical protein
Length = 1791
Score = 78.6 bits (192), Expect = 2e-14
Identities = 49/142 (34%), Positives = 73/142 (50%), Gaps = 1/142 (0%)
Query: 490 IDLVPGMSPIWITPYPMSASELGELKKQLEELLEK-QFIRPSVSPWGAPVLLVKKKDGSV 548
+++ P P+ + + +++E+LL+ Q I W A ++VKKK+G
Sbjct: 769 LNVDPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKW 828
Query: 549 RLCVDYRQLNKVTIKNRYPLLRIDDLMDQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVF 608
R+CVDY LNK K+ YPL ID L++ G + S +D SGY+ + +D KT F
Sbjct: 829 RVCVDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSF 888
Query: 609 RTRYGHYEYFVMQFGVYNVPGT 630
T G Y Y VM FG+ N T
Sbjct: 889 VTDRGTYCYKVMSFGLKNAGAT 910
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 77.4 bits (189), Expect = 5e-14
Identities = 47/111 (42%), Positives = 63/111 (56%), Gaps = 1/111 (0%)
Query: 517 QLEELLEKQFIRPSVSP-WGAPVLLVKKKDGSVRLCVDYRQLNKVTIKNRYPLLRIDDLM 575
++ LLE IR P W A ++VKKK+G R+CVD+ LNK K+ +PL ID L+
Sbjct: 451 EVVRLLEVGRIREVKYPEWLANPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLV 510
Query: 576 DQLVGAEVFSKIDFRSGYHHNRVKAEDISKTVFRTRYGHYEYFVMQFGVYN 626
+ G E+ S +D SGY+ + ED KT F T G Y Y VM FG+ N
Sbjct: 511 ESTTGHEMLSFMDAFSGYNQILMNPEDQEKTSFITECGTYYYKVMPFGLKN 561
>At4g03650 putative reverse transcriptase
Length = 839
Score = 75.9 bits (185), Expect = 2e-13
Identities = 48/148 (32%), Positives = 71/148 (47%), Gaps = 4/148 (2%)
Query: 15 RRLDRFLRNNPPTYNGRFDPEGAQTWVQGMERIFCAMVTSDDQKVRLTTHMLAEEAEYWW 74
R + + R ++G PE A +W +ER F + + +V LT H L +A WW
Sbjct: 143 RMMKQLQRIGTEYFSGGTSPEEADSWRSQVERNFGSSRCPAEYRVDLTVHFLEGDAHLWW 202
Query: 75 TNAKGRLEIVGEVVTWAKFKAEFLRKYFPEDLRTRKEVAFLNLK*GSISVAEYAAKFEKL 134
+ R ++WA F AEF KYFP + R E FL L G SV EY KF +L
Sbjct: 203 RSVTARRRQAD--MSWADFMAEFNAKYFPREALDRMEARFLELTQGVRSVREYDRKFNRL 260
Query: 135 SRFCPYIIAEDAMVSKCVKFESGLRPDI 162
+ + +D ++ +F GLRP++
Sbjct: 261 LVYAGRGMEDDQ--AQMRRFLRGLRPNL 286
Score = 68.2 bits (165), Expect = 3e-11
Identities = 46/144 (31%), Positives = 76/144 (51%), Gaps = 11/144 (7%)
Query: 388 DLVCLPLKHMDVIFGMDWLLFFGVSINCLTKSVTFSKPVEKLDRKFLTAEQVKKSLDGEA 447
DL+ ++ DVI GMDWL + V ++C V+F +P R+ + + L G+
Sbjct: 387 DLIISHVELYDVILGMDWLDHYRVHLDCHRGRVSFERPEGSAGRE--NDREGLRGLPGDD 444
Query: 448 CVFMMFASLKENSEKGVGDLPIVQEFP---EDITELPLEREVEFAIDLVPGMSPIWITPY 504
++A + + V D+ +VQEF + + LP R F I+L P +P+ PY
Sbjct: 445 ----IYAGVC--GQVAVSDIRVVQEFQYVFQSLQGLPPSRSDPFTIELEPKTAPLSKAPY 498
Query: 505 PMSASELGELKKQLEELLEKQFIR 528
M+ +++ ELKKQLE+LL + +R
Sbjct: 499 RMAPAKMAELKKQLEDLLAEADVR 522
Score = 43.9 bits (102), Expect = 7e-04
Identities = 17/27 (62%), Positives = 21/27 (76%)
Query: 602 DISKTVFRTRYGHYEYFVMQFGVYNVP 628
D+ KT FRTRYGH+E+ VM FG+ N P
Sbjct: 520 DVRKTAFRTRYGHFEFVVMPFGLTNAP 546
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.350 0.157 0.564
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,987,806
Number of Sequences: 26719
Number of extensions: 1173501
Number of successful extensions: 5307
Number of sequences better than 10.0: 111
Number of HSP's better than 10.0 without gapping: 73
Number of HSP's successfully gapped in prelim test: 38
Number of HSP's that attempted gapping in prelim test: 4872
Number of HSP's gapped (non-prelim): 315
length of query: 1419
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1307
effective length of database: 8,326,068
effective search space: 10882170876
effective search space used: 10882170876
T: 11
A: 40
X1: 14 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 38 (21.9 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144724.7


