

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144724.10 - phase: 0 /pseudo
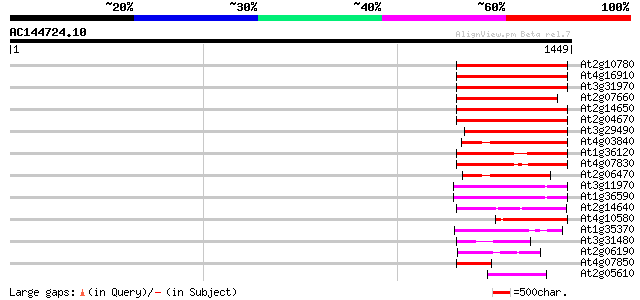
(1449 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g10780 pseudogene 330 3e-90
At4g16910 retrotransposon like protein 322 1e-87
At3g31970 hypothetical protein 314 3e-85
At2g07660 putative retroelement pol polyprotein 313 6e-85
At2g14650 putative retroelement pol polyprotein 304 3e-82
At2g04670 putative retroelement pol polyprotein 300 4e-81
At3g29490 hypothetical protein 270 3e-72
At4g03840 putative transposon protein 265 1e-70
At1g36120 putative reverse transcriptase gb|AAD22339.1 264 2e-70
At4g07830 putative reverse transcriptase 256 5e-68
At2g06470 putative retroelement pol polyprotein 233 6e-61
At3g11970 hypothetical protein 177 5e-44
At1g36590 hypothetical protein 177 5e-44
At2g14640 putative retroelement pol polyprotein 153 6e-37
At4g10580 putative reverse-transcriptase -like protein 144 3e-34
At1g35370 hypothetical protein 140 4e-33
At3g31480 hypothetical protein 137 5e-32
At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein 93 1e-18
At4g07850 putative polyprotein 91 6e-18
At2g05610 putative retroelement pol polyprotein 87 5e-17
>At2g10780 pseudogene
Length = 1611
Score = 330 bits (847), Expect = 3e-90
Identities = 155/285 (54%), Positives = 210/285 (73%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD RFTS+FWK+ Q+ALG+++ LS+AYHPQTD QSERTIQ+LED+LR CVL+ GG W
Sbjct: 1298 VSDRDTRFTSKFWKAFQKALGTRVNLSTAYHPQTDEQSERTIQTLEDMLRACVLDWGGNW 1357
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
+ +L L+EF YNNS+ +SIGM+P+EALYGR CRT LCW G R + GP IV +TTE++K
Sbjct: 1358 EKYLRLVEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERMK 1417
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
++ K+K +Q RQKSY +KRRK+LEFQ GD V+L+ G GR KKL+PR++GPY+
Sbjct: 1418 FLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYK 1477
Query: 1335 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLR 1394
+ ERVG VAY++ LPP L+ H+VFHVSQLRK + D ++ +++N+TVE P+R
Sbjct: 1478 VIERVGAVAYKLDLPPKLNAFHNVFHVSQLRKCLSDQEESVEDIPPGLKENMTVEAWPVR 1537
Query: 1395 IDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLESYPKLF 1439
I DR K RGK L++V+W+ E TWE E+KM ++P+ F
Sbjct: 1538 IMDRMTKGTRGKARDLLKVLWNCRGREEYTWETENKMKANFPEWF 1582
>At4g16910 retrotransposon like protein
Length = 687
Score = 322 bits (825), Expect = 1e-87
Identities = 150/285 (52%), Positives = 207/285 (72%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD RFTS+FWK Q+ LG+++ LS+AYHPQTDGQSERTIQ+LED+LR CVL+ GG W
Sbjct: 382 VSDRDTRFTSKFWKPFQKVLGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNW 441
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
+ +L L+EF YNNS+ +SIGM+P+EALYGR RT LCW G R + GP +V +TT+K+K
Sbjct: 442 EKYLRLVEFAYNNSFQASIGMSPYEALYGRAGRTPLCWTPVGERRLFGPAVVDETTKKMK 501
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
++ K+K +Q RQKSY +KRRK+LEFQ GD V+L+ G GR KKL PR++GPY+
Sbjct: 502 FLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLRPRYVGPYK 561
Query: 1335 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLR 1394
+ ERVG VAY++ LPP L H+VFHVSQLRK + + ++ +++N+TVE P+R
Sbjct: 562 VIERVGAVAYKLDLPPKLDAFHNVFHVSQLRKCLSEQEESMEDVPPGLKENMTVEAWPVR 621
Query: 1395 IDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLESYPKLF 1439
I D+ K RGK + L++++W+ E TWE E+KM ++P+ F
Sbjct: 622 IMDQMKKGTRGKSMDLLKILWNCGGREEYTWETETKMKANFPEWF 666
>At3g31970 hypothetical protein
Length = 1329
Score = 314 bits (804), Expect = 3e-85
Identities = 150/287 (52%), Positives = 211/287 (73%), Gaps = 4/287 (1%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD +FTS FW++ Q +G+K+++S+AYHPQTDGQSERTIQ+LED+LR+CVL++GG W
Sbjct: 1039 VSDRDSKFTSAFWRAFQGEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDRGGHW 1098
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
HL L+EF YNNSY +SI MAPFEALYGR CRT LCW + G R + G + V +TTE+++
Sbjct: 1099 ADHLSLVEFAYNNSYQASIRMAPFEALYGRPCRTPLCWTQVGERSIYGADYVLETTERIR 1158
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+++ MK +Q RQ+SY DKRR++LEF+ GD V+L++ + G R++ KLTPR++GP++
Sbjct: 1159 VLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLTPRYMGPFR 1218
Query: 1335 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLR 1394
I ERVG VAYR+ LP + H VFHVS LRK + V+ ++ N+T+E P+R
Sbjct: 1219 IVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKILEDLQPNMTLEARPVR 1278
Query: 1395 IDDRKVKTLRGKEIPLVRVVW--DGATGESLTWELESKMLESYPKLF 1439
I +R++K LR K+IPL++V+W DG T E TWE E++M S+ K F
Sbjct: 1279 ILERRIKELRRKKIPLIKVLWNCDGVTEE--TWEPEARMKASFKKWF 1323
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 313 bits (801), Expect = 6e-85
Identities = 147/260 (56%), Positives = 196/260 (74%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD RFTS+FW + Q+ALG+++ LS+AYHPQTDGQSERTIQ+LED+LR CVL+ GG W
Sbjct: 690 VSDRDTRFTSKFWNAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNW 749
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
+ +L LIEF YNNS+ +SIGM+P+EALYGR CRT LCW G R + GP IV +TTE++K
Sbjct: 750 EKYLRLIEFAYNNSFQASIGMSPYEALYGRACRTPLCWTPVGERRLFGPTIVDETTERMK 809
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
++ K+K +Q RQKSY +KRRK+LEFQ GD V+L+ G GR KKL+PR++GPY+
Sbjct: 810 FLKIKLKEAQDRQKSYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYK 869
Query: 1335 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLR 1394
+ ERVG VAY++ LPP L+ H+VFHVSQLRKY+ D ++ +++N+TVE P+R
Sbjct: 870 VIERVGAVAYKLDLPPKLNVFHNVFHVSQLRKYLSDQEESVEDIPPGLKENMTVEAWPVR 929
Query: 1395 IDDRKVKTLRGKEIPLVRVV 1414
I DR K RGK L++V+
Sbjct: 930 IMDRMSKGTRGKSRDLLKVL 949
>At2g14650 putative retroelement pol polyprotein
Length = 1328
Score = 304 bits (778), Expect = 3e-82
Identities = 146/287 (50%), Positives = 208/287 (71%), Gaps = 4/287 (1%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD +FTS FW++ Q +G+K+++S+AYHPQT GQSERTIQ+LED+LR+CVL+ GG W
Sbjct: 1041 VSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTYGQSERTIQTLEDMLRMCVLDWGGHW 1100
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
HL L+EF YNNSY +SIGMAPFEALY R CRT LC + G R + G + VQ+TTE+++
Sbjct: 1101 ADHLSLVEFAYNNSYPASIGMAPFEALYERPCRTPLCLTQVGERSIYGADYVQETTERIR 1160
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+++ MK +Q RQ+SY DKRR++LEF+ GD V+L++ + G R++ KL+PR++GP++
Sbjct: 1161 VLKLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSISETKLSPRYMGPFR 1220
Query: 1335 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLR 1394
I ERVG VAYR+ LP + H VFHVS LRK + V+ ++ N+T+E P+R
Sbjct: 1221 IVERVGPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVR 1280
Query: 1395 IDDRKVKTLRGKEIPLVRVVW--DGATGESLTWELESKMLESYPKLF 1439
+ +R++K LR K+IPL++V+W DG T E TWE E++M + K F
Sbjct: 1281 VLERRIKELRRKKIPLIKVLWDCDGVTKE--TWEPEARMKARFKKWF 1325
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 300 bits (768), Expect = 4e-81
Identities = 144/287 (50%), Positives = 207/287 (71%), Gaps = 4/287 (1%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD +FT FW++ Q +G+K+++S+AYHPQTDGQSERTIQ+LED+LR+CVL+ GG W
Sbjct: 1121 VSDRDSKFTFAFWRAFQAKMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWGGHW 1180
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
HL L+EF YNNSY +SIGMAPFEALYGR C T L W + R + G + VQ+TTE+++
Sbjct: 1181 ADHLSLVEFAYNNSYQASIGMAPFEALYGRPCWTPLRWTQVEERSIYGADYVQETTERIR 1240
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+++ MK +Q+RQ+SY DKRR++LEF+ GD V+L++ + G R++ KL+PR++GP++
Sbjct: 1241 VLKLNMKEAQARQRSYADKRRRELEFEVGDRVYLKMAMLRGPNRSILETKLSPRYMGPFR 1300
Query: 1335 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLR 1394
I ERVG VAYR+ LP + H VFHV LRK + V+ ++ N+T+E P+R
Sbjct: 1301 IVERVGPVAYRLELPDVMRAFHKVFHVLMLRKCLHKDDEVLVKIPEDLQPNMTLEARPVR 1360
Query: 1395 IDDRKVKTLRGKEIPLVRVVW--DGATGESLTWELESKMLESYPKLF 1439
+ +R++K LR K+IPL++V+W DG T E TWE E++M + K F
Sbjct: 1361 VLERRIKELRRKKIPLIKVLWDCDGVTEE--TWEPEARMKARFKKWF 1405
>At3g29490 hypothetical protein
Length = 438
Score = 270 bits (691), Expect = 3e-72
Identities = 130/268 (48%), Positives = 189/268 (70%), Gaps = 4/268 (1%)
Query: 1174 LGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNSYHSSI 1233
+G+K+++S+ YHPQTDGQ ERTIQ+LED+LR+CVL+ GG W HL L+EF YNNSY + I
Sbjct: 70 MGTKVQMSTPYHPQTDGQFERTIQTLEDMLRMCVLDWGGHWADHLSLVEFAYNNSYQAGI 129
Query: 1234 GMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVKMIQEKMKASQSRQKSYHDK 1293
GMAPFEALYGR CRT LCW + G R + G + VQ+TTE++++++ MK +Q RQ SY DK
Sbjct: 130 GMAPFEALYGRPCRTPLCWTQVGERSIYGADYVQETTERIRVLKLNMKEAQDRQWSYADK 189
Query: 1294 RRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQISERVGTVAYRVGLPPHLS 1353
RR++LEF+ GD V+L++ + G R++ KL+ R++GP++I ERVG VAY + LP +
Sbjct: 190 RRRELEFEVGDRVYLKMAMLRGPNRSISETKLSLRYMGPFRIVERVGPVAYMLELPDVMR 249
Query: 1354 NLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLRIDDRKVKTLRGKEIPLVRV 1413
H VFHVS LRK + V+ ++ N+T+E +R+ +R++K L+ K+I L++V
Sbjct: 250 AFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARQVRVLERRIKELQRKKISLIKV 309
Query: 1414 VW--DGATGESLTWELESKMLESYPKLF 1439
+W DG T E TW+ E++M + K F
Sbjct: 310 LWDCDGVTEE--TWQPEARMKARFKKWF 335
>At4g03840 putative transposon protein
Length = 973
Score = 265 bits (677), Expect = 1e-70
Identities = 131/274 (47%), Positives = 181/274 (65%), Gaps = 19/274 (6%)
Query: 1166 FWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTY 1225
FWK+ Q+ALG+++ LS+AYHPQTDGQSERTIQ+LED+LR C L+ GG W+ +L L
Sbjct: 690 FWKAFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACALDWGGNWEKYLRL----- 744
Query: 1226 NNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVKMIQEKMKASQS 1285
ALYGR CRT LCW G R + GP IV +TTE++K ++ K+K +Q
Sbjct: 745 --------------ALYGRACRTPLCWTPVGERRLFGPIIVDETTERMKFLKIKLKEAQD 790
Query: 1286 RQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQISERVGTVAYR 1345
RQKSY +KRRK+LEFQ D V+L+ G GR KKL+PR++GPY++ ERVG VAY+
Sbjct: 791 RQKSYANKRRKELEFQVEDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYK 850
Query: 1346 VGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLRIDDRKVKTLRG 1405
+ LPP L+ H+VFHVSQLRK + + ++ +++N+TVE P++I DR K RG
Sbjct: 851 LDLPPKLNAFHNVFHVSQLRKCLSNQEESVEDVPPGLKENMTVEAWPVQIMDRMTKGTRG 910
Query: 1406 KEIPLVRVVWDGATGESLTWELESKMLESYPKLF 1439
K L++V+W+ E TWE E+KM ++ + F
Sbjct: 911 KSRDLLKVLWNCGGREQYTWETENKMKANFSEWF 944
>At1g36120 putative reverse transcriptase gb|AAD22339.1
Length = 1235
Score = 264 bits (675), Expect = 2e-70
Identities = 132/287 (45%), Positives = 190/287 (65%), Gaps = 33/287 (11%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
LS RD +FTS FW++ Q +G+K+++S+AYHPQTDGQSERTIQ+LED+L++CVL+ GG W
Sbjct: 974 LSHRDSKFTSAFWRAFQVEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLQMCVLDWGGHW 1033
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
HL L++F YNNSY +SIGMAPFEALYGR CRT LCW + G + + G + VQ+TTE+++
Sbjct: 1034 ADHLSLVKFAYNNSYQASIGMAPFEALYGRPCRTLLCWTQVGEKSIYGADYVQETTERIR 1093
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+++ MK +Q RQ+SY DKRR++LEF+ G +
Sbjct: 1094 VLKLNMKEAQDRQRSYADKRRRELEFEVGT-----------------------------E 1124
Query: 1335 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLR 1394
I ERVG VAYR+ LP + H+VFHVS LRK + V+ ++ N+T+E P+R
Sbjct: 1125 IVERVGPVAYRLELPDVMRAFHNVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVR 1184
Query: 1395 IDDRKVKTLRGKEIPLVRVVW--DGATGESLTWELESKMLESYPKLF 1439
+ +R++K +R K+IP+++V+W DG T E TWE E+++ + K F
Sbjct: 1185 VLERRIKEVRRKKIPMIKVLWDCDGVTEE--TWEPEARIKARFKKWF 1229
>At4g07830 putative reverse transcriptase
Length = 611
Score = 256 bits (655), Expect = 5e-68
Identities = 134/287 (46%), Positives = 191/287 (65%), Gaps = 25/287 (8%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD +FTS FW++ Q +G+K+++S+AYHPQTDGQSERTIQ+LED+LR+CVL+ G W
Sbjct: 306 VSDRDSKFTSAFWRAFQAEMGTKVQMSTAYHPQTDGQSERTIQTLEDMLRMCVLDWRGHW 365
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
HL L+EF YNNSY +SIGMAPFE LYGR CRT LCW + G R + G + VQ+ TE+++
Sbjct: 366 ADHLSLVEFAYNNSYQASIGMAPFEVLYGRPCRT-LCWTQVGERSIYGADYVQEITERIR 424
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+++ MK +Q+RQ+SY DKRRK+LEF+ GD V+ G +S+
Sbjct: 425 VLKLNMKEAQNRQRSYADKRRKELEFEVGD-------SVSQDGHVARSE----------- 466
Query: 1335 ISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLR 1394
+RVG VA+R+ L + H VFHVS LRK + V+ ++ N+T+E P+R
Sbjct: 467 --QRVGPVAFRLELSDVMRAFHKVFHVSMLRKCLHKDDEVLAKIPEDLQPNMTLEARPVR 524
Query: 1395 IDDRKVKTLRGKEIPLVRVV--WDGATGESLTWELESKMLESYPKLF 1439
+ +R++K LR K+IPL++V+ DG T E TWE E+++ + K F
Sbjct: 525 VLERRIKELRRKKIPLIKVLRNCDGVTEE--TWEPEARLKARFKKWF 569
>At2g06470 putative retroelement pol polyprotein
Length = 899
Score = 233 bits (594), Expect = 6e-61
Identities = 115/229 (50%), Positives = 155/229 (67%), Gaps = 19/229 (8%)
Query: 1169 SLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWDSHLPLIEFTYNNS 1228
+ Q+ALG+++ LS+AYHPQTDGQSERTIQ+LED+LR CVL+ GG W+ +L L
Sbjct: 690 AFQKALGTRVNLSTAYHPQTDGQSERTIQTLEDMLRACVLDWGGNWEKYLTL-------- 741
Query: 1229 YHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVKMIQEKMKASQSRQK 1288
ALYGR CRT LCW G R + GP IV +TTE++K ++ K+K + RQK
Sbjct: 742 -----------ALYGRACRTPLCWTPVGERRLFGPTIVDETTERMKFLKIKLKEAHDRQK 790
Query: 1289 SYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQISERVGTVAYRVGL 1348
SY +KRRK+LEFQ GD V+L+ G GR KKL+PR++GPY++ ERVG VAY++ L
Sbjct: 791 SYANKRRKELEFQVGDLVYLKAMTYKGAGRFTSRKKLSPRYVGPYKVIERVGAVAYKLDL 850
Query: 1349 PPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVETLPLRIDD 1397
PP L+ H+VFHVSQLRK + D ++ +++N+TVE P+RI D
Sbjct: 851 PPKLNAFHNVFHVSQLRKCLSDQEESVEDVPPGLKENMTVEAWPVRIMD 899
>At3g11970 hypothetical protein
Length = 1499
Score = 177 bits (448), Expect = 5e-44
Identities = 100/296 (33%), Positives = 164/296 (54%), Gaps = 5/296 (1%)
Query: 1146 KVAWCSFEHLSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRV 1205
K+ C +SDRD FTS FW+ G L+L+SAYHPQ+DGQ+E + LE LR
Sbjct: 1208 KLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRC 1267
Query: 1206 CVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEI 1265
++ W L L E+ YN +YHSS M PFE +YG+ L + +V +
Sbjct: 1268 MCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARS 1327
Query: 1266 VQQTTEKVKMIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALK-SKK 1324
+Q+ + + ++ + +Q R K + D+ R + EF+ GD+V++++ P ++ ++K
Sbjct: 1328 LQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQK 1387
Query: 1325 LTPRFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRD 1384
L+P++ GPY+I +R G VAY++ LP + S +H VFHVSQL+ V + S + V ++D
Sbjct: 1388 LSPKYFGPYKIIDRCGEVAYKLALPSY-SQVHPVFHVSQLKVLVGNVSTTVHLPSV-MQD 1445
Query: 1385 NLTVETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLESYPKLFA 1440
E +P ++ +RK+ +GK + V V W E TWE + +++P+ A
Sbjct: 1446 --VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFPEFEA 1499
>At1g36590 hypothetical protein
Length = 1499
Score = 177 bits (448), Expect = 5e-44
Identities = 100/296 (33%), Positives = 164/296 (54%), Gaps = 5/296 (1%)
Query: 1146 KVAWCSFEHLSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRV 1205
K+ C +SDRD FTS FW+ G L+L+SAYHPQ+DGQ+E + LE LR
Sbjct: 1208 KLHGCPTSIVSDRDVVFTSEFWREFFTLQGVALKLTSAYHPQSDGQTEVVNRCLETYLRC 1267
Query: 1206 CVLEQGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEI 1265
++ W L L E+ YN +YHSS M PFE +YG+ L + +V +
Sbjct: 1268 MCHDRPQLWSKWLALAEYWYNTNYHSSSRMTPFEIVYGQVPPVHLPYLPGESKVAVVARS 1327
Query: 1266 VQQTTEKVKMIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALK-SKK 1324
+Q+ + + ++ + +Q R K + D+ R + EF+ GD+V++++ P ++ ++K
Sbjct: 1328 LQEREDMLLFLKFHLMRAQHRMKQFADQHRTEREFEIGDYVYVKLQPYRQQSVVMRANQK 1387
Query: 1325 LTPRFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRD 1384
L+P++ GPY+I +R G VAY++ LP + S +H VFHVSQL+ V + S + V ++D
Sbjct: 1388 LSPKYFGPYKIIDRCGEVAYKLALPSY-SQVHPVFHVSQLKVLVGNVSTTVHLPSV-MQD 1445
Query: 1385 NLTVETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLESYPKLFA 1440
E +P ++ +RK+ +GK + V V W E TWE + +++P+ A
Sbjct: 1446 --VFEKVPEKVVERKMVNRQGKAVTKVLVKWSNEPLEEATWEFLFDLQKTFPEFEA 1499
>At2g14640 putative retroelement pol polyprotein
Length = 945
Score = 153 bits (387), Expect = 6e-37
Identities = 100/288 (34%), Positives = 157/288 (53%), Gaps = 11/288 (3%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SD DP F S FW+ + +KL +S+AYHPQTDGQ+E + +E LR V W
Sbjct: 653 VSDCDPIFMSLFWQEFWKLSRTKLWMSTAYHPQTDGQTEVVNRCIEQFLRCFVHYHPKQW 712
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGP--EIVQQTTEK 1272
S +P E+ YN ++H+S GM PF+ALYGR + + +E G VV G E + E
Sbjct: 713 SSFIPWAEYWYNTTFHASTGMTPFQALYGRP-PSPIPAYELGS-VVCGELNEQMAARDEL 770
Query: 1273 VKMIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPV---TGVGRALKSKKLTPRF 1329
+ +++ + + + K D R +D+ FQ GD V LR+ P T R+ S+KL+ RF
Sbjct: 771 LAELKQHLVTANNCMKQQADSRLRDVSFQVGDWVLLRIQPYRQKTLFRRS--SQKLSHRF 828
Query: 1330 IGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTVE 1389
GP+Q++ + G VAYR+ LP + +H VFHVS L+ +V D +R+N ++
Sbjct: 829 YGPFQVASKHGEVAYRLTLPEG-TRIHPVFHVSLLKPWVGD-GEPDMGQLPPLRNNGELK 886
Query: 1390 TLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWELESKMLESYPK 1437
P + + + ++ K + + V W+G E TWE ++ S+P+
Sbjct: 887 LQPTAVLEVRWRSQDKKRVADLLVQWEGLHIEDATWEEYDQLAASFPE 934
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 144 bits (364), Expect = 3e-34
Identities = 76/188 (40%), Positives = 118/188 (62%), Gaps = 5/188 (2%)
Query: 1254 ESGGRVVLGPEIVQQTTEKVKMIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPV 1313
++ G VL + V + K+ + MK +Q RQ+SY DKRR++LEF+ GD V+L++ +
Sbjct: 1050 KTDGAAVLAKKFVSEIV-KLHGVPLNMKEAQDRQRSYADKRRRELEFEVGDRVYLKMAML 1108
Query: 1314 TGVGRALKSKKLTPRFIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSH 1373
G R++ KL+PR++GP++I ERV VAYR+ LP + H VFHVS LRK +
Sbjct: 1109 RGPNRSISETKLSPRYMGPFKIVERVEPVAYRLELPDVMRAFHKVFHVSMLRKCLHKDDE 1168
Query: 1374 VIQSDDVQVRDNLTVETLPLRIDDRKVKTLRGKEIPLVRVVW--DGATGESLTWELESKM 1431
+ ++ N+T+E P+R+ +R++K LR K+IPL++V+W DG T E TWE E++M
Sbjct: 1169 ALAKIPEDLQPNMTLEARPVRVLERRIKELRQKKIPLIKVLWDCDGVTEE--TWEPEARM 1226
Query: 1432 LESYPKLF 1439
+ K F
Sbjct: 1227 KARFKKWF 1234
>At1g35370 hypothetical protein
Length = 1447
Score = 140 bits (354), Expect = 4e-33
Identities = 88/278 (31%), Positives = 137/278 (48%), Gaps = 28/278 (10%)
Query: 1150 CSFEHLSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLE 1209
C +SDRD FTS FWK + G +LR+SSAYHPQ+DGQ+E + LE+ LR
Sbjct: 1183 CPTSIVSDRDVLFTSDFWKEFFKLQGVELRMSSAYHPQSDGQTEVVNRCLENYLRCMCHA 1242
Query: 1210 QGGTWDSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQT 1269
+ W+ LPL E+ YN +YHSS M PFE +YG+ L + +V + +Q+
Sbjct: 1243 RPHLWNKWLPLAEYWYNTNYHSSSQMTPFELVYGQAPPIHLPYLPGKSKVAVVARSLQER 1302
Query: 1270 TEKVKMIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALK-SKKLTPR 1328
+ ++ + +Q R K + D+ R + F GD V++++ P L+ ++KL+P+
Sbjct: 1303 ENMLLFLKFHLMRAQHRMKQFADQHRTERTFDIGDFVYVKLQPYRQQSVVLRVNQKLSPK 1362
Query: 1329 FIGPYQISERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPDPSHVIQSDDVQVRDNLTV 1388
+ GPY+I E+ G V + +V +QL +PD
Sbjct: 1363 YFGPYKIIEKCGEVM-----------VGNVTTSTQLPSVLPD----------------IF 1395
Query: 1389 ETLPLRIDDRKVKTLRGKEIPLVRVVWDGATGESLTWE 1426
E P I +RK+ +G+ +V V W G E TW+
Sbjct: 1396 EKAPEYILERKLVKRQGRAATMVLVKWIGEPVEEATWK 1433
>At3g31480 hypothetical protein
Length = 338
Score = 137 bits (345), Expect = 5e-32
Identities = 69/191 (36%), Positives = 111/191 (57%), Gaps = 42/191 (21%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SD+D +FTS FW + Q +G+K+++S+AYHP+TDG+ E+TIQ+LED+LR+
Sbjct: 60 VSDKDSKFTSAFWIAFQAEMGTKVQISTAYHPKTDGKFEKTIQTLEDMLRM--------- 110
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVK 1274
T LCW + G R + G V +TTE+++
Sbjct: 111 ---------------------------------TPLCWTQVGERSIYGANYVHETTERIQ 137
Query: 1275 MIQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQ 1334
+++ MK + RQ+SY D RR+++EF+ GD V+L++ + R + KL+P+++GP++
Sbjct: 138 VLKLNMKEAHDRQRSYADMRRREVEFEVGDRVYLKMDMLQSPKRFILETKLSPKYMGPFR 197
Query: 1335 ISERVGTVAYR 1345
I ERVG VAYR
Sbjct: 198 IVERVGPVAYR 208
>At2g06190 putative Ty3-gypsy-like retroelement pol polyprotein
Length = 280
Score = 93.2 bits (230), Expect = 1e-18
Identities = 62/215 (28%), Positives = 103/215 (47%), Gaps = 20/215 (9%)
Query: 1156 SDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTWD 1215
SDRD +F S FW +L G+ L SS H Q DGQ+E T ++L +++R + WD
Sbjct: 62 SDRDTKFLSHFWSTLWRMFGTALNRSSTPHTQIDGQTEVTNRTLGNMVRSICGDNPKQWD 121
Query: 1216 SHLPLIEFTYNNSYHSSIGMAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVKM 1275
LP IEF YN+ H + + G T + ++ ++
Sbjct: 122 LALPQIEFAYNSVVH-VVDLVKLPKALGASAET----------------MAEEILVVKEV 164
Query: 1276 IQEKMKASQSRQKSYHDKRRKDLEFQEGDHVFLRVTPVTGVGRALKSKKLTPRFIGPYQI 1335
++ K++A+ + K DKRR+ F+EGD V + + G K+ PR GP+++
Sbjct: 165 VKAKLEATGKKNKVAADKRRRFKVFKEGDDVMVLLR--KGRFAVGTYNKVKPRKYGPFKV 222
Query: 1336 SERVGTVAYRVGLPPHLSNLHDVFHVSQLRKYVPD 1370
++ AY V LP + N+ + F+V+ + +Y D
Sbjct: 223 LRKINDNAYVVALPKSM-NISNTFNVADIHEYHAD 256
>At4g07850 putative polyprotein
Length = 1138
Score = 90.5 bits (223), Expect = 6e-18
Identities = 42/89 (47%), Positives = 59/89 (66%)
Query: 1155 LSDRDPRFTSRFWKSLQEALGSKLRLSSAYHPQTDGQSERTIQSLEDLLRVCVLEQGGTW 1214
+SDRD +F S FWK+L LG+KL S+ HPQTDGQ+E ++L LLR + + TW
Sbjct: 895 VSDRDTKFLSYFWKTLWSKLGTKLLFSTTCHPQTDGQTEVVNRTLSTLLRALIKKNLKTW 954
Query: 1215 DSHLPLIEFTYNNSYHSSIGMAPFEALYG 1243
+ LP +EF YN+S HS+ +PF+ +YG
Sbjct: 955 EDCLPHVEFAYNHSVHSATKFSPFQIVYG 983
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 87.4 bits (215), Expect = 5e-17
Identities = 49/156 (31%), Positives = 89/156 (56%), Gaps = 5/156 (3%)
Query: 1235 MAPFEALYGRRCRTQLCWFESGGRVVLGPEIVQQTTEKVKMIQEKMKASQSRQKSYHDKR 1294
M P+EA+YG+ L + +V + +Q+ + ++ + +Q R K D+
Sbjct: 625 MTPYEAVYGQPPPLHLPYLPGESKVAVVARSMQERESMILFLKFHLMRAQHRMKQLADQH 684
Query: 1295 RKDLEFQEGDHVFLRVTPVTGVGRALKS-KKLTPRFIGPYQISERVGTVAYRVGLPPHLS 1353
+ EF+ GD+VF+++ P ++S +KL+P++ GPY++ +R G VAY++ LP + S
Sbjct: 685 ITEREFEVGDYVFVKLQPYRQQSVVMRSTQKLSPKYFGPYKVIDRCGEVAYKLQLPAN-S 743
Query: 1354 NLHDVFHVSQLRKY---VPDPSHVIQSDDVQVRDNL 1386
+H VFHVSQLR V +H+++ + +R+NL
Sbjct: 744 QVHPVFHVSQLRVLVGTVTTSTHLLRCYLMSLRENL 779
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.362 0.162 0.603
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 26,320,438
Number of Sequences: 26719
Number of extensions: 961354
Number of successful extensions: 3918
Number of sequences better than 10.0: 45
Number of HSP's better than 10.0 without gapping: 37
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 3839
Number of HSP's gapped (non-prelim): 56
length of query: 1449
length of database: 11,318,596
effective HSP length: 112
effective length of query: 1337
effective length of database: 8,326,068
effective search space: 11131952916
effective search space used: 11131952916
T: 11
A: 40
X1: 14 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (22.0 bits)
S2: 67 (30.4 bits)
Medicago: description of AC144724.10


