

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
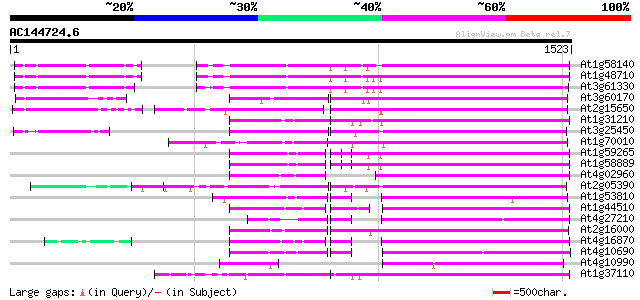
Query= AC144724.6 + phase: 0
(1523 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g58140 hypothetical protein 644 0.0
At1g48710 hypothetical protein 639 0.0
At3g61330 copia-type polyprotein 638 0.0
At3g60170 putative protein 500 e-141
At2g15650 putative retroelement pol polyprotein 496 e-140
At1g31210 putative reverse transcriptase 425 e-118
At3g25450 hypothetical protein 421 e-117
At1g70010 hypothetical protein 421 e-117
At1g59265 polyprotein, putative 419 e-117
At1g58889 polyprotein, putative 418 e-116
At4g02960 putative polyprotein of LTR transposon 407 e-113
At2g05390 putative retroelement pol polyprotein 405 e-112
At1g53810 403 e-112
At1g44510 polyprotein, putative 400 e-111
At4g27210 putative protein 396 e-110
At2g16000 putative retroelement pol polyprotein 391 e-108
At4g16870 retrotransposon like protein 388 e-107
At4g10690 retrotransposon like protein 387 e-107
At4g10990 putative retrotransposon polyprotein 383 e-106
At1g37110 362 e-100
>At1g58140 hypothetical protein
Length = 1320
Score = 644 bits (1661), Expect = 0.0
Identities = 382/1083 (35%), Positives = 562/1083 (51%), Gaps = 117/1083 (10%)
Query: 508 KIRVKCCFCDKYGHNESVCHV--KKKFIKQNNLNLSSERSHLNRSESSQKAEKAKKTCFY 565
K VKC C K+GH S C KKF ++ N +K ++
Sbjct: 276 KSSVKCYNCGKFGHYASECKAPSNKKFEEKANY-------------VEEKIQEEDMLLMA 322
Query: 566 CNKSDHKRQNVTFRKDLLEELTFKDPTLHGLDSGCSRHMTGDRDCFLTFEKKDGGLVTFG 625
K D + +N + LDSG S HM G + F ++ G V G
Sbjct: 323 SYKKDEQEENHKWY----------------LDSGASNHMCGRKSMFAELDESVRGNVALG 366
Query: 626 NNDKGKIRGKGTI----GNLNSARIENVQYVEGLKHNLLSISQLCDSGFEVIFKPNICEV 681
+ K +++GKG I N + I NV Y+ +K N+LS+ QL + G+++ K N +
Sbjct: 367 DESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSI 426
Query: 682 RQTSSNKLFFSGSRRKNLYVLELNDMPAESCFMSLEKDKWIWHKRAGHISMKTIAKLSQL 741
R SN + + ++VL + + A+ M +++ W+WH R GH++ + LS+
Sbjct: 427 RDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRK 486
Query: 742 DLVRGLPKISFEKDRICEACVKGKQVKSSFKTIEFISTQKPLELLHIDLFGPVQTASLTG 801
++VRGLP I+ +++CE C+ GKQ K SF QKPLEL+H D+ GP++ SL
Sbjct: 487 EMVRGLPCIN-HPNQVCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGK 545
Query: 802 KRYGFVIVDDFSRFTWVLFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENAS 861
Y + +DDFSR TWV FLK K E FE F+ F V+ E G I T+RSD GGEF +
Sbjct: 546 SNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKE 605
Query: 862 FKTFFDEN---------------------------------------------VFGCYCY 876
F + ++N C Y
Sbjct: 606 FLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAVY 665
Query: 877 ILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVY----------NLKTQTVEISMHIIFDE 926
+L N+ K+ + + G RV+ +++ + S IF
Sbjct: 666 LL-NRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIG 724
Query: 927 YDEHSKPKE--NEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSF-------QSPPRSWR 977
YD +SK + N DT+ + V + E E + + N +D +F + P
Sbjct: 725 YDNNSKGYKLYNPDTKKTIISRNIVFDEEG--EWDWNSNEEDYNFFPHFEEDKPEPTREE 782
Query: 978 MVGDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAMKEELSQF 1037
+ PT T+ + + EP EAI +W AM EE+
Sbjct: 783 PPSEEPTTPPTSPTSSQIE-------------EKCEPMDFQEAIEKKTWRNAMDEEIKSI 829
Query: 1038 ERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAP 1097
++N W L IG +WV++ K + +G+V R KARLVA+GY+Q+ GIDYDE FAP
Sbjct: 830 QKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVERYKARLVAKGYSQRAGIDYDEVFAP 889
Query: 1098 VARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLT 1157
VARLE +R++++ AA K+ QMDVKSAFLNG L EEVY+ QP G+I K + + V +L
Sbjct: 890 VARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYIEQPQGYIVKGEEDKVLRLK 949
Query: 1158 KALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATK 1217
KALYGLKQAPRAW R+ + E F + + L+ K D+LI +YVDD+IF
Sbjct: 950 KALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQKEDILIACLYVDDLIFTGNN 1009
Query: 1218 IKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKYVKDILKKYKMNEAKI 1277
M EEF M EFEM+ +G + ++LG+++KQ NGIFI+QE Y K++LKK+KM+++
Sbjct: 1010 PSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFITQEGYAKEVLKKFKMDDSNP 1069
Query: 1278 MSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGLCARFQTCAKESH 1337
+ TPM L K E G+ + ++ ++GSL YLT +RPDI++AVG+ +R+ +H
Sbjct: 1070 VCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPDILYAVGVVSRYMEHPTTTH 1129
Query: 1338 LTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDKVERKSTSGSCQFLGQALIG 1397
A KRI RY+ GT + GL Y S + LV Y D+D+ GD +RKSTSG ++G
Sbjct: 1130 FKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDVDDRKSTSGFVFYIGDTAFT 1189
Query: 1398 WSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSL-RYTSVPIYCDNTSAINLSK 1456
W +KQ + LST EAEYV+A SC +W+RN L++ SL + I+ DN SAI L+K
Sbjct: 1190 WMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELSLPQEEPTKIFVDNKSAIALAK 1249
Query: 1457 NPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKPLVEDRFNFLKEKLLI 1516
NP+ H RSKHI+ ++H+IR+ V KK++ L +V T +Q+ADIFTKPL + F ++ L +
Sbjct: 1250 NPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADIFTKPLKREDFIKMRSLLGV 1309
Query: 1517 MKN 1519
K+
Sbjct: 1310 AKS 1312
Score = 113 bits (283), Expect = 7e-25
Identities = 90/351 (25%), Positives = 158/351 (44%), Gaps = 27/351 (7%)
Query: 12 SSNRP---PLFDGSNYYFWKGKMELFLRSQDNDMWAVITNGDFVPTNKEGAVKAKSAWST 68
S+N P P+ SNY W +M+ L + +D+W ++ G P N+ + +
Sbjct: 3 SNNVPFQVPVLTKSNYDNWSLRMKAILGA--HDVWEIVEKGFIEPENEGSLSQTQKDGLR 60
Query: 69 DEKAQVLLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGV 128
D + + + KA + L + E+V E T+AK W+ L+ ++G VK+ R+
Sbjct: 61 DSRKR---DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLR 117
Query: 129 RKFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAIT 188
+FE +M E E + + ++R T+ N ++ G+ + K+LR L + +VT I
Sbjct: 118 GEFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIE 177
Query: 189 QAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEE 248
+ KDL++M +E L+GSL+A+E +K KK + + Q + ++ + + +
Sbjct: 178 ETKDLEAMTIEQLLGSLQAYE-----EKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQ 232
Query: 249 VHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKN 308
V R + + K+ DKS V CY C K GH+ +
Sbjct: 233 VRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYAS 292
Query: 309 ECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMA--QSDDEEE 357
EC K F++KA + ++E DM LMA + D++EE
Sbjct: 293 EC----KAPSNKKFEEKANYVEEKIQEE--------DMLLMASYKKDEQEE 331
>At1g48710 hypothetical protein
Length = 1352
Score = 639 bits (1649), Expect = 0.0
Identities = 385/1102 (34%), Positives = 569/1102 (50%), Gaps = 123/1102 (11%)
Query: 508 KIRVKCCFCDKYGHNESVCHV--KKKFIKQNNLNLSSERSHLNRSESSQKAEKAKKTCFY 565
K VKC C K+GH S C KKF ++ N +K ++
Sbjct: 276 KSSVKCYNCGKFGHYASECKAPSNKKFEEKANY-------------VEEKIQEEDMLLMA 322
Query: 566 CNKSDHKRQNVTFRKDLLEELTFKDPTLHGLDSGCSRHMTGDRDCFLTFEKKDGGLVTFG 625
K D + +N + LDSG S HM G + F ++ G V G
Sbjct: 323 SYKKDEQEENHKWY----------------LDSGASNHMCGRKSMFAELDESVRGNVALG 366
Query: 626 NNDKGKIRGKGTI----GNLNSARIENVQYVEGLKHNLLSISQLCDSGFEVIFKPNICEV 681
+ K +++GKG I N + I NV Y+ +K N+LS+ QL + G+++ K N +
Sbjct: 367 DESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSI 426
Query: 682 RQTSSNKLFFSGSRRKNLYVLELNDMPAESCFMSLEKDKWIWHKRAGHISMKTIAKLSQL 741
R SN + + ++VL + + A+ M +++ W+WH R GH++ + LS+
Sbjct: 427 RDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRK 486
Query: 742 DLVRGLPKISFEKDRICEACVKGKQVKSSFKTIEFISTQKPLELLHIDLFGPVQTASLTG 801
++VRGLP I+ +++CE C+ GKQ K SF QK LEL+H D+ GP++ SL
Sbjct: 487 EMVRGLPCIN-HPNQVCEGCLLGKQFKMSFPKESSSRAQKSLELIHTDVCGPIKPKSLGK 545
Query: 802 KRYGFVIVDDFSRFTWVLFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENAS 861
Y + +DDFSR TWV FLK K E FE F+ F V+ E G I T+RSD GGEF +
Sbjct: 546 SNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKE 605
Query: 862 FKTFFDEN---------------------------------------------VFGCYCY 876
F + ++N C Y
Sbjct: 606 FLKYCEDNGIRRQLTVPRSPQQNGVAERKNRTILEMARSMLKSKRLPKELWAEAVACAVY 665
Query: 877 ILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVY----------NLKTQTVEISMHIIFDE 926
+L N+ K+ + + G + RV+ +++ + S IF
Sbjct: 666 LL-NRSPTKSVSGKTPQEAWSGRKSGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIG 724
Query: 927 YDEHSKPKE--NEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSF-------QSPPRSWR 977
YD +SK + N DT+ + V + E E + + N +D +F + P
Sbjct: 725 YDNNSKGYKLYNPDTKKTIISRNIVFDEEG--EWDWNSNEEDYNFFPHFEEDEPEPTREE 782
Query: 978 MVGDHPT--------DQIIGSTTDGVRTRLSFQD-----------NNMAMISQMEPKSIN 1018
+ PT QI S+++ S Q+ + ++ EP
Sbjct: 783 PPSEEPTTPPTSPTSSQIEESSSERTPRFRSIQELYEVTENQENLTLFCLFAECEPMDFQ 842
Query: 1019 EAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARL 1078
EAI +W AM EE+ ++N W L IG +WV++ K + +G+V R KARL
Sbjct: 843 EAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKTIGVKWVYKAKKNSKGEVERYKARL 902
Query: 1079 VAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYV 1138
VA+GY Q+ GIDYDE FAPVARLE +R++++ AA K+ QMDVKSAFLNG L EEVY+
Sbjct: 903 VAKGYIQRAGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYI 962
Query: 1139 SQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHN 1198
QP G+I K + + V +L KALYGLKQAPRAW R+ + E F + + L+ K
Sbjct: 963 EQPQGYIVKGEEDKVLRLKKALYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQK 1022
Query: 1199 TDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFIS 1258
D+LI +YVDD+IF M EEF M EFEM+ +G + ++LG+++KQ NGIFI+
Sbjct: 1023 EDILIACLYVDDLIFTGNNPSMFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFIT 1082
Query: 1259 QEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPD 1318
QE Y K++LKK+KM+++ + TPM L K E G+ + ++ ++GSL YLT +RPD
Sbjct: 1083 QEGYAKEVLKKFKMDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPD 1142
Query: 1319 IVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDK 1378
I++AVG+ +R+ +H A KRI RY+ GT + GL Y S + LV Y D+D+ GD
Sbjct: 1143 ILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDV 1202
Query: 1379 VERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSL- 1437
+RKSTSG ++G W +KQ + LST EAEYV+A SC +W+RN L++ SL
Sbjct: 1203 DDRKSTSGFVFYIGDTAFTWMSKKQPIVVLSTCEAEYVAATSCVCHAIWLRNLLKELSLP 1262
Query: 1438 RYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADI 1497
+ I+ DN SAI L+KNP+ H RSKHI+ ++H+IR+ V KK++ L +V T +Q+ADI
Sbjct: 1263 QEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADI 1322
Query: 1498 FTKPLVEDRFNFLKEKLLIMKN 1519
FTKPL + F ++ L + K+
Sbjct: 1323 FTKPLKREDFIKMRSLLGVAKS 1344
Score = 114 bits (284), Expect = 6e-25
Identities = 90/351 (25%), Positives = 158/351 (44%), Gaps = 27/351 (7%)
Query: 12 SSNRP---PLFDGSNYYFWKGKMELFLRSQDNDMWAVITNGDFVPTNKEGAVKAKSAWST 68
S+N P P+ SNY W +M+ L + +D+W ++ G P N+ + +
Sbjct: 3 SNNVPFQVPVLTKSNYDNWSLRMKAILGA--HDVWEIVEKGFIEPENEGSLSQTQKDGLR 60
Query: 69 DEKAQVLLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGV 128
D + + + KA + L + E+V E T+AK W+ L+ ++G VK+ R+
Sbjct: 61 DSRKR---DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLR 117
Query: 129 RKFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAIT 188
+FE +M E E + + ++R T+ N ++ G+ + K+LR L + +VT I
Sbjct: 118 GEFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIE 177
Query: 189 QAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEE 248
+ KDL++M +E L+GSL+A+E +K KK + + Q + ++ + + +
Sbjct: 178 ETKDLEAMTIEQLLGSLQAYE-----EKKKKKEDIIEQVLNMQITKEENGQSYQRRGGGQ 232
Query: 249 VHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKN 308
V R + + K+ DKS V CY C K GH+ +
Sbjct: 233 VRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYAS 292
Query: 309 ECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMA--QSDDEEE 357
EC K F++KA + ++E DM LMA + D++EE
Sbjct: 293 EC----KAPSNKKFEEKANYVEEKIQEE--------DMLLMASYKKDEQEE 331
>At3g61330 copia-type polyprotein
Length = 1352
Score = 638 bits (1645), Expect = 0.0
Identities = 385/1099 (35%), Positives = 572/1099 (52%), Gaps = 125/1099 (11%)
Query: 508 KIRVKCCFCDKYGHNESVCHV--KKKFIKQNNLNLSSERSHLNRSESSQKAEKAKKTCFY 565
K VKC C K+GH S C KKF E++H +K ++
Sbjct: 276 KSSVKCYNCGKFGHYASECKAPSNKKF---------EEKAHY----VEEKIQEEDMLLMA 322
Query: 566 CNKSDHKRQNVTFRKDLLEELTFKDPTLHGLDSGCSRHMTGDRDCFLTFEKKDGGLVTFG 625
K D +++N + LDSG S HM G + F ++ G V G
Sbjct: 323 SYKKDEQKENHKWY----------------LDSGASNHMCGRKSMFAELDESVRGNVALG 366
Query: 626 NNDKGKIRGKGTI----GNLNSARIENVQYVEGLKHNLLSISQLCDSGFEVIFKPNICEV 681
+ K +++GKG I N + I NV Y+ +K N+LS+ QL + G+++ K N +
Sbjct: 367 DESKMEVKGKGNILIRLKNGDHQFISNVYYIPSMKTNILSLGQLLEKGYDIRLKDNNLSI 426
Query: 682 RQTSSNKLFFSGSRRKNLYVLELNDMPAESCFMSLEKDKWIWHKRAGHISMKTIAKLSQL 741
R SN + + ++VL + + A+ M +++ W+WH R GH++ + LS+
Sbjct: 427 RDQESNLITKVPMSKNRMFVLNIRNDIAQCLKMCYKEESWLWHLRFGHLNFGGLELLSRK 486
Query: 742 DLVRGLPKISFEKDRICEACVKGKQVKSSFKTIEFISTQKPLELLHIDLFGPVQTASLTG 801
++VRGLP I+ +++CE C+ GKQ K SF QKPLEL+H D+ GP++ SL
Sbjct: 487 EMVRGLPCIN-HPNQVCEGCLLGKQFKMSFPKESSSRAQKPLELIHTDVCGPIKPKSLGK 545
Query: 802 KRYGFVIVDDFSRFTWVLFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENAS 861
Y + +DDFSR TWV FLK K E FE F+ F V+ E G I T+RSD GGEF +
Sbjct: 546 SNYFLLFIDDFSRKTWVYFLKEKSEVFEIFKKFKAHVEKESGLVIKTMRSDRGGEFTSKE 605
Query: 862 FKTFFDEN---------------------------------------------VFGCYCY 876
F + ++N C Y
Sbjct: 606 FLKYCEDNGIRRQLTVPRSPQQNGVVERKNRTILEMARSMLKSKRLPKELWAEAVACAVY 665
Query: 877 ILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVY----------NLKTQTVEISMHIIFDE 926
+L N+ K+ + + G RV+ +++ + S IF
Sbjct: 666 LL-NRSPTKSVSGKTPQEAWSGRKPGVSHLRVFGSIAHAHVPDEKRSKLDDKSEKYIFIG 724
Query: 927 YDEHSKPKE--NEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSF-------QSPPRSWR 977
YD +SK + N DT+ + V + E E + + N +D +F + P
Sbjct: 725 YDNNSKGYKLYNPDTKKTIISRNIVFDEEG--EWDWNSNEEDYNFFPHFEEDEPEPTREE 782
Query: 978 MVGDHPT--------DQIIGSTTDGVRTRLSFQD--------NNMAM---ISQMEPKSIN 1018
+ PT QI S+++ S Q+ N+ + ++ EP
Sbjct: 783 PPSEEPTTPPTSPTSSQIEESSSERTPRFRSIQELYEVTENQENLTLFCLFAECEPMDFQ 842
Query: 1019 EAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARL 1078
+AI +W AM EE+ ++N W L IG +WV++ K + +G+V R KARL
Sbjct: 843 KAIEKKTWRNAMDEEIKSIQKNDTWELTSLPNGHKAIGVKWVYKAKKNSKGEVERYKARL 902
Query: 1079 VAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYV 1138
VA+GY+Q+ GIDYDE FAPVARLE +R++++ AA K+ QMDVKSAFLNG L EEVY+
Sbjct: 903 VAKGYSQRVGIDYDEVFAPVARLETVRLIISLAAQNKWKIHQMDVKSAFLNGDLEEEVYI 962
Query: 1139 SQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHN 1198
QP G+I K + + V +L K LYGLKQAPRAW R+ + E F + + L+ K
Sbjct: 963 EQPQGYIVKGEEDKVLRLKKVLYGLKQAPRAWNTRIDKYFKEKDFIKCPYEHALYIKIQK 1022
Query: 1199 TDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFIS 1258
D+LI +YVDD+IF + EEF M EFEM+ +G + ++LG+++KQ NGIFI+
Sbjct: 1023 EDILIACLYVDDLIFTGNNPSIFEEFKKEMTKEFEMTDIGLMSYYLGIEVKQEDNGIFIT 1082
Query: 1259 QEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPD 1318
QE Y K++LKK+K++++ + TPM L K E G+ + ++ ++GSL YLT +RPD
Sbjct: 1083 QEGYAKEVLKKFKIDDSNPVCTPMECGIKLSKKEEGEGVDPTTFKSLVGSLRYLTCTRPD 1142
Query: 1319 IVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDK 1378
I++AVG+ +R+ +H A KRI RY+ GT + GL Y S + LV Y D+D+ GD
Sbjct: 1143 ILYAVGVVSRYMEHPTTTHFKAAKRILRYIKGTVNFGLHYSTTSDYKLVGYSDSDWGGDV 1202
Query: 1379 VERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSL- 1437
+RKSTSG ++G W +KQ + LST EAEYV+A SC +W+RN L++ SL
Sbjct: 1203 DDRKSTSGFVFYIGDTAFTWMSKKQPIVTLSTCEAEYVAATSCVCHAIWLRNLLKELSLP 1262
Query: 1438 RYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADI 1497
+ I+ DN SAI L+KNP+ H RSKHI+ ++H+IR+ V KK++ L +V T +Q+AD
Sbjct: 1263 QEEPTKIFVDNKSAIALAKNPVFHDRSKHIDTRYHYIRECVSKKDVQLEYVKTHDQVADF 1322
Query: 1498 FTKPLVEDRFNFLKEKLLI 1516
FTKPL R NF+K + L+
Sbjct: 1323 FTKPL--KRENFIKMRSLL 1339
Score = 111 bits (278), Expect = 3e-24
Identities = 83/329 (25%), Positives = 147/329 (44%), Gaps = 17/329 (5%)
Query: 12 SSNRP---PLFDGSNYYFWKGKMELFLRSQDNDMWAVITNGDFVPTNKEGAVKAKSAWST 68
S+N P P+ SNY W +M+ L + +D+W ++ G P N+ + +
Sbjct: 3 SNNVPFQVPVLTKSNYDNWSLRMKAILGA--HDVWEIVEKGFIEPENEGSLSQTQKDGLR 60
Query: 69 DEKAQVLLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGV 128
D + + + KA + L + E+V E T+AK W+ L+ ++G VK+ R+
Sbjct: 61 DSRKR---DKKALCLIYQGLDEDTFEKVVEATSAKEAWEKLRTSYKGADQVKKVRLQTLR 117
Query: 129 RKFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAIT 188
+FE +M E E + + ++R T+ N ++ G+ + K+LR L + +VT I
Sbjct: 118 GEFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIE 177
Query: 189 QAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEE 248
+ KDL++M +E L+GSL+A+E +K KK + Q + ++ + + +
Sbjct: 178 ETKDLEAMTIEQLLGSLQAYE-----EKKKKKEDIAEQVLNMQITKEENGQSYQRRGGGQ 232
Query: 249 VHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKN 308
V R + + K+ DKS V CY C K GH+ +
Sbjct: 233 VRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGKGHPKSRYDKSSVKCYNCGKFGHYAS 292
Query: 309 ECPDIKKVQRKPPFKKKAMITWDDMEESD 337
EC K F++KA + ++E D
Sbjct: 293 EC----KAPSNKKFEEKAHYVEEKIQEED 317
>At3g60170 putative protein
Length = 1339
Score = 500 bits (1288), Expect = e-141
Identities = 268/667 (40%), Positives = 401/667 (59%), Gaps = 22/667 (3%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDEYDE 929
VFGC Y+ +K K D KS K +FLG S SK +R+Y+ + + IS ++FDE
Sbjct: 664 VFGCIGYVHIPDQKRSKLDDKSKKCVFLGVSEESKAWRLYDPVMKKIVISKDVVFDEDKS 723
Query: 930 HSKPKENEDTEVPTLQNVPVQNTENTVEKE-----------DDQNAQDQSFQSP------ 972
+ + + + TL+ + +N+ E D N +P
Sbjct: 724 WDWDQADVEAKEVTLECGDEDDEKNSEVVEPIAVASPNHVGSDNNVSSSPILAPSSPAPS 783
Query: 973 PRSWRMVGDH--PTDQIIGSTTDGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAM 1030
P + ++ + P T +G + + M+++ +P ++A+ D W +AM
Sbjct: 784 PVAAKVTRERRPPGWMADYETGEGEEIEENLSVMLLMMMTEADPIQFDDAVKDKIWREAM 843
Query: 1031 KEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGID 1090
+ E+ +N W L + T IG +WV++ KL+E+G+V + KARLVA+GY Q GID
Sbjct: 844 EHEIESIVKNNTWELTTLPKGFTPIGVKWVYKTKLNEDGEVDKYKARLVAKGYAQCYGID 903
Query: 1091 YDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKP 1150
Y E FAPVARL+ +R +LA ++ + ++FQ+DVKSAFL+G L EEVYV QP GFI + +
Sbjct: 904 YTEVFAPVARLDTVRTILAISSQFNWEIFQLDVKSAFLHGELKEEVYVRQPEGFIREGEE 963
Query: 1151 NHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDD 1210
V+KL KALYGLKQAPRAWY R+ + ++ F R + TLF KT ++LIV +YVDD
Sbjct: 964 EKVYKLRKALYGLKQAPRAWYSRIEAYFLKEEFERCPSEHTLFTKTRVGNILIVSLYVDD 1023
Query: 1211 IIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKYVKDILKKY 1270
+IF + MC+EF M EFEMS +G++ FLG+++KQ GIFI Q +Y +++L ++
Sbjct: 1024 LIFTGSDKAMCDEFKKSMMLEFEMSDLGKMKHFLGIEVKQSDGGIFICQRRYAREVLARF 1083
Query: 1271 KMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGLCARFQ 1330
M+E+ + P+ P + L KDE+G+ + E ++ ++GSL+YLT +RPD+++ V L +RF
Sbjct: 1084 GMDESNAVKNPIVPGTKLTKDENGEKVDETMFKQLVGSLMYLTVTRPDLMYGVCLISRFM 1143
Query: 1331 TCAKESHLTAVKRIFRYLVGTTDLGLWY--RKGSSFDLVAYCDADYAGDKVERKSTSGSC 1388
+ + SH A KRI RYL GT +LG++Y RK S L+A+ D+DYAGD +R+STSG
Sbjct: 1144 SNPRMSHWLAAKRILRYLKGTVELGIFYRRRKNRSLKLMAFTDSDYAGDLNDRRSTSGFV 1203
Query: 1389 QFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSVP-IYCD 1447
+ I W+ +KQ +ALSTTEAEY++AA C Q +W+R LE S I CD
Sbjct: 1204 FLMASGAICWASKKQPVVALSTTEAEYIAAAFCACQCVWLRKVLEKLGAEEKSATVINCD 1263
Query: 1448 NTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKPLVEDRF 1507
N+S I LSK+P+ H +SKHIE++ H++RD V + L + TE+Q+ADIFTKPL ++F
Sbjct: 1264 NSSTIQLSKHPVLHGKSKHIEVRFHYLRDLVNGDVVKLEYCPTEDQVADIFTKPLKLEQF 1323
Query: 1508 NFLKEKL 1514
L+ L
Sbjct: 1324 EKLRALL 1330
Score = 158 bits (399), Expect = 3e-38
Identities = 95/285 (33%), Positives = 148/285 (51%), Gaps = 24/285 (8%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNNDKGKIRGKGTIG---NLNSARIENVQYV 652
LDSGCS HMTG ++ F E+ V GN+ + + GKG++ N + I V YV
Sbjct: 302 LDSGCSNHMTGSKEWFSELEEGFNRTVKLGNDTRMSVVGKGSVKVKVNGVTQVIPEVYYV 361
Query: 653 EGLKHNLLSISQLCDSGFEVIFKPNICEV----------RQTSSNKLFF--SGSRRKNLY 700
L++NLLS+ QL + G ++ + C+V S N++FF + +KN
Sbjct: 362 PELRNNLLSLGQLQERGLAILIRDGTCKVYHPSKGAIMETNMSGNRMFFLLASKPQKNSL 421
Query: 701 VLELNDMPAESCFMSLEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEA 760
L+ ++ ++K+ +WH R GH++ + + L+ +V GLP + K+ IC
Sbjct: 422 CLQTEEV--------MDKENHLWHCRFGHLNQEGLKLLAHKKMVIGLPILKATKE-ICAI 472
Query: 761 CVKGKQVKSSFKTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLF 820
C+ GKQ + S + L+L+H D+ GP+ S +GKRY +DDF+R TWV F
Sbjct: 473 CLTGKQHRESMSKKTSWKSSTQLQLVHSDICGPITPISHSGKRYILSFIDDFTRKTWVYF 532
Query: 821 LKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTF 865
L K E+F F+ F V+ E G + +R+D GGEF + F F
Sbjct: 533 LHEKSEAFATFKIFKASVEKEIGAFLTCLRTDRGGEFTSNEFGEF 577
Score = 117 bits (293), Expect = 5e-26
Identities = 84/299 (28%), Positives = 144/299 (48%), Gaps = 41/299 (13%)
Query: 17 PLFDGSNYYFWKGKMELFLRSQDNDMWAVITNGDFVPTNKEGAVKAKSAWSTDEKAQVLL 76
P FDG Y FW ME FLRS++ +W ++ G +P G A + + L
Sbjct: 13 PRFDGY-YDFWSMTMENFLRSRE--LWRLVEEG--IPAIVVGTTPVSEAQRSAVEEAKLK 67
Query: 77 NSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGVRKFEVFEM 136
+ K + FL A+ E E + + + +K +W+++K ++G++ VK ++ ++FE+ M
Sbjct: 68 DLKVKNFLFQAIDREILETILDKSTSKAIWESMKKKYQGSTKVKRAQLQALRKEFELLAM 127
Query: 137 SENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQAKDLKSM 196
E E ID R T+VN+M++ G+ + KILR L + +V +I ++ DL ++
Sbjct: 128 KEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEESNDLSTL 187
Query: 197 NLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEEVHEEEAED 256
++++L GSL HE L G VQE Q L+ HEE
Sbjct: 188 SIDELHGSLLVHEQRLNG------------------------HVQEEQALKVTHEERP-- 221
Query: 257 ELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECPDIKK 315
S+ R + R ++ R + ++ +++ V CY C+ GHF+ ECP+ +K
Sbjct: 222 -----SQGRGRGVFRGSRGRGRGRG-----RSGTNRAIVECYKCHNLGHFQYECPEWEK 270
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 496 bits (1278), Expect = e-140
Identities = 260/666 (39%), Positives = 392/666 (58%), Gaps = 21/666 (3%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDEYD- 928
+FG CY+ +K K D K+ I +GYS +KGYRV+ L+ + VE+S ++F E
Sbjct: 675 IFGSICYVHIPDQKRRKLDAKAKCGILIGYSNQTKGYRVFLLEDEKVEVSRDVVFQEDKK 734
Query: 929 -EHSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRSWRMVGDHPTDQI 987
+ K +E + T V ++ ++ + D D + + V DQ
Sbjct: 735 WDWDKQEEVKKTFVMSINDIQESRDQQETSSHDLSQIDDHANNGEGETSSHVLSQVNDQE 794
Query: 988 IGSTTDGVRTRLSFQD---------NNMA-------MISQMEPKSINEAIIDDSWIKAMK 1031
T++ + S ++ N+ A +++ EP++ +EA D W +AM
Sbjct: 795 ERETSESPKKYKSMKEILEKAPRMENDEAAQGIEACLVANEEPQTYDEARGDKEWEEAMN 854
Query: 1032 EELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDY 1091
EE+ E+N+ W LV + K +I +W+++ K D G V++KARLVA+G++Q+ GIDY
Sbjct: 855 EEIKVIEKNRTWKLVDKPEKKNVISVKWIYKIKTDASGNHVKHKARLVARGFSQEYGIDY 914
Query: 1092 DETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPN 1151
ETFAPV+R + IR LLAYAA +L+QMDVKSAFLNG L EEVYV+QPPGF+ + K
Sbjct: 915 LETFAPVSRYDTIRALLAYAAQMKWRLYQMDVKSAFLNGELEEEVYVTQPPGFVIEGKEE 974
Query: 1152 HVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDDI 1211
V +L KALYGLKQAPRAWY+R+ ++ I+NGF+R D L+ K D+LIV +YVDD+
Sbjct: 975 KVLRLYKALYGLKQAPRAWYERIDSYFIQNGFARSMNDAALYSKKKGEDVLIVSLYVDDL 1034
Query: 1212 IFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKYVKDILKKYK 1271
I + F M+ EFEM+ +G L +FLG+++ Q +GIF+SQEKY ++ K+
Sbjct: 1035 IITGNNTHLINTFKKNMKDEFEMTDLGLLNYFLGMEVNQDDSGIFLSQEKYANKLIDKFG 1094
Query: 1272 MNEAKIMSTPMHPSSSLD--KDESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGLCARF 1329
M E+K +STP+ P + + + +YR ++G LLYL ASRPD+++A +R+
Sbjct: 1095 MKESKSVSTPLTPQGKRKGVEGDDKEFADPTKYRRIVGGLLYLCASRPDVMYASSYLSRY 1154
Query: 1330 QTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDKVERKSTSGSCQ 1389
+ H KR+ RY+ GT++ G+ + + LV Y D+D+ G ++KST+G
Sbjct: 1155 MSSPSIQHYQEAKRVLRYVKGTSNFGVLFTSKETPRLVGYSDSDWGGSLEDKKSTTGYVF 1214
Query: 1390 FLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRY-TSVPIYCDN 1448
LG A+ W KQ T+A ST EAEY++ + +Q +W++ ED+ L++ +PI CDN
Sbjct: 1215 TLGLAMFCWQSCKQQTVAQSTAEAEYIAVCAATNQAIWLQRLFEDFGLKFKEGIPILCDN 1274
Query: 1449 TSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKPLVEDRFN 1508
SAI + +NP+QH R+KHIEIK+HF+R+ K I L + E+QLAD+ TK L RF
Sbjct: 1275 KSAIAIGRNPVQHRRTKHIEIKYHFVREAEHKGLIQLEYCKGEDQLADVLTKALSVSRFE 1334
Query: 1509 FLKEKL 1514
L+ KL
Sbjct: 1335 GLRRKL 1340
Score = 159 bits (401), Expect = 2e-38
Identities = 131/486 (26%), Positives = 224/486 (45%), Gaps = 46/486 (9%)
Query: 394 ELSEIKEEKEILQNKYDESRKTIK----ILQDSHFDMSEKQREINRKQKGIMSVPSEVQK 449
+L ++ E E L+ +++ KT +L+ EK+ QK ++S+P++
Sbjct: 114 KLQSLRREYENLKMYDNDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDS 173
Query: 450 ENILLK--KEVETLK-KDLTGFIKSTETFQNIVGSQNKSTKKSGLGFKDPSKIIGSFVPK 506
+L+ ++++ L +L G +K+ E V ++ +STK+ + + G
Sbjct: 174 IVSVLEQTRDLDALTMSELLGILKAQEAR---VTAREESTKEGAFYVRSKGRESGFKQDN 230
Query: 507 AKIRVK-----CCFCDKYGHNESVCHVKKKFIKQNNLNLSSERSHLNRSESSQKAEKAKK 561
RV C F H E C K K H S+ K K K
Sbjct: 231 TNNRVNQDKKWCGFHKSSKHTEEECREKPK-----------NDDHGKNKRSNIKCYKCGK 279
Query: 562 TCFY---CNKSDHKRQNVTFRKDLL-----------EELTFKDPTLHGLDSGCSRHMTGD 607
Y C + +R +VT ++ + EE T + +DSGC+ HMT +
Sbjct: 280 IGHYANECRSKNKERAHVTLEEEDVNEDHMLFSASEEESTTLREDVWLVDSGCTNHMTKE 339
Query: 608 RDCFLTFEKKDGGLVTFGNNDKGKIRGKG--TIGNLNSARI-ENVQYVEGLKHNLLSISQ 664
F K + N D GKG T+ + RI +NV V GL+ NLLS+ Q
Sbjct: 340 ERYFSNINKSIKVPIRVRNGDIVMTAGKGDITVMTRHGKRIIKNVFLVPGLEKNLLSVPQ 399
Query: 665 LCDSGFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAESCFMSLEKDKWIWH 724
+ SG+ V F+ C ++ + ++ K+ + ++L+ + E+ +++ ++ WH
Sbjct: 400 IISSGYWVRFQDKRCIIQDANGKEIMNIEMTDKS-FKIKLSSVEEEAMTANVQTEE-TWH 457
Query: 725 KRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEACVKGKQVKSSFKTIEFISTQKPLE 784
KR GH+S K + ++ +LV GLP+ K+ C+AC GKQ + SF T++ LE
Sbjct: 458 KRLGHVSNKRLQQMQDKELVNGLPRFKVTKET-CKACNLGKQSRKSFPKESQTKTREKLE 516
Query: 785 LLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESFEAFQNFCKRVQNEKGY 844
++H D+ GP+Q S+ G RY + +DD++ WV FLK K E+F F+ F V+ +
Sbjct: 517 IVHTDVCGPMQHQSIDGSRYYVLFLDDYTHMCWVYFLKQKSETFATFKKFKALVEKQSNC 576
Query: 845 NIITVR 850
+I T+R
Sbjct: 577 SIKTLR 582
Score = 105 bits (261), Expect = 3e-22
Identities = 84/352 (23%), Positives = 163/352 (45%), Gaps = 33/352 (9%)
Query: 9 EGGSSNRPPLFDGSNYYFWKGKMELFLRSQDNDMWAVITNGDFV-PTNKEGAVKAKSAWS 67
+G S P+FDG Y FW KM R++ +W+V+ G V P E + A +
Sbjct: 2 QGSSHQVIPIFDGEKYDFWSIKMATIFRTRK--LWSVVEEGVPVEPVQAEETPETARAKT 59
Query: 68 TDEKAQVLLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIG 127
E+A V ++ A L A+T + R+ +++K WD LK ++G+ V+ ++
Sbjct: 60 LREEA-VTNDTMALQILQTAVTDQIFSRIAAASSSKEAWDVLKDEYQGSPQVRLVKLQSL 118
Query: 128 VRKFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAI 187
R++E +M +N+ I + + ++ G+ + I+KIL LP+ + +V+ +
Sbjct: 119 RREYENLKMYDNDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVL 178
Query: 188 TQAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELE 247
Q +DL ++ + +L+G L+A E + + K +++ + S ++
Sbjct: 179 EQTRDLDALTMSELLGILKAQEARVTAREESTKEGAFYVRSKGRESGFKQDNTN-----N 233
Query: 248 EVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFK 307
V++++ SK + + R+K PK + K + +S + CY C K GH+
Sbjct: 234 RVNQDKKWCGFHKSSKHTE------EECREK-PKNDDHGKNK--RSNIKCYKCGKIGHYA 284
Query: 308 NECPDIKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQSDDEEEVI 359
NEC + K++A +T + +ED + D L + S++E +
Sbjct: 285 NEC--------RSKNKERAHVTLE-------EEDVNEDHMLFSASEEESTTL 321
>At1g31210 putative reverse transcriptase
Length = 1415
Score = 425 bits (1092), Expect = e-118
Identities = 247/689 (35%), Positives = 373/689 (53%), Gaps = 42/689 (6%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDEYD- 928
VFG CY KFDP+S + +FLGY++ KGYR + T V IS ++IF+E +
Sbjct: 681 VFGSACYPCLRPLAQNKFDPRSLQCVFLGYNSQYKGYRCFYPPTGKVYISRNVIFNESEL 740
Query: 929 ---------------------EHSKPKENEDTEVPT-LQNVPV--------QNTENTVEK 958
+H+K E P L + P+ Q TE +
Sbjct: 741 PFKEKYQSLVPQYSTPLLQAWQHNKISEISVPAAPVQLFSKPIDLNTYAGSQVTEQLTDP 800
Query: 959 EDDQNAQDQSFQSPPRSWRMVGDHPTDQIIGSTTDGVRTRLSFQDNNM------AMISQM 1012
E N + + P + + + +Q+I S R++ Q N + ++
Sbjct: 801 EPTSNNEGSDEEVNPVAEEIAANQ--EQVINSHAMTTRSKAGIQKPNTRYALITSRMNTA 858
Query: 1013 EPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVV 1072
EPK++ A+ W +A+ EE+++ W+LVP D I+ ++WVF+ KL +G +
Sbjct: 859 EPKTLASAMKHPGWNEAVHEEINRVHMLHTWSLVPPTDDMNILSSKWVFKTKLHPDGSID 918
Query: 1073 RNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFL 1132
+ KARLVA+G++Q+EG+DY ETF+PV R IR++L + K + Q+DV +AFL+G L
Sbjct: 919 KLKARLVAKGFDQEEGVDYLETFSPVVRTATIRLVLDVSTSKGWPIKQLDVSNAFLHGEL 978
Query: 1133 NEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTL 1192
E V++ QP GFI+ +KP HV +LTKA+YGLKQAPRAW+D S FL++ GF K D +L
Sbjct: 979 QEPVFMYQPSGFIDPQKPTHVCRLTKAIYGLKQAPRAWFDTFSNFLLDYGFVCSKSDPSL 1038
Query: 1193 FRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHS 1252
F + +L + +YVDDI+ + + E+ +++ F M +G +FLG+QI+ ++
Sbjct: 1039 FVCHQDGKILYLLLYVDDILLTGSDQSLLEDLLQALKNRFSMKDLGPPRYFLGIQIEDYA 1098
Query: 1253 NGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYL 1312
NG+F+ Q Y DIL++ M++ M TP+ LD S +R + G L YL
Sbjct: 1099 NGLFLHQTAYATDILQQAGMSDCNPMPTPL--PQQLDNLNSELFAEPTYFRSLAGKLQYL 1156
Query: 1313 TASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDA 1372
T +RPDI FAV + S +KRI RY+ GT +GL ++ S+ L AY D+
Sbjct: 1157 TITRPDIQFAVNFICQRMHSPTTSDFGLLKRILRYIKGTIGMGLPIKRNSTLTLSAYSDS 1216
Query: 1373 DYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQL 1432
D+AG K R+ST+G C LG LI WS ++Q T++ S+TEAEY + +I W+ L
Sbjct: 1217 DHAGCKNTRRSTTGFCILLGSNLISWSAKRQPTVSNSSTEAEYRALTYAAREITWISFLL 1276
Query: 1433 EDYSL-RYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTE 1491
D + +Y +YCDN SA+ LS NP H+RSKH + +H+IR+ V I +
Sbjct: 1277 RDLGIPQYLPTQVYCDNLSAVYLSANPALHNRSKHFDTDYHYIREQVALGLIETQHISAT 1336
Query: 1492 NQLADIFTKPLVEDRFNFLKEKLLIMKNP 1520
QLAD+FTK L F L+ KL + +P
Sbjct: 1337 FQLADVFTKSLPRRAFVDLRSKLGVSGSP 1365
Score = 112 bits (281), Expect = 1e-24
Identities = 86/281 (30%), Positives = 125/281 (43%), Gaps = 15/281 (5%)
Query: 597 DSGCSRHMTGDRDCFLT---FEKKDGGLVTFGNNDKGKIRGKGTIGNLNSA-RIENVQYV 652
DS + H+T + + +E D LV G G TI + N + V V
Sbjct: 325 DSAATAHVTSSTNGLQSATEYEGDDAVLVGDGTYLPITHTGSTTIKSSNGKIPLNEVLVV 384
Query: 653 EGLKHNLLSISQLCDS-GFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAES 711
++ +LLS+S+LCD V F N + + K+ +G RR LYVLE + A
Sbjct: 385 PNIQKSLLSVSKLCDDYPCGVYFDANKVCIIDLQTQKVVTTGPRRNGLYVLENQEFVALY 444
Query: 712 CFMSLEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDR---ICEACVKGKQVK 768
+ +WH R GH + K + L + I K R +CE C GK +
Sbjct: 445 SNRQCAATEEVWHHRLGHANSKALQHL------QNSKAIQINKSRTSPVCEPCQMGKSSR 498
Query: 769 SSFKTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESF 828
F I PL+ +H DL+GP S G +Y + VDD+SR++W L +K E
Sbjct: 499 LPF-LISDSRVLHPLDRIHCDLWGPSPVVSNQGLKYYAIFVDDYSRYSWFYPLHNKSEFL 557
Query: 829 EAFQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTFFDEN 869
F +F K V+N+ I +SD GGEF + KT E+
Sbjct: 558 SVFISFQKLVENQLNTKIKVFQSDGGGEFVSNKLKTHLSEH 598
Score = 34.3 bits (77), Expect = 0.57
Identities = 45/205 (21%), Positives = 83/205 (39%), Gaps = 28/205 (13%)
Query: 22 SNYYFWKGKMELFLRSQD-----------NDMWAVITNGDFVPTNKEGAVKAKSAWSTDE 70
SNY WK + E L SQ ++ NG+ T++E +S + TD+
Sbjct: 24 SNYLLWKTQFESLLSSQKLIGFVNGAVNAPSQSRLVVNGEV--TSEEPNPLYESWFCTDQ 81
Query: 71 KAQVLLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGVRK 130
R +L L+ E V + ++ +W +L + +S +E + ++
Sbjct: 82 --------LVRSWLFGTLSEEVLGHVHNLSTSRQIWVSLAENFNKSSVAREFSLRQNLQL 133
Query: 131 FEVFEMSENETIDEMYAR-FTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAI-T 188
+S+ E +Y R F TI + + S+GK +I L L + P+ T I +
Sbjct: 134 -----LSKKEKPFSVYCREFKTICDALSSIGKPVDESMKIFGFLNGLGRDYDPITTVIQS 188
Query: 189 QAKDLKSMNLEDLIGSLRAHEVVLQ 213
L + D++ ++ + LQ
Sbjct: 189 SLSKLPTPTFNDVVSEVQGFDSKLQ 213
>At3g25450 hypothetical protein
Length = 1343
Score = 421 bits (1081), Expect = e-117
Identities = 243/675 (36%), Positives = 368/675 (54%), Gaps = 41/675 (6%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE--- 926
VFGC Y L K D +S ++LG SK YR+ + + + +S ++FDE
Sbjct: 654 VFGCVSYAKVEVPNLKKLDDRSRMLVYLGTEPGSKAYRLLDPTKRRIFVSRDVVFDENRS 713
Query: 927 --YDEHSKPKENE---------------------DTEVPTLQNVPVQNTENTVEKEDDQN 963
+ E S + E TE + + + + +E +
Sbjct: 714 WMWQESSSETDKESGTFTITLSEFGNNGVTENDISTEPEETEEAEINGEDENIIEEAETE 773
Query: 964 AQDQSFQSPP---RSWRMV--GDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQMEPKSIN 1018
DQS + P RS R V ++ D ++ + + L+ D EP
Sbjct: 774 EHDQSQEEPQPVRRSQRQVIRPNYLKDYVLCAEIEAEHLLLAVND---------EPWDFK 824
Query: 1019 EAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARL 1078
EA W A KEE+ E+N+ W+LV IG +WVF+ K + +G + + KARL
Sbjct: 825 EANKSKEWRDACKEEIQSIEKNRTWSLVDLPVGSKAIGVKWVFKLKHNSDGSINKYKARL 884
Query: 1079 VAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYV 1138
VA+GY Q+ G+D++E FAPVAR+E +R+++A AA ++ +DVK+AFL+G L E+VYV
Sbjct: 885 VAKGYVQRHGVDFEEVFAPVARIETVRLIIALAASNGWEIHHLDVKTAFLHGELREDVYV 944
Query: 1139 SQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHN 1198
SQP GF NKE V+KL KALYGL+QAPRAW +L+ L E F + + +L+RK
Sbjct: 945 SQPEGFTNKESKEKVYKLHKALYGLRQAPRAWNTKLNEILKELKFEKCHKEPSLYRKQEG 1004
Query: 1199 TDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFIS 1258
++L+V VYVDD++ + + + F M +FEMS +G+L ++LG+++ Q +GI +
Sbjct: 1005 ENILVVAVYVDDLLVTGSNLDIILNFKKGMVGKFEMSDLGKLTYYLGIEVLQSKDGITLK 1064
Query: 1259 QEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPD 1318
QE+Y K IL++ M++ ++TPM S L K + K I E +YR IG L YL +RPD
Sbjct: 1065 QERYAKKILEEAGMSKCNTVNTPMIASLELSKAQDEKRIDETDYRRNIGCLRYLLHTRPD 1124
Query: 1319 IVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDK 1378
+ + VG+ +R+ +ESH A+K+I RYL GTT GL+++KG + L+ Y D+ + D
Sbjct: 1125 LSYNVGILSRYLQEPRESHGAALKQILRYLQGTTSHGLYFKKGENAGLIGYSDSSHNVDL 1184
Query: 1379 VERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQL-EDYSL 1437
+ KST G +L I W +KQ + LS+ EAE+++A Q +W++ L E
Sbjct: 1185 DDGKSTGGHIFYLNDCPITWCSQKQQVVTLSSCEAEFMAATEAAKQAIWLQELLAEVIGT 1244
Query: 1438 RYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADI 1497
V I DN SAI L+KNP+ H RSKHI ++HFIR+ V+ I + V Q ADI
Sbjct: 1245 ECEKVTIRVDNKSAIALTKNPVFHGRSKHIHRRYHFIRECVENGQIEVEHVPGVRQKADI 1304
Query: 1498 FTKPLVEDRFNFLKE 1512
TK L + +F ++E
Sbjct: 1305 LTKALGKIKFLEMRE 1319
Score = 165 bits (418), Expect = 2e-40
Identities = 93/274 (33%), Positives = 146/274 (52%), Gaps = 4/274 (1%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNNDKGKIRGKGTIGNLNSAR----IENVQY 651
LD+G S HMTG+R F ++ G V FG++ I+GKG+I ++ + +V Y
Sbjct: 294 LDNGASNHMTGNRAWFCKLDEMITGKVRFGDDSCINIKGKGSIPFISKGGERKILFDVYY 353
Query: 652 VEGLKHNLLSISQLCDSGFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAES 711
+ LK N+LS+ Q +SG ++ + + + N L + R LY + L ++
Sbjct: 354 IPDLKSNILSLGQATESGCDIRMREDYLTLHDREGNLLIKAQRSRNRLYKVSLEVENSKC 413
Query: 712 CFMSLEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEACVKGKQVKSSF 771
++ + IWH R GHIS +TI + + +LV G+ ++ C +C+ GKQ + SF
Sbjct: 414 LQLTTTNESTIWHARLGHISFETIKAMIKKELVIGISSSVPQEKETCGSCLFGKQARHSF 473
Query: 772 KTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESFEAF 831
+ LEL+H DL GP+ ++ KRY FV++DD SR+ W + LK K E+F F
Sbjct: 474 PKATSYRAAQVLELIHGDLCGPISPSTAAKKRYVFVLIDDHSRYMWSILLKEKSEAFGKF 533
Query: 832 QNFCKRVQNEKGYNIITVRSDHGGEFENASFKTF 865
+ F V+ E G I T R+D GGEF + F+ F
Sbjct: 534 KEFKALVEQECGAIIKTFRTDRGGEFLSHEFQEF 567
Score = 80.5 bits (197), Expect = 7e-15
Identities = 66/261 (25%), Positives = 115/261 (43%), Gaps = 33/261 (12%)
Query: 10 GGSSNRPPLFDGSNYYFWKGKMELFLRSQDNDMWAVITNGDFVPTNKEGAVKAKSAWSTD 69
G +S + P+ + NY W +ME LR + +W I G S D
Sbjct: 16 GSASIQCPMLNSVNYTVWTMRMEAVLRV--HKLWGTIEPG-----------------SAD 56
Query: 70 EKAQVLLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGVR 129
E+ N AR L ++ +V + + VW+ +K + G VKE R+ +
Sbjct: 57 EEK----NDMARALLFQSIPESLILQVGKQKTSSAVWEAIKSRNLGAERVKEARLQTLMA 112
Query: 130 KFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLP-SMWRPMVTAIT 188
+F+ +M ++ETID+ R + I + +LG+ ++K L+ LP + +V A+
Sbjct: 113 EFDKLKMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALE 172
Query: 189 QAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEE 248
Q DLK+ ED+ G ++ +E + D + + + V++E V +LEE
Sbjct: 173 QVLDLKTTTFEDIAGRIKTYEDRVWDDDDSHE------DQGKLMTEVEEEVV---DDLEE 223
Query: 249 VHEEEAEDELALISKRIQRMM 269
EE E+ S I R++
Sbjct: 224 EEEEVINKEIKAKSHVIDRLL 244
>At1g70010 hypothetical protein
Length = 1315
Score = 421 bits (1081), Expect = e-117
Identities = 246/684 (35%), Positives = 375/684 (53%), Gaps = 33/684 (4%)
Query: 864 TFFDENVFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHII 923
T+ VFGC CY + + KF P++ F+GY + KGY++ +L+T ++ +S H++
Sbjct: 623 TYDHIKVFGCLCYASTSPKDRHKFSPRAKACAFIGYPSGFKGYKLLDLETHSIIVSRHVV 682
Query: 924 FDEY------DEHSKPKEN---EDTEVPTLQ-----NVPVQNTENTVE----KEDDQNAQ 965
F E + S+ ++N + P +Q +V ++ ++VE N
Sbjct: 683 FHEELFPFLGSDLSQEEQNFFPDLNPTPPMQRQSSDHVNPSDSSSSVEILPSANPTNNVP 742
Query: 966 DQSFQSPPRSWRM---VGDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQM-------EPK 1015
+ S Q+ R + + D+ ++ ST +R LS+ N ++ + EP
Sbjct: 743 EPSVQTSHRKAKKPAYLQDYYCHSVVSSTPHEIRKFLSYDRINDPYLTFLACLDKTKEPS 802
Query: 1016 SINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNK 1075
+ EA W AM E E W + DK IG RW+F+ K + +G V R K
Sbjct: 803 NYTEAEKLQVWRDAMGAEFDFLEGTHTWEVCSLPADKRCIGCRWIFKIKYNSDGSVERYK 862
Query: 1076 ARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEE 1135
ARLVAQGY Q+EGIDY+ETF+PVA+L ++++LL AA + L Q+D+ +AFLNG L+EE
Sbjct: 863 ARLVAQGYTQKEGIDYNETFSPVAKLNSVKLLLGVAARFKLSLTQLDISNAFLNGDLDEE 922
Query: 1136 VYVSQPPGFINKE----KPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTT 1191
+Y+ P G+ +++ PN V +L K+LYGLKQA R WY + S+ L+ GF + D T
Sbjct: 923 IYMRLPQGYASRQGDSLPPNAVCRLKKSLYGLKQASRQWYLKFSSTLLGLGFIQSYCDHT 982
Query: 1192 LFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQH 1251
F K + L V VY+DDII + + + M+S F++ +GEL +FLGL+I +
Sbjct: 983 CFLKISDGIFLCVLVYIDDIIIASNNDAAVDILKSQMKSFFKLRDLGELKYFLGLEIVRS 1042
Query: 1252 SNGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLY 1311
GI ISQ KY D+L + K S PM PS D G + YR +IG L+Y
Sbjct: 1043 DKGIHISQRKYALDLLDETGQLGCKPSSIPMDPSMVFAHDSGGDFVEVGPYRRLIGRLMY 1102
Query: 1312 LTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCD 1371
L +RPDI FAV A+F +++HL AV +I +Y+ GT GL+Y S L Y +
Sbjct: 1103 LNITRPDITFAVNKLAQFSMAPRKAHLQAVYKILQYIKGTIGQGLFYSATSELQLKVYAN 1162
Query: 1372 ADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQ 1431
ADY + R+STSG C FLG +LI W RKQ+ ++ S+ EAEY S + +++W+ N
Sbjct: 1163 ADYNSCRDSRRSTSGYCMFLGDSLICWKSRKQDVVSKSSAEAEYRSLSVATDELVWLTNF 1222
Query: 1432 LEDYSLRYTS-VPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDT 1490
L++ + + ++CDN +AI+++ N + H R+KHIE H +R+ + K L ++T
Sbjct: 1223 LKELQVPLSKPTLLFCDNEAAIHIANNHVFHERTKHIESDCHSVRERLLKGLFELYHINT 1282
Query: 1491 ENQLADIFTKPLVEDRFNFLKEKL 1514
E Q+AD FTKPL F+ L K+
Sbjct: 1283 ELQIADPFTKPLYPSHFHRLISKM 1306
Score = 105 bits (261), Expect = 3e-22
Identities = 115/460 (25%), Positives = 186/460 (40%), Gaps = 70/460 (15%)
Query: 430 QREINRKQKGIMSVPS--EVQKENILLKKEVETLKKDLTGFIKSTETFQNIVGSQNKSTK 487
+RE NR +M + + + IL+KK + +L ++ I ET ++ S
Sbjct: 126 ERETNRVIDFLMGLNDCYDTVRSQILMKKTLPSLS-EVFNMIDQDETQRSARISTTPGMT 184
Query: 488 KSGLGFKDPSK--IIGSFVPKAKIRVKCCFCDKYGHNESVCHVK----------KKFIKQ 535
S + S + + K R C +C + GH E C+ K +KF+K
Sbjct: 185 SSVFPVSNQSSQSALNGDTYQKKERPVCSYCSRPGHVEDTCYKKHGYPTSFKSKQKFVKP 244
Query: 536 N---NLNLSSERSHLNRSESSQKAEKAKKTCFYCNKSDHKRQNVTFRKDLLEE-LTFKDP 591
+ N + SE N S S+ ++ +Q V+F L+ T P
Sbjct: 245 SISANAAIGSEEVVNNTSVSTGDLTTSQI-----------QQLVSFLSSKLQPPSTPVQP 293
Query: 592 TLHGLDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNNDKGKIRGKGTIGNLNSARIENVQY 651
+H + S S + C ++ G V G + + +V +
Sbjct: 294 EVHSI-SVSSDPSSSSTVCPIS------GSVHLGRH----------------LILNDVLF 330
Query: 652 VEGLKHNLLSISQLCDS-GFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAE 710
+ K NLLS+S L S G + F C ++ + + G + NLY+++L+ +
Sbjct: 331 IPQFKFNLLSVSSLTKSMGCRIWFDETSCVLQDATRELMVGMGKQVANLYIVDLDSLSHP 390
Query: 711 SCFMSLE----KDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEACVKGKQ 766
S+ +WHKR GH S++ + +S L PK D C C KQ
Sbjct: 391 GTDSSITVASVTSHDLWHKRLGHPSVQKLQPMSSL---LSFPKQKNNTDFHCRVCHISKQ 447
Query: 767 VKSSFKTIEFIS----TQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLK 822
K + F+S + +P +L+HID +GP + G RY IVDD+SR TWV L+
Sbjct: 448 -----KHLPFVSHNNKSSRPFDLIHIDTWGPFSVQTHDGYRYFLTIVDDYSRATWVYLLR 502
Query: 823 HKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASF 862
+K + F V+N+ I VRSD+ E F
Sbjct: 503 NKSDVLTVIPTFVTMVENQFETTIKGVRSDNAPELNFTQF 542
>At1g59265 polyprotein, putative
Length = 1466
Score = 419 bits (1078), Expect = e-117
Identities = 241/664 (36%), Positives = 358/664 (53%), Gaps = 43/664 (6%)
Query: 900 STTSKGYRVYNLKTQTVEISMHIIFDEYDEHSKPKEN--EDTEVPTLQNVPVQNTENTVE 957
S+ S +R + + ++ S F E + P++N + T PT +++NT +
Sbjct: 801 SSPSAPFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQ 860
Query: 958 KEDDQNAQDQSFQS---------------------------------PPRSWRMVGDHPT 984
+ Q QS PP + ++
Sbjct: 861 NNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNN 920
Query: 985 DQIIGSTTDGVRTRLSFQDNN------MAMISQMEPKSINEAIIDDSWIKAMKEELSQFE 1038
+ + + G R + N +++ ++ EP++ +A+ D+ W AM E++
Sbjct: 921 QAPLNTHSMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQI 980
Query: 1039 RNKVWNLVPNNQDK-TIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAP 1097
N W+LVP TI+G RW+F K + +G + R KARLVA+GYNQ+ G+DY ETF+P
Sbjct: 981 GNHTWDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARLVAKGYNQRPGLDYAETFSP 1040
Query: 1098 VARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLT 1157
V + +IRI+L A +S + Q+DV +AFL G L ++VY+SQPPGFI+K++PN+V KL
Sbjct: 1041 VIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLR 1100
Query: 1158 KALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATK 1217
KALYGLKQAPRAWY L +L+ GF DT+LF ++ + VYVDDI+
Sbjct: 1101 KALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGND 1160
Query: 1218 IKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKYVKDILKKYKMNEAKI 1277
+ + + F + EL +FLG++ K+ G+ +SQ +Y+ D+L + M AK
Sbjct: 1161 PTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKP 1220
Query: 1278 MSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGLCARFQTCAKESH 1337
++TPM PS L K EYRG++GSL YL +RPDI +AV ++F E H
Sbjct: 1221 VTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEH 1280
Query: 1338 LTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDKVERKSTSGSCQFLGQALIG 1397
L A+KRI RYL GT + G++ +KG++ L AY DAD+AGDK + ST+G +LG I
Sbjct: 1281 LQALKRILRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPIS 1340
Query: 1398 WSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSVP-IYCDNTSAINLSK 1456
WS +KQ + S+TEAEY S A+ S++ W+ + L + +R T P IYCDN A L
Sbjct: 1341 WSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCA 1400
Query: 1457 NPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKPLVEDRFNFLKEKLLI 1516
NP+ HSR KHI I +HFIR+ VQ + + V T +QLAD TKPL F K+ +
Sbjct: 1401 NPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
Query: 1517 MKNP 1520
+ P
Sbjct: 1461 TRVP 1464
Score = 84.3 bits (207), Expect = 5e-16
Identities = 75/270 (27%), Positives = 116/270 (42%), Gaps = 13/270 (4%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGG---LVTFGNNDKGKIRGKGTIGNLNSA-RIENVQY 651
LDSG + H+T D + + GG +V G+ G ++ + + N+ Y
Sbjct: 333 LDSGATHHITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKSRPLNLHNILY 392
Query: 652 VEGLKHNLLSISQLCDS-GFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAE 710
V + NL+S+ +LC++ G V F P +V+ ++ G + LY +
Sbjct: 393 VPNIHKNLISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASSQPV 452
Query: 711 SCFMSLEKDKW--IWHKRAGHISMKTI-AKLSQLDLVRGLPKISFEKDRICEACVKGKQV 767
S F S WH R GH + + + +S L P F C C+ K
Sbjct: 453 SLFASPSSKATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLS---CSDCLINKSN 509
Query: 768 KSSFKTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDES 827
K F ST +PLE ++ D++ S RY + VD F+R+TW+ LK K +
Sbjct: 510 KVPFSQSTINST-RPLEYIYSDVWSS-PILSHDNYRYYVIFVDHFTRYTWLYPLKQKSQV 567
Query: 828 FEAFQNFCKRVQNEKGYNIITVRSDHGGEF 857
E F F ++N I T SD+GGEF
Sbjct: 568 KETFITFKNLLENRFQTRIGTFYSDNGGEF 597
Score = 50.8 bits (120), Expect = 6e-06
Identities = 25/57 (43%), Positives = 31/57 (53%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE 926
VFGC CY K D KS + +FLGYS T Y +L+T + IS H+ FDE
Sbjct: 690 VFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDE 746
Score = 41.6 bits (96), Expect = 0.004
Identities = 25/106 (23%), Positives = 52/106 (48%), Gaps = 5/106 (4%)
Query: 87 ALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGVRKFEVFEMSENETIDEMY 146
A++M V T A +W+TL+ + S+ T++ ++++ +TID+
Sbjct: 91 AISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWT----KGTKTIDDYM 146
Query: 147 ARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQAKD 192
T +++ LGK +++ ++L LP ++P++ I AKD
Sbjct: 147 QGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIA-AKD 191
>At1g58889 polyprotein, putative
Length = 1466
Score = 418 bits (1074), Expect = e-116
Identities = 240/664 (36%), Positives = 357/664 (53%), Gaps = 43/664 (6%)
Query: 900 STTSKGYRVYNLKTQTVEISMHIIFDEYDEHSKPKEN--EDTEVPTLQNVPVQNTENTVE 957
S+ S +R + + ++ S F E + P++N + T PT +++NT +
Sbjct: 801 SSPSAPFRNSQVSSSNLDSSFSSSFPSSPEPTAPRQNGPQPTTQPTQTQTQTHSSQNTSQ 860
Query: 958 KEDDQNAQDQSFQS---------------------------------PPRSWRMVGDHPT 984
+ Q QS PP + ++
Sbjct: 861 NNPTNESPSQLAQSLSTPAQSSSSSPSPTTSASSSSTSPTPPSILIHPPPPLAQIVNNNN 920
Query: 985 DQIIGSTTDGVRTRLSFQDNN------MAMISQMEPKSINEAIIDDSWIKAMKEELSQFE 1038
+ + + G R + N +++ ++ EP++ +A+ D+ W AM E++
Sbjct: 921 QAPLNTHSMGTRAKAGIIKPNPKYSLAVSLAAESEPRTAIQALKDERWRNAMGSEINAQI 980
Query: 1039 RNKVWNLVPNNQDK-TIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAP 1097
N W+LVP TI+G RW+F K + +G + R KAR VA+GYNQ+ G+DY ETF+P
Sbjct: 981 GNHTWDLVPPPPSHVTIVGCRWIFTKKYNSDGSLNRYKARFVAKGYNQRPGLDYAETFSP 1040
Query: 1098 VARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLT 1157
V + +IRI+L A +S + Q+DV +AFL G L ++VY+SQPPGFI+K++PN+V KL
Sbjct: 1041 VIKSTSIRIVLGVAVDRSWPIRQLDVNNAFLQGTLTDDVYMSQPPGFIDKDRPNYVCKLR 1100
Query: 1158 KALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATK 1217
KALYGLKQAPRAWY L +L+ GF DT+LF ++ + VYVDDI+
Sbjct: 1101 KALYGLKQAPRAWYVELRNYLLTIGFVNSVSDTSLFVLQRGKSIVYMLVYVDDILITGND 1160
Query: 1218 IKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKYVKDILKKYKMNEAKI 1277
+ + + F + EL +FLG++ K+ G+ +SQ +Y+ D+L + M AK
Sbjct: 1161 PTLLHNTLDNLSQRFSVKDHEELHYFLGIEAKRVPTGLHLSQRRYILDLLARTNMITAKP 1220
Query: 1278 MSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGLCARFQTCAKESH 1337
++TPM PS L K EYRG++GSL YL +RPDI +AV ++F E H
Sbjct: 1221 VTTPMAPSPKLSLYSGTKLTDPTEYRGIVGSLQYLAFTRPDISYAVNRLSQFMHMPTEEH 1280
Query: 1338 LTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDKVERKSTSGSCQFLGQALIG 1397
L A+KRI RYL GT + G++ +KG++ L AY DAD+AGDK + ST+G +LG I
Sbjct: 1281 LQALKRILRYLAGTPNHGIFLKKGNTLSLHAYSDADWAGDKDDYVSTNGYIVYLGHHPIS 1340
Query: 1398 WSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSVP-IYCDNTSAINLSK 1456
WS +KQ + S+TEAEY S A+ S++ W+ + L + +R T P IYCDN A L
Sbjct: 1341 WSSKKQKGVVRSSTEAEYRSVANTSSEMQWICSLLTELGIRLTRPPVIYCDNVGATYLCA 1400
Query: 1457 NPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKPLVEDRFNFLKEKLLI 1516
NP+ HSR KHI I +HFIR+ VQ + + V T +QLAD TKPL F K+ +
Sbjct: 1401 NPVFHSRMKHIAIDYHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRTAFQNFASKIGV 1460
Query: 1517 MKNP 1520
+ P
Sbjct: 1461 TRVP 1464
Score = 84.3 bits (207), Expect = 5e-16
Identities = 75/270 (27%), Positives = 116/270 (42%), Gaps = 13/270 (4%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGG---LVTFGNNDKGKIRGKGTIGNLNSA-RIENVQY 651
LDSG + H+T D + + GG +V G+ G ++ + + N+ Y
Sbjct: 333 LDSGATHHITSDFNNLSLHQPYTGGDDVMVADGSTIPISHTGSTSLSTKSRPLNLHNILY 392
Query: 652 VEGLKHNLLSISQLCDS-GFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAE 710
V + NL+S+ +LC++ G V F P +V+ ++ G + LY +
Sbjct: 393 VPNIHKNLISVYRLCNANGVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASSQPV 452
Query: 711 SCFMSLEKDKW--IWHKRAGHISMKTI-AKLSQLDLVRGLPKISFEKDRICEACVKGKQV 767
S F S WH R GH + + + +S L P F C C+ K
Sbjct: 453 SLFASPSSKATHSSWHARLGHPAPSILNSVISNYSLSVLNPSHKFLS---CSDCLINKSN 509
Query: 768 KSSFKTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDES 827
K F ST +PLE ++ D++ S RY + VD F+R+TW+ LK K +
Sbjct: 510 KVPFSQSTINST-RPLEYIYSDVWSS-PILSHDNYRYYVIFVDHFTRYTWLYPLKQKSQV 567
Query: 828 FEAFQNFCKRVQNEKGYNIITVRSDHGGEF 857
E F F ++N I T SD+GGEF
Sbjct: 568 KETFITFKNLLENRFQTRIGTFYSDNGGEF 597
Score = 50.8 bits (120), Expect = 6e-06
Identities = 25/57 (43%), Positives = 31/57 (53%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE 926
VFGC CY K D KS + +FLGYS T Y +L+T + IS H+ FDE
Sbjct: 690 VFGCACYPWLRPYNQHKLDDKSRQCVFLGYSLTQSAYLCLHLQTSRLYISRHVRFDE 746
Score = 41.6 bits (96), Expect = 0.004
Identities = 25/106 (23%), Positives = 52/106 (48%), Gaps = 5/106 (4%)
Query: 87 ALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGVRKFEVFEMSENETIDEMY 146
A++M V T A +W+TL+ + S+ T++ ++++ +TID+
Sbjct: 91 AISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQLRTQLKQWT----KGTKTIDDYM 146
Query: 147 ARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQAKD 192
T +++ LGK +++ ++L LP ++P++ I AKD
Sbjct: 147 QGLVTRFDQLALLGKPMDHDEQVERVLENLPEEYKPVIDQIA-AKD 191
>At4g02960 putative polyprotein of LTR transposon
Length = 1456
Score = 407 bits (1045), Expect = e-113
Identities = 213/530 (40%), Positives = 317/530 (59%), Gaps = 2/530 (0%)
Query: 993 DGVRTRLSFQDNNMAMISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDK 1052
DG+R ++ + EP++ +A+ DD W +AM E++ N W+LVP
Sbjct: 918 DGIRKPNQKYSYATSLAANSEPRTAIQAMKDDRWRQAMGSEINAQIGNHTWDLVPPPPPS 977
Query: 1053 -TIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYA 1111
TI+G RW+F K + +G + R KARLVA+GYNQ+ G+DY ETF+PV + +IRI+L A
Sbjct: 978 VTIVGCRWIFTKKFNSDGSLNRYKARLVAKGYNQRPGLDYAETFSPVIKSTSIRIVLGVA 1037
Query: 1112 AHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWY 1171
+S + Q+DV +AFL G L +EVY+SQPPGF++K++P++V +L KA+YGLKQAPRAWY
Sbjct: 1038 VDRSWPIRQLDVNNAFLQGTLTDEVYMSQPPGFVDKDRPDYVCRLRKAIYGLKQAPRAWY 1097
Query: 1172 DRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSE 1231
L T+L+ GF DT+LF ++ + VYVDDI+ + + + +
Sbjct: 1098 VELRTYLLTVGFVNSISDTSLFVLQRGRSIIYMLVYVDDILITGNDTVLLKHTLDALSQR 1157
Query: 1232 FEMSMMGELGFFLGLQIKQHSNGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKD 1291
F + +L +FLG++ K+ G+ +SQ +Y D+L + M AK ++TPM S L
Sbjct: 1158 FSVKEHEDLHYFLGIEAKRVPQGLHLSQRRYTLDLLARTNMLTAKPVATPMATSPKLTLH 1217
Query: 1292 ESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGT 1351
K EYRG++GSL YL +RPD+ +AV +++ + H A+KR+ RYL GT
Sbjct: 1218 SGTKLPDPTEYRGIVGSLQYLAFTRPDLSYAVNRLSQYMHMPTDDHWNALKRVLRYLAGT 1277
Query: 1352 TDLGLWYRKGSSFDLVAYCDADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTT 1411
D G++ +KG++ L AY DAD+AGD + ST+G +LG I WS +KQ + S+T
Sbjct: 1278 PDHGIFLKKGNTLSLHAYSDADWAGDTDDYVSTNGYIVYLGHHPISWSSKKQKGVVRSST 1337
Query: 1412 EAEYVSAASCCSQILWVRNQLEDYSLRYTSVP-IYCDNTSAINLSKNPIQHSRSKHIEIK 1470
EAEY S A+ S++ W+ + L + ++ + P IYCDN A L NP+ HSR KHI +
Sbjct: 1338 EAEYRSVANTSSELQWICSLLTELGIQLSHPPVIYCDNVGATYLCANPVFHSRMKHIALD 1397
Query: 1471 HHFIRDHVQKKNIALSFVDTENQLADIFTKPLVEDRFNFLKEKLLIMKNP 1520
+HFIR+ VQ + + V T +QLAD TKPL F K+ ++K P
Sbjct: 1398 YHFIRNQVQSGALRVVHVSTHDQLADTLTKPLSRVAFQNFSRKIGVIKVP 1447
Score = 89.0 bits (219), Expect = 2e-17
Identities = 79/270 (29%), Positives = 121/270 (44%), Gaps = 13/270 (4%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGGL-VTFGNNDKGKIRGKGTIGNLNSAR---IENVQY 651
LDSG + H+T D + + GG V + I G+ S+R + V Y
Sbjct: 312 LDSGATHHITSDFNNLSFHQPYTGGDDVMIADGSTIPITHTGSASLPTSSRSLDLNKVLY 371
Query: 652 VEGLKHNLLSISQLCDSG-FEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAE 710
V + NL+S+ +LC++ V F P +V+ ++ G + LY + A
Sbjct: 372 VPNIHKNLISVYRLCNTNRVSVEFFPASFQVKDLNTGVPLLQGKTKDELYEWPIASSQAV 431
Query: 711 SCFMS--LEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKIS-FEKDRICEACVKGKQV 767
S F S + WH R GH S+ A L+ + LP ++ K C C K
Sbjct: 432 SMFASPCSKATHSSWHSRLGHPSL---AILNSVISNHSLPVLNPSHKLLSCSDCFINKSH 488
Query: 768 KSSFKTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDES 827
K F I++ KPLE ++ D++ S+ RY + VD F+R+TW+ LK K +
Sbjct: 489 KVPFSN-STITSSKPLEYIYSDVWSS-PILSIDNYRYYVIFVDHFTRYTWLYPLKQKSQV 546
Query: 828 FEAFQNFCKRVQNEKGYNIITVRSDHGGEF 857
+ F F V+N I T+ SD+GGEF
Sbjct: 547 KDTFIIFKSLVENRFQTRIGTLYSDNGGEF 576
Score = 43.1 bits (100), Expect = 0.001
Identities = 21/57 (36%), Positives = 28/57 (48%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE 926
VFGC CY K + KS + F+GYS T Y ++ T + S H+ FDE
Sbjct: 669 VFGCACYPWLRPYNRHKLEDKSKQCAFMGYSLTQSAYLCLHIPTGRLYTSRHVQFDE 725
Score = 34.7 bits (78), Expect = 0.43
Identities = 30/134 (22%), Positives = 56/134 (41%), Gaps = 33/134 (24%)
Query: 87 ALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGVRKFEVFEMSENETIDEMY 146
A++M V T A +W+TL+ + S+ T++
Sbjct: 91 AISMSVQPAVSRATTAAQIWETLRKIYANPSYGHVTQL---------------------- 128
Query: 147 ARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQAKDL---------KSMN 197
RF T +++ LGK +++ ++L LP ++P++ I AKD + +N
Sbjct: 129 -RFITRFDQLALLGKPMDHDEQVERVLENLPDDYKPVIDQIA-AKDTPPSLTEIHERLIN 186
Query: 198 LEDLIGSLRAHEVV 211
E + +L + EVV
Sbjct: 187 RESKLLALNSAEVV 200
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 405 bits (1040), Expect = e-112
Identities = 241/671 (35%), Positives = 368/671 (53%), Gaps = 40/671 (5%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVY-----------NLKTQTVEI 918
VFGC Y L K D +S ++LG SK YR+ N ++T +I
Sbjct: 622 VFGCIGYAKIEGPHLRKLDDRSKMLVYLGTEPGSKAYRLLDPTNRKIIKWNNSDSETRDI 681
Query: 919 S--MHIIFDEYDEHSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQ--------S 968
S + E+ + E ++ T +N + E E++ N Q+Q S
Sbjct: 682 SGTFSLTLGEFGNNGI---QESDDIETEKNGEESENSHEEEGENEHNEQEQIDAEETQPS 738
Query: 969 FQSP----PRSWRMVG--DHPTDQIIGSTTDGVRTRLSFQDNNMAMISQMEPKSINEAII 1022
+P RS R VG ++ D ++ + +G + L+ D EP EA
Sbjct: 739 HATPLPTLRRSTRQVGKPNYLDDYVLMAEIEGEQVLLAIND---------EPWDFKEANK 789
Query: 1023 DDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQG 1082
W A KEE+ E+NK W+L+ + +IG +WVF+ K + +G + + KARLVA+G
Sbjct: 790 LKEWRDACKEEILSIEKNKTWSLIDLPVRRKVIGLKWVFKIKRNSDGSINKYKARLVAKG 849
Query: 1083 YNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPP 1142
Y Q+ GIDYDE FA VAR+E IR+++A AA ++ +DVK+AFL+G L E+VYV+QP
Sbjct: 850 YVQRHGIDYDEVFAHVARIETIRVIIALAASNGWEVHHLDVKTAFLHGELREDVYVTQPE 909
Query: 1143 GFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLL 1202
GF NK+ V+KL KALYGLKQAPRAW +L+ L E F + + +++R+ LL
Sbjct: 910 GFTNKDNEGKVYKLHKALYGLKQAPRAWNTKLNKILQELNFVKCSKEPSVYRRQEEKKLL 969
Query: 1203 IVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKY 1262
IV +YVDD++ + + + F M +FEMS +G+L ++LG+++ NGI + QE+Y
Sbjct: 970 IVAIYVDDLLVTGSSLDLILCFKKDMAGKFEMSDLGQLTYYLGIEVLHRKNGIILRQERY 1029
Query: 1263 VKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPDIVFA 1322
I+++ M+ + PM L K + K I+E++YR MIG L Y+ +RPD+ +
Sbjct: 1030 AMKIIEEAGMSNCNPVLIPMAAGLELCKAQEEKCITERDYRRMIGCLRYIVHTRPDLSYC 1089
Query: 1323 VGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDKVERK 1382
VG+ +R+ +ESH A+K++ RYL GT GL+ ++G LV Y D+ ++ D + K
Sbjct: 1090 VGVLSRYLQQPRESHGNALKQVLRYLKGTMSHGLYLKRGFKSGLVGYSDSSHSADLDDGK 1149
Query: 1383 STSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQL-EDYSLRYTS 1441
ST+G +L Q I W +KQ +ALS+ EAE+++A Q +W+++ E
Sbjct: 1150 STAGHIFYLHQCPITWCSQKQQVVALSSCEAEFMAATEAAKQAIWLQDLFAEVCGTTSEK 1209
Query: 1442 VPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKP 1501
V I DN SAI L+KN + H RSKHI ++HFIR+ V+ + + V Q ADI TKP
Sbjct: 1210 VMIRVDNKSAIALTKNLVFHGRSKHIHRRYHFIRECVENNLVEVDHVPGVEQRADILTKP 1269
Query: 1502 LVEDRFNFLKE 1512
L +F ++E
Sbjct: 1270 LGRIKFREMRE 1280
Score = 141 bits (355), Expect = 3e-33
Identities = 141/575 (24%), Positives = 244/575 (41%), Gaps = 89/575 (15%)
Query: 330 WDDMEE-SDSQEDADTDMGLMAQSDDEEEVI-------------IYKTDSLYKDL--ENK 373
W+ ++ SD E D L+ QS E ++ KT +L + E K
Sbjct: 11 WETIDPGSDDMEKNDMARALLFQSVPESTILQVGKHKTSKAMWEAIKTRNLGAERVKEAK 70
Query: 374 IDSLLYDSNFLTNRCHSLIKE----LSEIKEEKEILQNKYDESRKTIKILQDS------H 423
+ +L+ + + L + + I E +SEI + E L + +ES+ K L+ H
Sbjct: 71 LQTLMAEFDRLNMKDNETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIH 130
Query: 424 FDMSEKQREINRKQKGIMSVPSEVQKENILLKKEVETLKKDLTGFIKSTETFQNIVGSQN 483
++ ++ ++ G + ++ + E ++ ++ ++E+ + G +
Sbjct: 131 I-IAALEQILDLNTTGFEDIVGRMKTYEDRVCDEDDSPEEQGKLMYANSESSYDTRGGRG 189
Query: 484 KSTKKS------GLGF--KDPSKIIGSFVPKAKIRVKCCFCDKYGHNESVCHVKK-KFIK 534
+ +S G G+ +D SK+I C CDK GH S C + K IK
Sbjct: 190 RGRGRSSGRGRGGYGYQQRDKSKVI------------CYRCDKTGHYASECLDRLLKLIK 237
Query: 535 QNNLNLSSERSHLNRSESSQKAEKAKKTCFYCNKSDHKRQNVTFRKDLLEELTFKDPTLH 594
++ N + + Y N+ K + D
Sbjct: 238 -------AQEQQQNNEDDDEIESLMMHEVVYLNERSVKPKEFEACSD----------NSW 280
Query: 595 GLDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNNDKGKIRGKGTIGNLNSARIE----NVQ 650
LD+G S HMTG+ F + G V FG++ + I+GKG+I + I +V
Sbjct: 281 YLDNGASNHMTGNLQWFSKLNEMITGKVRFGDDSRIDIKGKGSIVLITKGGIRKTLTDVY 340
Query: 651 YVEGLKHNLLSISQLCDSGFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAE 710
++ LK N++S+ Q ++G +V K + + L + R LY ++LN
Sbjct: 341 FIPDLKSNIISLGQATEAGCDVRMKDDQLTLHDREGCLLLRATRSRNRLYKVDLN----- 395
Query: 711 SCFMSLEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEACVKGKQVKSS 770
+E K + A + + +LV G+ I EK+ C +C+ GKQ +
Sbjct: 396 -----VENVKCL---------QLEAATMVRKELVIGISNIPKEKET-CGSCLLGKQARQP 440
Query: 771 FKTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESFEA 830
F + LEL+H DL GP+ ++ KRY V++DD +R+ W + LK K E+FE
Sbjct: 441 FPKATTYRASQVLELVHGDLCGPITQSTTAKKRYILVLIDDHTRYMWSMLLKEKSEAFEK 500
Query: 831 FQNFCKRVQNEKGYNIITVRSDHGGEFENASFKTF 865
F++F +V+ E G I T R+D GGEF + F+ F
Sbjct: 501 FRDFKTKVEQESGVKIKTFRTDKGGEFVSQEFQDF 535
Score = 87.8 bits (216), Expect = 4e-17
Identities = 86/366 (23%), Positives = 147/366 (39%), Gaps = 53/366 (14%)
Query: 57 EGAVKAKSAWSTDEKAQ--VLLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHE 114
E ++ W T + + N AR L ++ +V + +K +W+ +K +
Sbjct: 2 EATLRVHKVWETIDPGSDDMEKNDMARALLFQSVPESTILQVGKHKTSKAMWEAIKTRNL 61
Query: 115 GTSHVKETRIDIGVRKFEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILR 174
G VKE ++ + +F+ M +NETIDE R + I + SLG+ ++K L+
Sbjct: 62 GAERVKEAKLQTLMAEFDRLNMKDNETIDEFVGRISEISTKSESLGEEIEESKIVKKFLK 121
Query: 175 CLP-SMWRPMVTAITQAKDLKSMNLEDLIGSLRAHEVVLQGDKPVKKVRTLALKASQQTS 233
LP + ++ A+ Q DL + ED++G ++ +E
Sbjct: 122 SLPRKKYIHIIAALEQILDLNTTGFEDIVGRMKTYE-----------------------D 158
Query: 234 SVDDEDVQEPQELEEVHEEEAEDELALISKRIQRMMLRRNQIRKKFPKTNISIKTEADKS 293
V DED P+E ++ +E R R + R + DKS
Sbjct: 159 RVCDED-DSPEEQGKLMYANSESSYDTRGGR------GRGRGRSSGRGRGGYGYQQRDKS 211
Query: 294 QVTCYGCNKTGHFKNECPD--IKKVQRKPPFKKKAMITWDDMEESDSQEDADTDMGLMAQ 351
+V CY C+KTGH+ +EC D +K ++ + E+ + ED D LM
Sbjct: 212 KVICYRCDKTGHYASECLDRLLKLIKAQ--------------EQQQNNEDDDEIESLMM- 256
Query: 352 SDDEEEVIIYKTDSLYKDLENKIDSLLYDSNFLTNRCHSLIKELSEIKEEKEILQNKYDE 411
E V + + K+ E D+ Y N +N ++ S++ E D+
Sbjct: 257 ---HEVVYLNERSVKPKEFEACSDNSWYLDNGASNHMTGNLQWFSKLNEMITGKVRFGDD 313
Query: 412 SRKTIK 417
SR IK
Sbjct: 314 SRIDIK 319
>At1g53810
Length = 1522
Score = 403 bits (1036), Expect = e-112
Identities = 208/516 (40%), Positives = 306/516 (58%), Gaps = 10/516 (1%)
Query: 1009 ISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEE 1068
+S EPK++ EA+ W AM+EE+ + + W LVP + + ++G+ WVFR KL +
Sbjct: 911 VSIPEPKTVTEALKHPGWNNAMQEEMGNCKETETWTLVPYSPNMNVLGSMWVFRTKLHAD 970
Query: 1069 GKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFL 1128
G + + KARLVA+G+ Q+EGIDY ET++PV R +R++L A +L QMDVK+AFL
Sbjct: 971 GSLDKLKARLVAKGFKQEEGIDYLETYSPVVRTPTVRLILHVATVLKWELKQMDVKNAFL 1030
Query: 1129 NGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKI 1188
+G L E VY+ QP GF++K KP+HV L K+LYGLKQ+PRAW+DR S FL+E GF
Sbjct: 1031 HGDLTETVYMRQPAGFVDKSKPDHVCLLHKSLYGLKQSPRAWFDRFSNFLLEFGFICSLF 1090
Query: 1189 DTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQI 1248
D +LF + N D++++ +YVDD++ + + EF M MG++ +FLG+QI
Sbjct: 1091 DPSLFVYSSNNDVILLLLYVDDMVITGNNSQSLTHLLAALNKEFRMKDMGQVHYFLGIQI 1150
Query: 1249 KQHSNGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGS 1308
+ + G+F+SQ+KY +D+L M M TP+ ++ +R + G
Sbjct: 1151 QTYDGGLFMSQQKYAEDLLITASMANCSPMPTPLPLQLDRVSNQDEVFSDPTYFRSLAGK 1210
Query: 1309 LLYLTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSS----- 1363
L YLT +RPDI FAV + S +KRI RY+ GT +G+ Y SS
Sbjct: 1211 LQYLTLTRPDIQFAVNFVCQKMHQPSVSDFNLLKRILRYIKGTVSMGIQYNSNSSSVVSA 1270
Query: 1364 ----FDLVAYCDADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAA 1419
+DL AY D+DYA K R+S G C F+GQ +I WS +KQ T++ S+TEAEY S +
Sbjct: 1271 YESDYDLSAYSDSDYANCKETRRSVGGYCTFMGQNIISWSSKKQPTVSRSSTEAEYRSLS 1330
Query: 1420 SCCSQILWVRNQLEDYSLRYTSVP-IYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHV 1478
S+I W+ + L + + P ++CDN SA+ L+ NP H+R+KH ++ HH+IR+ V
Sbjct: 1331 ETASEIKWMSSILREIGVSLPDTPELFCDNLSAVYLTANPAFHARTKHFDVDHHYIRERV 1390
Query: 1479 QKKNIALSFVDTENQLADIFTKPLVEDRFNFLKEKL 1514
K + + + QLADIFTK L + F L+ KL
Sbjct: 1391 ALKTLVVKHIPGHLQLADIFTKSLPFEAFTRLRFKL 1426
Score = 118 bits (295), Expect = 3e-26
Identities = 94/331 (28%), Positives = 143/331 (42%), Gaps = 24/331 (7%)
Query: 552 SSQKAEKAKKTCFYCNKSDHKRQNVTFRKD----------LLEELTFKDPTLHG-----L 596
S +A + C C K+ H R D L + D T H
Sbjct: 269 SGSQAGNSGLVCQICGKAGHHALKCWHRFDNSYQHEDLPMALATMRITDVTDHHGHEWIP 328
Query: 597 DSGCSRHMTGDRDCFLT---FEKKDGGLVTFGNNDKGKIRGKGTIGNLNSA-RIENVQYV 652
DS S H+T +R + D +V GN G G+I + + ++ V
Sbjct: 329 DSAASAHVTNNRHVLQQSQPYHGSDSIMVADGNFLPITHTGSGSIASSSGKIPLKEVLVC 388
Query: 653 EGLKHNLLSISQLC-DSGFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAES 711
+ +LLS+S+L D V F + + ++ KL G R LY LE +
Sbjct: 389 PDIVKSLLSVSKLTSDYPCSVEFDADSVRINDKATKKLLVMGRNRDGLYSLEEPKLQVLY 448
Query: 712 CFMSLEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEACVKGKQVKSSF 771
+WH+R GH + + + +L+ + + K+ +CEAC GK + F
Sbjct: 449 STRQNSASSEVWHRRLGHANAEVLHQLASSKSIIIINKVV---KTVCEACHLGKSTRLPF 505
Query: 772 KTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESFEAF 831
F + +PLE +H DL+GP T+S+ G RY V +D +SRFTW LK K + F F
Sbjct: 506 MLSTF-NASRPLERIHCDLWGPSPTSSVQGFRYYVVFIDHYSRFTWFYPLKLKSDFFSTF 564
Query: 832 QNFCKRVQNEKGYNIITVRSDHGGEFENASF 862
F K V+N+ G+ I + D GGEF ++ F
Sbjct: 565 VMFQKLVENQLGHKIKIFQCDGGGEFISSQF 595
Score = 56.2 bits (134), Expect = 1e-07
Identities = 27/57 (47%), Positives = 33/57 (57%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE 926
VFGC CY KFDP+S K +FLGY+ KGYR T + IS H++FDE
Sbjct: 686 VFGCACYPTLRDYASTKFDPRSLKCVFLGYNEKYKGYRCLYPPTGRIYISRHVVFDE 742
Score = 37.7 bits (86), Expect = 0.051
Identities = 49/204 (24%), Positives = 82/204 (40%), Gaps = 28/204 (13%)
Query: 23 NYYFWKGKMELFLRSQDNDMWAVITNGDFVP-----------TNKEGAVKAKSAWSTDEK 71
NY WK + E FL Q + +T P T++E + + TD+
Sbjct: 23 NYILWKSQFESFLSGQ--GLLGFVTGSISAPAQTRSVTHNNVTSEEPNPEFYTWHQTDQV 80
Query: 72 AQV-LLNSKARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTSHVKETRIDIGVRK 130
+ LL S A LS V C + VW TL H + V +R+ R+
Sbjct: 81 VKSWLLGSFAEDILSV---------VVNCFTSHQVWLTLANHF---NRVSSSRLFELQRR 128
Query: 131 FEVFEMSENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQA 190
+ E +N T++ I +++ S+G +I L L + P+ T I +
Sbjct: 129 LQTLEKKDN-TMEVFLKDLKHICDQLASVGSPVPEKMKIFSALNGLGREYEPIKTTIENS 187
Query: 191 KDLK-SMNLEDLIGSLRAHEVVLQ 213
D S++L+++ LR ++ LQ
Sbjct: 188 VDSNPSLSLDEVASKLRGYDDRLQ 211
>At1g44510 polyprotein, putative
Length = 1459
Score = 400 bits (1029), Expect = e-111
Identities = 208/509 (40%), Positives = 304/509 (58%), Gaps = 1/509 (0%)
Query: 1013 EPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVV 1072
EP ++ +A+ D W AM +E +RN W+LVP N + ++G RWVF+ K G +
Sbjct: 949 EPNTVTQALKDKKWRFAMSDEFDAQQRNHTWDLVPPNPTQHLVGCRWVFKLKYLPNGLID 1008
Query: 1073 RNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFL 1132
+ KARLVA+G+NQQ G+DY ETF+PV + IR++L A K+ L Q+DV +AFL G L
Sbjct: 1009 KYKARLVAKGFNQQYGVDYAETFSPVIKATTIRVVLDVAVKKNWPLKQLDVNNAFLQGTL 1068
Query: 1133 NEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTL 1192
EEVY++QPPGF++K++P+HV +L KA+YGLKQAPRAWY L L+ GF DT+L
Sbjct: 1069 TEEVYMAQPPGFVDKDRPSHVCRLRKAIYGLKQAPRAWYMELKQHLLNIGFVNSLADTSL 1128
Query: 1193 FRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHS 1252
F +H T LL + VYVDDII + K + + F + +L +FLG++ + +
Sbjct: 1129 FIYSHGTTLLYLLVYVDDIIVTGSDHKSVSAVLSSLAERFSIKDPTDLHYFLGIEATRTN 1188
Query: 1253 NGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYL 1312
G+ + Q KY+ D+L K+ M +AK ++TP+ S L K EYR ++GSL YL
Sbjct: 1189 TGLHLMQRKYMTDLLAKHNMLDAKPVATPLPTSPKLTLHGGTKLNDASEYRSVVGSLQYL 1248
Query: 1313 TASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDA 1372
+RPDI FAV ++F H A KR+ RYL GTT G++ S L A+ DA
Sbjct: 1249 AFTRPDIAFAVNRLSQFMHQPTSDHWQAAKRVLRYLAGTTTHGIFLNSSSPIHLHAFSDA 1308
Query: 1373 DYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQL 1432
D+AGD + ST+ +LG+ I WS +KQ ++ S+TE+EY + A+ S+I W+ + L
Sbjct: 1309 DWAGDSADYVSTNAYVIYLGRNPISWSSKKQRGVSRSSTESEYRAVANAASEIRWLCSLL 1368
Query: 1433 EDYSLRYTSVP-IYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTE 1491
+ +R P I+CDN A + NP+ HSR KHI + +HF+R +Q + + +S V T
Sbjct: 1369 TELHIRLPHGPTIFCDNIGATYICANPVFHSRMKHIALDYHFVRGMIQSRALRVSHVSTN 1428
Query: 1492 NQLADIFTKPLVEDRFNFLKEKLLIMKNP 1520
+QLAD TK L F + K+ + + P
Sbjct: 1429 DQLADALTKSLSRPHFLSARSKIGVRQLP 1457
Score = 92.4 bits (228), Expect = 2e-18
Identities = 76/271 (28%), Positives = 124/271 (45%), Gaps = 15/271 (5%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGG---LVTFGNNDKGKIRGKGTIGNLN-SARIENVQY 651
LDSG + H+T D + + +GG ++ G K G + + N + V Y
Sbjct: 339 LDSGATHHITSDLNALSLHQPYNGGEYVMIADGTGLTIKQTGSTFLPSQNRDLALHKVLY 398
Query: 652 VEGLKHNLLSISQLCDSG-FEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAE 710
V ++ NL+S+ +LC++ V F P +V+ ++ L G + +LY + + PA
Sbjct: 399 VPDIRKNLISVYRLCNTNQVSVEFFPASFQVKDLNTGTLLLQGRTKDDLYEWPVTNPPAT 458
Query: 711 SCFMSLEKDKWI--WHKRAGHISMKTI-AKLSQLDLVRGLPKISFEKDRICEACVKGKQV 767
+ F S + WH R GH S + LS+ L ++ C C+ K
Sbjct: 459 ALFTSPSPKTTLSSWHSRLGHPSASILNTLLSKFSLP---VSVASSNKTSCSDCLINKSH 515
Query: 768 KSSFKTIEFISTQKPLELLHIDLF-GPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDE 826
K F T I + PLE + D++ P+ S +Y V+VD ++R+TW+ L+ K +
Sbjct: 516 KLPFATSS-IHSSSPLEYIFTDVWTSPI--ISHDNYKYYLVLVDHYTRYTWLYPLQQKSQ 572
Query: 827 SFEAFQNFCKRVQNEKGYNIITVRSDHGGEF 857
F F V+N I T+ SD+GGEF
Sbjct: 573 VKATFIAFKALVENRFQAKIRTLYSDNGGEF 603
Score = 45.1 bits (105), Expect = 3e-04
Identities = 28/108 (25%), Positives = 47/108 (42%), Gaps = 6/108 (5%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE--Y 927
VFGC C+ K + +S + +FLGYS T Y ++ + S H++FDE Y
Sbjct: 696 VFGCLCFPWLRPYTRNKLEERSKRCVFLGYSLTQTAYLCLDVDNNRLYTSRHVMFDESTY 755
Query: 928 DEHSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPRS 975
+ +E + + T P +++ ++ QSPP S
Sbjct: 756 PFAASIREQSQSSLVT----PPESSSSSSPANSGFPCSVLRLQSPPAS 799
Score = 42.7 bits (99), Expect = 0.002
Identities = 51/216 (23%), Positives = 90/216 (41%), Gaps = 20/216 (9%)
Query: 22 SNYYFWKGKMELFLRSQDNDMWAVITNGDFVP---TNKEGAVKAKSAWSTDEKAQVLLNS 78
+NY W ++ L D + + N +P T V A +++ ++ L+ S
Sbjct: 36 TNYLMWSIQIHALLDGYD--LAGYLDNSVVIPPETTTINSVVSANPSFTLWKRQDKLIFS 93
Query: 79 KARLFLSCALTMEESERVDECTNAKGVWDTLKIHHEGTS--HVKETRIDIGVRKFEVFEM 136
L A++ V TN+ +W TL + S H+K+ R I
Sbjct: 94 A----LIGAISPAVQSLVSRATNSSQIWSTLNNTYAKPSYGHIKQLRQQIQRLT------ 143
Query: 137 SENETIDEMYARFTTIVNEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQAKDLKSM 196
+TIDE TT ++++ LGK +++ IL+ LP ++ +V I + KD +
Sbjct: 144 KGTKTIDEYVQSHTTRLDQLAILGKPMEHEEQVEHILKGLPEEYKTVVDQI-EGKD-NTP 201
Query: 197 NLEDLIGSLRAHEVVLQGDK-PVKKVRTLALKASQQ 231
+ ++ L HE L D+ P ++ A QQ
Sbjct: 202 TITEIHERLINHESKLLSDEVPPSSSFPMSANAVQQ 237
>At4g27210 putative protein
Length = 1318
Score = 396 bits (1018), Expect = e-110
Identities = 204/515 (39%), Positives = 309/515 (59%), Gaps = 5/515 (0%)
Query: 1009 ISQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEE 1068
+S EPK++ A+ W AM EE+ + W+LVP D ++G++WVFR KL +
Sbjct: 709 VSYPEPKTVTAALKHPGWTGAMTEEIGNCSETQTWSLVPYKSDMHVLGSKWVFRTKLHAD 768
Query: 1069 GKVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFL 1128
G + + KAR+VA+G+ Q+EGIDY ET++PV R +R++L A + + QMDVK+AFL
Sbjct: 769 GTLNKLKARIVAKGFLQEEGIDYLETYSPVVRTPTVRLVLHLATALNWDIKQMDVKNAFL 828
Query: 1129 NGFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKI 1188
+G L E VY++QP GF++ KP+HV L K++YGLKQ+PRAW+D+ STFL+E GF K
Sbjct: 829 HGDLKETVYMTQPAGFVDPSKPDHVCLLHKSIYGLKQSPRAWFDKFSTFLLEFGFFCSKS 888
Query: 1189 DTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQI 1248
D +LF HN +L+++ +YVDD++ + + EF M+ MG+L +FLG+Q+
Sbjct: 889 DPSLFIYAHNNNLILLLLYVDDMVITGNSSQTLTSLLAALNKEFRMTDMGQLHYFLGIQV 948
Query: 1249 KQHSNGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGS 1308
++ NG+F+SQ+KY +D+L M + TP+ + +R + G
Sbjct: 949 QRQQNGLFMSQQKYAEDLLIAASMEHCTPLPTPLPVQLDRVPHQEELFSDPTYFRSIAGK 1008
Query: 1309 LLYLTASRPDIVFAVG-LCARF-QTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDL 1366
L YLT +RPDI FAV +C + Q + HL +KRI RY+ GT +G+ Y + S L
Sbjct: 1009 LQYLTLTRPDIQFAVNFVCQKMHQPTISDFHL--LKRILRYIKGTITMGISYSRDSPTLL 1066
Query: 1367 VAYCDADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQIL 1426
AY D+D+ K R+S G C F+G L+ WS +K T++ S+TEAEY S + S+IL
Sbjct: 1067 QAYSDSDWGNCKQTRRSVGGLCTFMGTNLVSWSSKKHPTVSRSSTEAEYKSLSDAASEIL 1126
Query: 1427 WVRNQLEDYSLRYTSVP-IYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIAL 1485
W+ L + + P ++CDN SA+ L+ NP H+R+KH +I HF+R+ V K + +
Sbjct: 1127 WLSTLLRELRIPLPDTPELFCDNLSAVYLTANPAFHARTKHFDIDFHFVRERVALKALVV 1186
Query: 1486 SFVDTENQLADIFTKPLVEDRFNFLKEKLLIMKNP 1520
+ Q+ADIFTK L + F L+ KL + +P
Sbjct: 1187 KHIPGSEQIADIFTKSLPYEAFIHLRGKLGVTLSP 1221
Score = 73.9 bits (180), Expect = 6e-13
Identities = 58/220 (26%), Positives = 93/220 (41%), Gaps = 49/220 (22%)
Query: 646 IENVQYVEGLKHNLLSISQLC-DSGFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLEL 704
+ +V + +LLS+S+L D V F+ + V ++ KL GS R LY L+
Sbjct: 233 LTDVLVCPSITKSLLSMSKLTQDFPCTVEFEYDGVRVNDKATKKLLLMGSNRDGLYCLK- 291
Query: 705 NDMPAESCFMSLEKDKW--IWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEACV 762
+D ++ F + ++ +WH+R GH + +
Sbjct: 292 DDKQFQAFFSTRQRSASDEVWHRRLGHPHPQIL--------------------------- 324
Query: 763 KGKQVKSSFKTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLK 822
+PLE +H DL+GP S+ G RY V +D +SRF+W+ LK
Sbjct: 325 ------------------QPLERVHCDLWGPTTITSVQGFRYYAVFIDHYSRFSWIYPLK 366
Query: 823 HKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFENASF 862
K + + F F K V+N+ I + D GGEF + F
Sbjct: 367 LKSDFYNIFLAFHKLVENQLSQKISVFQCDGGGEFVSHKF 406
Score = 45.4 bits (106), Expect = 2e-04
Identities = 24/56 (42%), Positives = 30/56 (52%)
Query: 871 FGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE 926
FG C+ KF+P S K +FLGY+ KGYR T + IS H+IFDE
Sbjct: 498 FGSACFPTLRDYAENKFNPCSLKCVFLGYNEKYKGYRCLYPPTGRLYISRHVIFDE 553
>At2g16000 putative retroelement pol polyprotein
Length = 1454
Score = 391 bits (1004), Expect = e-108
Identities = 222/667 (33%), Positives = 369/667 (55%), Gaps = 20/667 (2%)
Query: 871 FGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIF-DEYDE 929
FGC CY + ++ KF P+S +FLGY + KGY++ +L++ TV IS ++ F +E
Sbjct: 781 FGCLCYSSTSPKQRHKFQPRSRACLFLGYPSGYKGYKLMDLESNTVFISRNVQFHEEVFP 840
Query: 930 HSKPKENEDTEVPTLQNVPVQNTENTVEKEDDQNAQDQSFQSPPR--SWRM------VGD 981
+K +E + VPV + + + Q PP+ S R+ + D
Sbjct: 841 LAKNPGSESSLKLFTPMVPVSSGIISDTTHSPSSLPSQISDLPPQISSQRVRKPPAHLND 900
Query: 982 HPTDQIIGSTTDGVRTRLSFQD---NNMAMISQME----PKSINEAIIDDSWIKAMKEEL 1034
+ + + + + +S+ ++M I+ + P + EA W +A+ E+
Sbjct: 901 YHCNTMQSDHKYPISSTISYSKISPSHMCYINNITKIPIPTNYAEAQDTKEWCEAVDAEI 960
Query: 1035 SQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVVRNKARLVAQGYNQQEGIDYDET 1094
E+ W + + K +G +WVF K +G + R KARLVA+GY Q+EG+DY +T
Sbjct: 961 GAMEKTNTWEITTLPKGKKAVGCKWVFTLKFLADGNLERYKARLVAKGYTQKEGLDYTDT 1020
Query: 1095 FAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNEEVYVSQPPGFINKE----KP 1150
F+PVA++ I++LL +A K L Q+DV +AFLNG L EE+++ P G+ ++
Sbjct: 1021 FSPVAKMTTIKLLLKVSASKKWFLKQLDVSNAFLNGELEEEIFMKIPEGYAERKGIVLPS 1080
Query: 1151 NHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFRKTHNTDLLIVQVYVDD 1210
N V +L +++YGLKQA R W+ + S+ L+ GF + D TLF K ++ + +IV VYVDD
Sbjct: 1081 NVVLRLKRSIYGLKQASRQWFKKFSSSLLSLGFKKTHGDHTLFLKMYDGEFVIVLVYVDD 1140
Query: 1211 IIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHSNGIFISQEKYVKDILKKY 1270
I+ +T + + + F++ +G+L +FLGL++ + + GI I Q KY ++L+
Sbjct: 1141 IVIASTSEAAAAQLTEELDQRFKLRDLGDLKYFLGLEVARTTAGISICQRKYALELLQST 1200
Query: 1271 KMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSLLYLTASRPDIVFAVGLCARFQ 1330
M K +S PM P+ + KD+ ++YR ++G L+YLT +RPDI FAV +F
Sbjct: 1201 GMLACKPVSVPMIPNLKMRKDDGDLIEDIEQYRRIVGKLMYLTITRPDITFAVNKLCQFS 1260
Query: 1331 TCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYCDADYAGDKVERKSTSGSCQF 1390
+ + +HLTA R+ +Y+ GT GL+Y S L + D+D+A + R+ST+ F
Sbjct: 1261 SAPRTTHLTAAYRVLQYIKGTVGQGLFYSASSDLTLKGFADSDWASCQDSRRSTTSFTMF 1320
Query: 1391 LGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRNQLEDYSLRYTSVPIYCDNTS 1450
+G +LI W +KQ+T++ S+ EAEY + A +++W+ L +Y D+T+
Sbjct: 1321 VGDSLISWRSKKQHTVSRSSAEAEYRALALATCEMVWLFTLLVSLQASPPVPILYSDSTA 1380
Query: 1451 AINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVDTENQLADIFTKPLVEDRFNFL 1510
AI ++ NP+ H R+KHI++ H +R+ + + L V TE+Q+ADI TKPL +F L
Sbjct: 1381 AIYIATNPVFHERTKHIKLDCHTVRERLDNGELKLLHVRTEDQVADILTKPLFPYQFEHL 1440
Query: 1511 KEKLLIM 1517
K K+ I+
Sbjct: 1441 KSKMSIL 1447
Score = 138 bits (348), Expect = 2e-32
Identities = 86/268 (32%), Positives = 135/268 (50%), Gaps = 8/268 (2%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNNDKGKIRGKGTIGNLNSARIENVQYVEGL 655
+DSG + H++ DR F + + V KI G GT+ + ++NV ++
Sbjct: 433 IDSGATHHVSHDRSLFSSLDTSVLSAVNLPTGPTVKISGVGTLKLNDDILLKNVLFIPEF 492
Query: 656 KHNLLSISQLCDS-GFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAESCFM 714
+ NL+SIS L D G VIF N CE++ ++ G R NLY+L++ D +S +
Sbjct: 493 RLNLISISSLTDDIGSRVIFDKNSCEIQDLIKGRMLGQGRRVANLYLLDVGD---QSISV 549
Query: 715 SLEKDKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEACVKGKQVKSSFKTI 774
+ D +WH+R GH S++ + +S G + + C C KQ K SF T
Sbjct: 550 NAVVDISMWHRRLGHASLQRLDAISDS---LGTTRHKNKGSDFCHVCHLAKQRKLSFPTS 606
Query: 775 EFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESFEAFQNF 834
+ ++ +LLHID++GP ++ G +Y IVDD SR TW+ LK K E F F
Sbjct: 607 NKVC-KEIFDLLHIDVWGPFSVETVEGYKYFLTIVDDHSRATWMYLLKTKSEVLTVFPAF 665
Query: 835 CKRVQNEKGYNIITVRSDHGGEFENASF 862
++V+N+ + VRSD+ E + SF
Sbjct: 666 IQQVENQYKVKVKAVRSDNAPELKFTSF 693
>At4g16870 retrotransposon like protein
Length = 1474
Score = 388 bits (997), Expect = e-107
Identities = 206/512 (40%), Positives = 300/512 (58%), Gaps = 2/512 (0%)
Query: 1010 SQMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEG 1069
S EP ++ +A+ D W AM +E +RN W+LVP+ + + ++G +WVF+ K G
Sbjct: 962 SPSEPTNVTQALKDKKWRFAMSDEFDAQQRNHTWDLVPH-ESQLLVGCKWVFKLKYLPNG 1020
Query: 1070 KVVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLN 1129
+ + KARLVA+G+NQQ G+DY ETF+PV + IR++L A K ++ Q+DV +AFL
Sbjct: 1021 AIDKYKARLVAKGFNQQYGVDYAETFSPVIKSTTIRLVLDVAVKKDWEIKQLDVNNAFLQ 1080
Query: 1130 GFLNEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKID 1189
G L EEVY++QPPGFI+K++P HV +L KA+YGLKQAPRAWY L L GF D
Sbjct: 1081 GTLTEEVYMAQPPGFIDKDRPTHVCRLRKAIYGLKQAPRAWYMELKQHLFNIGFVNSLSD 1140
Query: 1190 TTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIK 1249
+LF H T + V VYVDDII + + + F + +L +FLG++
Sbjct: 1141 ASLFIYCHGTTFVYVLVYVDDIIVTGSDKSSIDAVLTSLAERFSIKDPTDLHYFLGIEAT 1200
Query: 1250 QHSNGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRGMIGSL 1309
+ G+ + Q KY+KD+L K+ M +AK + TP+ S L K EYR ++GSL
Sbjct: 1201 RTKQGLHLMQRKYIKDLLAKHNMADAKPVLTPLPTSPKLTLHGGTKLNDASEYRSVVGSL 1260
Query: 1310 LYLTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAY 1369
YL +RPDI +AV ++ E H A KR+ RYL GT+ G++ S +L A+
Sbjct: 1261 QYLAFTRPDIAYAVNRLSQLMPQPTEDHWQAAKRVLRYLAGTSTHGIFLDTTSPLNLHAF 1320
Query: 1370 CDADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVR 1429
DAD+AGD + ST+ +LG+ I WS +KQ +A S+TE+EY + A+ S++ W+
Sbjct: 1321 SDADWAGDSDDYVSTNAYVIYLGKNPISWSSKKQRGVARSSTESEYRAVANAASEVKWLC 1380
Query: 1430 NQLEDYSLRYTSVP-IYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFV 1488
+ L +R P I+CDN A L NP+ HSR KHI I +HF+R+ +Q + +S V
Sbjct: 1381 SLLSKLHIRLPIRPSIFCDNIGATYLCANPVFHSRMKHIAIDYHFVRNMIQSGALRVSHV 1440
Query: 1489 DTENQLADIFTKPLVEDRFNFLKEKLLIMKNP 1520
T +QLAD TKPL F + K+ + + P
Sbjct: 1441 STRDQLADALTKPLSRAHFQSARFKIGVRQLP 1472
Score = 90.1 bits (222), Expect = 9e-18
Identities = 76/270 (28%), Positives = 119/270 (43%), Gaps = 14/270 (5%)
Query: 596 LDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNNDKGKIRGKGTI---GNLNSARIENVQYV 652
LDSG + H+T D + + +G V + KI G+ N + V YV
Sbjct: 335 LDSGATHHITSDLNALALHQPYNGDDVMIADGTSLKITKTGSTFLPSNARDLTLNKVLYV 394
Query: 653 EGLKHNLLSISQLCDSG-FEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAES 711
++ NL+S+ +LC++ V F P +V+ ++ L G + LY + + A +
Sbjct: 395 PDIQKNLVSVYRLCNTNQVSVEFFPASFQVKDLNTGTLLLQGRTKDELYEWPVTNPKATA 454
Query: 712 CFMSLEKDKWI--WHKRAGHISMKTIAKLSQLDLVRGLP-KISFEKDRICEACVKGKQVK 768
F + + WH R GH S L+ L LP +S C C K K
Sbjct: 455 LFTTPSPKTTLSSWHSRLGHPSSSI---LNTLISKFSLPVSVSASNKLACSDCFINKSHK 511
Query: 769 SSFKTIEFISTQKPLELLHIDLF-GPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDES 827
F +I I + PLE + D++ P+ S +Y V+VD +R+TW+ L+ K +
Sbjct: 512 LPF-SISSIKSTSPLEYIFSDVWMSPI--LSPDNYKYYLVLVDHHTRYTWLYPLQQKSQV 568
Query: 828 FEAFQNFCKRVQNEKGYNIITVRSDHGGEF 857
F F V+N I T+ SD+GGEF
Sbjct: 569 KSTFIAFKALVENRFQAKIRTLYSDNGGEF 598
Score = 45.4 bits (106), Expect = 2e-04
Identities = 19/57 (33%), Positives = 32/57 (55%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE 926
VFGC C+ K + +S + +FLGYS T Y ++++ + + S H++FDE
Sbjct: 691 VFGCLCFPWLRPYTHNKLEERSRRCVFLGYSLTQTAYLCFDVEHKRLYTSRHVVFDE 747
Score = 45.1 bits (105), Expect = 3e-04
Identities = 50/237 (21%), Positives = 88/237 (37%), Gaps = 30/237 (12%)
Query: 96 VDECTNAKGVWDTLKIHHEGTS--HVKETRIDIGVRKFEVFEMSENETIDEMYARFTTIV 153
V T A +W TL + +S H+K+ R I K +TIDE TT++
Sbjct: 108 VSRATKASQIWKTLTNTYAKSSYDHIKQLRTQIKQLK------KGTKTIDEYVLSHTTLL 161
Query: 154 NEMRSLGKAYSTHDRIRKILRCLPSMWRPMVTAITQAKDLKSMNLEDLIGSLRAHEVVLQ 213
+++ LGK +++ +IL LP ++ +V I + KD + ++ ++ L HE
Sbjct: 162 DQLAILGKPMEHEEQVERILEGLPEDYKTVVDQI-EGKD-NTPSITEIHERLINHE---- 215
Query: 214 GDKPVKKVRTLALKASQQTSSVDDEDVQEPQELEEVHEEEAEDELALISKRIQRMMLRRN 273
K + T AL +S S + + ++ + + + Q ++
Sbjct: 216 ----AKLLSTAALSSSSLPMSANVAQQRHHNNNRNNNQNKNRTQGNTYTNNWQPSANNKS 271
Query: 274 QIRKKFPKTNISIKTEADKSQVTCYGCNKTGHFKNECPDIKKVQRKPPFKKKAMITW 330
R P C CN GH CP ++ +Q W
Sbjct: 272 GQRPFKPYLG------------KCQICNVQGHSARRCPQLQAMQPSSSSSASTFTPW 316
>At4g10690 retrotransposon like protein
Length = 1515
Score = 387 bits (994), Expect = e-107
Identities = 204/512 (39%), Positives = 304/512 (58%), Gaps = 7/512 (1%)
Query: 1013 EPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGKVV 1072
EPKS+ EA+ D+ W AM EE+ W+LVP ++G +WVF+ KL+ +G +
Sbjct: 919 EPKSVKEALKDEGWTNAMGEEMGTMHETDTWDLVPPEMVDRLLGCKWVFKTKLNSDGSLD 978
Query: 1073 RNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFL 1132
R KARLVA+GY Q+EG+DY ET++PV R +R +L A L Q+DVK+AFL+ L
Sbjct: 979 RLKARLVARGYEQEEGVDYVETYSPVVRSATVRSILHVATINKWSLKQLDVKNAFLHDEL 1038
Query: 1133 NEEVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTL 1192
E V+++QPPGF + +P++V KL KA+Y LKQAPRAW+D+ S++L++ GF D +L
Sbjct: 1039 KETVFMTQPPGFEDPSRPDYVCKLKKAIYDLKQAPRAWFDKFSSYLLKYGFICSFSDPSL 1098
Query: 1193 FRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHS 1252
F D++ + +YVDD+I + ++ N++ +EF M MG L +FLG+Q H+
Sbjct: 1099 FVYLKGRDVMFLLLYVDDMILTGNNDVLLQQLLNILSTEFRMKDMGALHYFLGIQAHYHN 1158
Query: 1253 NGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLD-KDESGKSISEKEY-RGMIGSLL 1310
+G+F+SQEKY D+L M++ M TP+ LD + K E Y R + G L
Sbjct: 1159 DGLFLSQEKYTSDLLVNAGMSDCSSMPTPL----QLDLLQGNNKPFPEPTYFRRLAGKLQ 1214
Query: 1311 YLTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDLVAYC 1370
YLT +RPDI FAV + S +KRI YL GT +G+ + L Y
Sbjct: 1215 YLTLTRPDIQFAVNFVCQKMHAPTMSDFHLLKRILHYLKGTMTMGINLSSNTDSVLRCYS 1274
Query: 1371 DADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQILWVRN 1430
D+D+AG K R+ST G C FLG +I WS ++ T++ S+TEAEY + + S++ W+
Sbjct: 1275 DSDWAGCKDTRRSTGGFCTFLGYNIISWSAKRHPTVSKSSTEAEYRTLSFAASEVSWIGF 1334
Query: 1431 QLEDYSLRYTSVP-IYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALSFVD 1489
L++ L +P +YCDN SA+ LS NP HSRSKH ++ ++++R+ V + + +
Sbjct: 1335 LLQEIGLPQQQIPEMYCDNLSAVYLSANPALHSRSKHFQVDYYYVRERVALGALTVKHIP 1394
Query: 1490 TENQLADIFTKPLVEDRFNFLKEKLLIMKNPN 1521
QLADIFTK L + F L+ KL ++ P+
Sbjct: 1395 ASQQLADIFTKSLPQAPFCDLRFKLGVVLPPD 1426
Score = 121 bits (303), Expect = 4e-27
Identities = 84/274 (30%), Positives = 138/274 (49%), Gaps = 16/274 (5%)
Query: 597 DSGCSRHMTGDRDCFLTFEKKDGG-LVTFGNNDKGKIRGKGTIGNLNSAR----IENVQY 651
DS + H+T D + G V GN D I GTI LN ++ +E+V
Sbjct: 327 DSAATAHITNTTDGLQNSQTYSGDDSVIVGNGDFLPITHIGTIP-LNISQGTLPLEDVLV 385
Query: 652 VEGLKHNLLSISQLCDS-GFEVIFKPNICEVRQTSSNKLFFSGSRRKNLYVLELNDMPAE 710
G+ +LLS+S+L D F + ++ + +L G++ K LYVL+ D+P +
Sbjct: 386 CPGITKSLLSVSKLTDDYPCSFTFDSDSVVIKDKRTQQLLTQGNKHKGLYVLK--DVPFQ 443
Query: 711 SCFMSLEK--DKWIWHKRAGHISMKTIAKLSQLDLVRGLPKISFEKDRICEACVKGKQVK 768
+ + + ++ D +WH+R GH + + + L + + ++ +CEAC GK +
Sbjct: 444 TYYSTRQQSSDDEVWHQRLGHPNKEVLQHLIKTKAI----VVNKTSSNMCEACQMGKVCR 499
Query: 769 SSFKTIEFISTQKPLELLHIDLFGPVQTASLTGKRYGFVIVDDFSRFTWVLFLKHKDESF 828
F EF+S+ +PLE +H DL+GP S G +Y + +D++SRFTW LK K + F
Sbjct: 500 LPFVASEFVSS-RPLERIHCDLWGPAPVTSAQGFQYYVIFIDNYSRFTWFYPLKLKSDFF 558
Query: 829 EAFQNFCKRVQNEKGYNIITVRSDHGGEFENASF 862
F F + V+N+ + I + D GGEF + F
Sbjct: 559 SVFVLFQQLVENQYQHKIAMFQCDGGGEFVSYKF 592
Score = 51.2 bits (121), Expect = 4e-06
Identities = 26/57 (45%), Positives = 31/57 (53%)
Query: 870 VFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDE 926
VFG CY KFDPKS +FLGY+ KGYR + T V I H++FDE
Sbjct: 683 VFGSACYPYLRPYAKNKFDPKSLLCVFLGYNNKYKGYRCLHPPTGKVYICRHVLFDE 739
>At4g10990 putative retrotransposon polyprotein
Length = 1203
Score = 383 bits (983), Expect = e-106
Identities = 201/499 (40%), Positives = 297/499 (59%), Gaps = 9/499 (1%)
Query: 1011 QMEPKSINEAIIDDSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEGK 1070
+ EPK+ +A+ + W +A EEL E+NK W + + K ++G +WVF K + +G
Sbjct: 522 ETEPKAFTQAMKSEKWTRAANEELHALEQNKTWIVESLTEGKNVVGCKWVFTIKYNPDGS 581
Query: 1071 VVRNKARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNG 1130
+ R KARLVAQG+ QQEGIDY ETF+PVA+ ++++LL AA L QMDV +AFL+G
Sbjct: 582 IERYKARLVAQGFTQQEGIDYMETFSPVAKFGSVKLLLGLAAATGWSLTQMDVSNAFLHG 641
Query: 1131 FLNEEVYVSQPPGFINK------EKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFS 1184
L+EE+Y+S P G+ KP V +L K+LYGLKQA R WY RLS+ + F
Sbjct: 642 ELDEEIYMSLPQGYTPPTGISLPSKP--VCRLLKSLYGLKQASRQWYKRLSSVFLGANFI 699
Query: 1185 RGKIDTTLFRKTHNTDLLIVQVYVDDIIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFL 1244
+ D T+F K T +++V VYVDD++ + E L++SEF++ +G FFL
Sbjct: 700 QSPADNTMFVKVSCTSIIVVLVYVDDLMIASNDSSAVENLKELLRSEFKIKDLGPARFFL 759
Query: 1245 GLQIKQHSNGIFISQEKYVKDILKKYKMNEAKIMSTPMHPSSSLDKDESGKSISEKEYRG 1304
GL+I + S GI + Q KY +++L+ ++ K S PM P+ L K+ + YR
Sbjct: 760 GLEIARSSEGISVCQRKYAQNLLEDVGLSGCKPSSIPMDPNLHLTKEMGTLLPNATSYRE 819
Query: 1305 MIGSLLYLTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSF 1364
++G LLYL +RPDI FAV ++F + + H+ A ++ RYL G GL Y S
Sbjct: 820 LVGRLLYLCITRPDITFAVHTLSQFLSAPTDIHMQAAHKVLRYLKGNPGQGLMYSASSEL 879
Query: 1365 DLVAYCDADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQ 1424
L + DAD+ K R+S +G C +LG +LI W +KQ+ ++ S+TE+EY S A +
Sbjct: 880 CLNGFSDADWGTCKDSRRSVTGFCIYLGTSLITWKSKKQSVVSRSSTESEYRSLAQATCE 939
Query: 1425 ILWVRNQLEDYSLRYT-SVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNI 1483
I+W++ L+D + T ++CDN SA++L+ NP+ H R+KHIEI H +RD ++ +
Sbjct: 940 IIWLQQLLKDLHVTMTCPAKLFCDNKSALHLATNPVFHERTKHIEIDCHTVRDQIKAGKL 999
Query: 1484 ALSFVDTENQLADIFTKPL 1502
V T NQLADI TKPL
Sbjct: 1000 KTLHVPTGNQLADILTKPL 1018
Score = 58.2 bits (139), Expect = 4e-08
Identities = 45/167 (26%), Positives = 72/167 (42%), Gaps = 9/167 (5%)
Query: 571 HKRQNVTFRKDLLEEL-TFKDPTLHGLDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNNDK 629
++ N+TF+ L L +DSG S H+ D F G VT N +
Sbjct: 75 YENNNLTFQNHTLSSLQNVLSSDAWIIDSGASSHVCSDLTMFRELIHVSGVTVTLPNGTR 134
Query: 630 GKIRGKGTIGNLNSARIENVQYVEGLKHNLLSISQLCDS-GFEVIFKPNICEVRQTSSNK 688
I GTI ++ + NV V K NL+S+ L + + F + C +++ +
Sbjct: 135 VAITHTGTICITSTLILHNVLLVPDFKFNLISVCCLVKTLSYSAHFFADCCYIQELTRGL 194
Query: 689 LFFSGSRRKNLYVLELN------DMPAESCFM-SLEKDKWIWHKRAG 728
+ G NLY+LE +PA S F +++ D +WH+R G
Sbjct: 195 MIGRGKTYNNLYILETQRTSFSPSLPAASSFTGTVQDDCLLWHQRLG 241
Score = 30.4 bits (67), Expect = 8.2
Identities = 17/60 (28%), Positives = 26/60 (43%)
Query: 853 HGGEFENASFKTFFDENVFGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLK 912
H + N F T D+ + Y++ NK ++ P K IF Y+ K R N+K
Sbjct: 244 HYLHYRNLYFLTLVDDCTRTTWVYMMKNKSEVSNIFPVFVKLIFTQYNAKIKAIRSDNVK 303
>At1g37110
Length = 1356
Score = 362 bits (929), Expect = e-100
Identities = 219/692 (31%), Positives = 362/692 (51%), Gaps = 48/692 (6%)
Query: 871 FGCYCYILNNKEKLGKFDPKSDKAIFLGYSTTSKGYRVYNLKTQTVEISMHIIFDEY--- 927
FG Y+ ++ GK P++ K FLGY +KGY+V+ L+ + IS +++F E
Sbjct: 667 FGSIAYVHQDQ---GKLKPRALKGFFLGYPAGTKGYKVWLLEEEKCVISRNVVFQESVVY 723
Query: 928 -------DEHSKPKENEDTEVPTLQNV------------------PVQNTENTVEKEDDQ 962
D+ + E T QN P+ E + + E++
Sbjct: 724 RDLKVKEDDTDNLNQKETTSSEVEQNKFAEASGSGGVIQLQSDSEPITEGEQSSDSEEEV 783
Query: 963 NAQDQSFQSPPRS----WRMVGDHPTDQIIGSTTDGVRTRLSFQDNNMAMISQMEPKSIN 1018
+++ ++P R+ +++ D I T + ++F + EP+S
Sbjct: 784 EYSEKTQETPKRTGLTTYKLARDRVRRNINPPTRFTEESSVTFALVVVENCIVQEPQSYQ 843
Query: 1019 EAIID---DSWIKAMKEELSQFERNKVWNLVPNNQDKTIIGTRWVFRNKLDEEG-KVVRN 1074
EA+ + W A +E+ +N W+LV +D+ IIG RW+F+ K G + R
Sbjct: 844 EAMESQDCEKWDMATHDEMDSLMKNGTWDLVDKPKDRKIIGCRWLFKLKSGIPGVEPTRF 903
Query: 1075 KARLVAQGYNQQEGIDYDETFAPVARLEAIRILLAYAAHKSIKLFQMDVKSAFLNGFLNE 1134
KARLVA+GY Q+EG+DY E FAPV + +IRIL++ K ++L QMDVK+ FL+G L E
Sbjct: 904 KARLVAKGYTQREGVDYQEIFAPVVKHVSIRILMSLVVDKDLELEQMDVKTTFLHGDLEE 963
Query: 1135 EVYVSQPPGFINKEKPNHVFKLTKALYGLKQAPRAWYDRLSTFLIENGFSRGKIDTTLFR 1194
E+Y+ QP GF++ N V +L K+LYGLKQ+PR W R F+ F R + D ++
Sbjct: 964 ELYMEQPEGFVSDSSENKVCRLKKSLYGLKQSPRQWNKRFDRFMSSQQFIRSEHDACVYV 1023
Query: 1195 K-THNTDLLIVQVYVDD-IIFGATKIKMCEEFSNLMQSEFEMSMMGELGFFLGLQIKQHS 1252
K D + + +YVDD +I GA+K ++ + +EFEM MG LG+ I +
Sbjct: 1024 KHVSEHDFIYLLLYVDDMLIAGASKAEI-NRVKEQLSTEFEMKDMGGASRILGIDIYRDR 1082
Query: 1253 NG--IFISQEKYVKDILKKYKMNEAKIMSTPMHPS---SSLDKDESGKSISEKEYRGMIG 1307
G + +SQE Y++ +L ++ M+ AK+ + P+ +++ +++ Y +G
Sbjct: 1083 KGGVLKLSQEIYIRKVLDRFNMSGAKMTNAPVGAHFKLAAVREEDECVDTDVVPYSSAVG 1142
Query: 1308 SLLY-LTASRPDIVFAVGLCARFQTCAKESHLTAVKRIFRYLVGTTDLGLWYRKGSSFDL 1366
S++Y + +RPD+ +A+ L +R+ + H AVK + RYL G DL L + K F +
Sbjct: 1143 SIMYAMLGTRPDLAYAICLISRYMSKPGSMHWEAVKWVMRYLKGAQDLNLVFTKEKDFTV 1202
Query: 1367 VAYCDADYAGDKVERKSTSGSCQFLGQALIGWSCRKQNTIALSTTEAEYVSAASCCSQIL 1426
YCD++YA D R+S SG +G + W Q +A+STTEAEY++ A + +
Sbjct: 1203 TGYCDSNYAADLDRRRSISGYVFTIGGNTVSWKASLQPVVAMSTTEAEYIALAEAAKEAM 1262
Query: 1427 WVRNQLEDYSLRYTSVPIYCDNTSAINLSKNPIQHSRSKHIEIKHHFIRDHVQKKNIALS 1486
W++ L+D ++ V I+CD+ SAI LSKN + H R+KHI+++ ++IRD V+ ++ +
Sbjct: 1263 WIKGLLQDMGMQQDKVKIWCDSQSAICLSKNSVYHERTKHIDVRFNYIRDVVESGDVDVL 1322
Query: 1487 FVDTENQLADIFTKPLVEDRFNFLKEKLLIMK 1518
+ T D TK + ++F L +MK
Sbjct: 1323 KIHTSRNPVDALTKCIPVNKFKSALGVLKLMK 1354
Score = 162 bits (411), Expect = 1e-39
Identities = 143/490 (29%), Positives = 227/490 (46%), Gaps = 63/490 (12%)
Query: 392 IKELSEIKEEKEILQNKYDESRKTIKILQD---SHFDMSEKQREINRKQKGIMSVPSEVQ 448
+ E I E L+ + DE + I IL SH + + N K + V S +
Sbjct: 145 VDEFLRIVAELGSLEIQVDEEVQAILILNSLPASHIQLKHTLKYGN-KTLTVQDVTSSAK 203
Query: 449 KENILLKKEVETLKKDLTGFIKSTETFQNIVGSQNKSTKKSGLGFKDPSKIIGSFVPKAK 508
L + V+ L K + +TE + +V ++ +K G G G +K
Sbjct: 204 SLERELAEAVD-LDKGQAAVLYTTERGRPLV----RNNQKGGQGK-------GRSRSNSK 251
Query: 509 IRVKCCFCDKYGHNESVCHVKKKFIKQNNLNLSSERSHLNRSESSQKAEKAKKTCFYCNK 568
+V C +C K GH + C+ +KK + ES + E T
Sbjct: 252 TKVPCWYCKKEGHVKKDCYSRKK-----------------KMESEGQGEAGVIT------ 288
Query: 569 SDHKRQNVTFRKDL-LEELTFKDPTLHGLDSGCSRHMTGDRDCFLTFEKKDGGLVTFGNN 627
+ + F + L + E KD L LDSGC+ HMT RD F++F++K + G++
Sbjct: 289 -----EKLVFSEALSVNEQMVKD--LWILDSGCTSHMTSRRDWFISFQEKGNTTILLGDD 341
Query: 628 DKGKIRGKGTI------GNLNSARIENVQYVEGLKHNLLSISQLCDSGFEVIFKPNICEV 681
+ +G+GTI G + +ENV+YV L+ NL+S L G+ + +V
Sbjct: 342 HSVESQGQGTIRIDTHGGTIKI--LENVKYVPHLRRNLISTGTLDKLGYR--HEGGEGKV 397
Query: 682 RQTSSNKLFFSGSRRKNLYVLELNDMPAESCFMSLEKDKW-IWHKRAGHISMKTIAKLSQ 740
R +NK GS LYVL+ + + +E C +K K +WH R GH+SM + L+
Sbjct: 398 RYFKNNKTALRGSLSNGLYVLDGSTVMSELCNAETDKVKTALWHSRLGHMSMNNLKVLAG 457
Query: 741 LDLVRGLPKISFEKDRICEACVKGKQVKSSFKTIEFISTQKPLELLHIDLFG-PVQTASL 799
L+ + + CE CV GK K SF + S + L +H DL+G P T S+
Sbjct: 458 KGLI---DRKEINELEFCEHCVMGKSKKVSFNVGKHTS-EDALSYVHADLWGSPNVTPSI 513
Query: 800 TGKRYGFVIVDDFSRFTWVLFLKHKDESFEAFQNFCKRVQNEKGYNIITVRSDHGGEFEN 859
+GK+Y I+DD +R W+ FLK KDE+F+ F + V+N+ + +R+D+G EF N
Sbjct: 514 SGKQYFLSIIDDKTRKVWLYFLKSKDETFDKFCEWKSLVENQVNKKVKCLRTDNGLEFCN 573
Query: 860 ASFKTFFDEN 869
+ F ++ E+
Sbjct: 574 SRFDSYCKEH 583
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.386
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 34,619,534
Number of Sequences: 26719
Number of extensions: 1559698
Number of successful extensions: 7844
Number of sequences better than 10.0: 287
Number of HSP's better than 10.0 without gapping: 138
Number of HSP's successfully gapped in prelim test: 153
Number of HSP's that attempted gapping in prelim test: 6630
Number of HSP's gapped (non-prelim): 899
length of query: 1523
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1411
effective length of database: 8,326,068
effective search space: 11748081948
effective search space used: 11748081948
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC144724.6


