

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.9 + phase: 0
(259 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
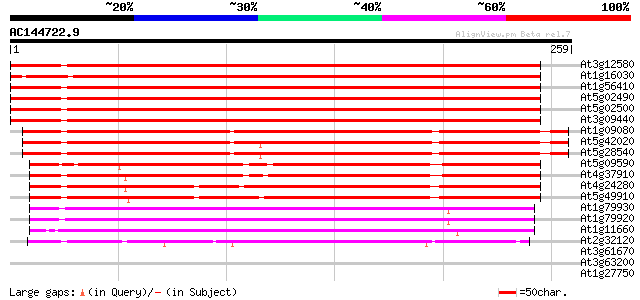
Score E
Sequences producing significant alignments: (bits) Value
At3g12580 putative protein 358 2e-99
At1g16030 unknown protein 357 4e-99
At1g56410 hypothetical protein 354 2e-98
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 353 7e-98
At5g02500 dnaK-type molecular chaperone hsc70.1 352 9e-98
At3g09440 heat-shock protein (At-hsc70-3) 350 4e-97
At1g09080 putative luminal binding protein 308 1e-84
At5g42020 luminal binding protein (dbj|BAA13948.1) 297 3e-81
At5g28540 luminal binding protein 297 3e-81
At5g09590 heat shock protein 70 (Hsc70-5) 228 2e-60
At4g37910 heat shock protein 70 like protein 226 7e-60
At4g24280 hsp 70-like protein 204 5e-53
At5g49910 heat shock protein 70 (Hsc70-7) 200 6e-52
At1g79930 putative heat-shock protein 159 1e-39
At1g79920 putative heat-shock protein (At1g79920) 155 2e-38
At1g11660 heat-shock protein like 150 5e-37
At2g32120 70kD heat shock protein 125 2e-29
At3g61670 putative protein 30 1.7
At3g63200 unknown protein 28 4.9
At1g27750 unknown protein 28 4.9
>At3g12580 putative protein
Length = 650
Score = 358 bits (918), Expect = 2e-99
Identities = 176/245 (71%), Positives = 208/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR++SDP VQ D WPFKV+SG +KPMI V +KG+EK
Sbjct: 59 KNQVAMNPTNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEA+L SPVKNAVVTVPAYFNDSQR+AT +AG I+GLNVMRIIN
Sbjct: 119 SAEEISSMVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKASSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>At1g16030 unknown protein
Length = 646
Score = 357 bits (916), Expect = 4e-99
Identities = 177/245 (72%), Positives = 208/245 (84%), Gaps = 3/245 (1%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K + A+GIDLGTTYSCVGVW+ + RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MATKSE-KAIGIDLGTTYSCVGVWMND--RVEIIPNDQGNRTTPSYVAFTDTERLIGDAA 57
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NPQNTVFDAKRLIGRKFSDP VQ DI+ WPFKV+SG +KPMI V YK +EK
Sbjct: 58 KNQVALNPQNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQF 117
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
EEISS++L MK++AEA+L VKNAVVTVPAYFNDSQR+AT +AGAI+GLNV+RIIN
Sbjct: 118 SPEEISSMVLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIIN 177
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD KG GE+N+ +FDLGGGTFDVSLLTI+ VFEVKATAG+THLGGED
Sbjct: 178 EPTAAAIAYGLDKKGTKAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGED 237
Query: 241 FDNRM 245
FDNR+
Sbjct: 238 FDNRL 242
>At1g56410 hypothetical protein
Length = 617
Score = 354 bits (909), Expect = 2e-98
Identities = 176/245 (71%), Positives = 205/245 (82%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ D+ FWPFKV G DKPMI V YKG+EK
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDASVQSDMKFWPFKVTPGQADKPMIFVNYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL S +KNAVVTVPAYFNDSQR+AT +AG IAGLNV+RIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K G +N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGIKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 353 bits (905), Expect = 7e-98
Identities = 174/245 (71%), Positives = 206/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ D WPF +ISG +KPMI V+YKG+EK
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDASVQSDRQLWPFTIISGTAEKPMIVVEYKGEEKQF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEA+L + VKNAVVTVPAYFNDSQR+AT +AG IAGLNV+RIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAFLGTTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 352 bits (904), Expect = 9e-98
Identities = 174/245 (71%), Positives = 206/245 (84%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
M+ K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MSGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+FSD VQ D+ WPFK+ +G DKPMI V+YKG+EK
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDSSVQSDMKLWPFKIQAGPADKPMIYVEYKGEEKEF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS++L M++IAEAYL +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 350 bits (898), Expect = 4e-97
Identities = 174/245 (71%), Positives = 205/245 (83%), Gaps = 2/245 (0%)
Query: 1 MARKHDGHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAA 60
MA K +G A+GIDLGTTYSCVGVW + +RVEII ND+GN+ TPS+VAFTD +RL+G AA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KDQAAINPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNL 120
K+Q A+NP NTVFDAKRLIGR+F+D VQ DI WPF + SG +KPMI V YKG++K
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKSGPAEKPMIVVNYKGEDKEF 118
Query: 121 CAEEISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIIN 180
AEEISS+IL M++IAEAYL + +KNAVVTVPAYFNDSQR+AT +AG IAGLNVMRIIN
Sbjct: 119 SAEEISSMILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 DPTAAAIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGED 240
+PTAAAIAYGLD K GE+N+ +FDLGGGTFDVSLLTI+ +FEVKATAG+THLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRM 245
FDNRM
Sbjct: 239 FDNRM 243
>At1g09080 putative luminal binding protein
Length = 678
Score = 308 bits (790), Expect = 1e-84
Identities = 161/252 (63%), Positives = 199/252 (78%), Gaps = 10/252 (3%)
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV+ VEII ND+GN+ITPS+VAFTD +RL+G AAK+QAA
Sbjct: 50 GTVIGIDLGTTYSCVGVY--HNKHVEIIANDQGNRITPSWVAFTDTERLIGEAAKNQAAK 107
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEIS 126
NP+ T+FD KRLIGRKF DP VQ+DI F P+KV++ + KP I VK KG+EK EEIS
Sbjct: 108 NPERTIFDPKRLIGRKFDDPDVQRDIKFLPYKVVNK-DGKPYIQVKVKGEEKLFSPEEIS 166
Query: 127 SIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAA 186
++IL MK+ AEA+L +K+AV+TVPAYFND+QR+AT +AGAIAGLNV+RIIN+PT AA
Sbjct: 167 AMILTKMKETAEAFLGKKIKDAVITVPAYFNDAQRQATKDAGAIAGLNVVRIINEPTGAA 226
Query: 187 IAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRMC 246
IAYGLD KG GE NI V+DLGGGTFDVS+LTI VFEV +T+G+THLGGEDFD+R+
Sbjct: 227 IAYGLDKKG---GESNILVYDLGGGTFDVSILTIDNGVFEVLSTSGDTHLGGEDFDHRV- 282
Query: 247 WNKMCFTMNLIK 258
M + + L+K
Sbjct: 283 ---MDYFIKLVK 291
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 297 bits (761), Expect = 3e-81
Identities = 160/253 (63%), Positives = 195/253 (76%), Gaps = 11/253 (4%)
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV+ + VEII ND+GN+ITPS+V FTD +RL+G AAK+QAA+
Sbjct: 35 GSVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAV 92
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEI 125
NP+ TVFD KRLIGRKF D VQKD P+++++ + KP I VK K G+ K EEI
Sbjct: 93 NPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVNK-DGKPYIQVKIKDGETKVFSPEEI 151
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
S++IL MK+ AEAYL +K+AVVTVPAYFND+QR+AT +AG IAGLNV RIIN+PTAA
Sbjct: 152 SAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 211
Query: 186 AIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
AIAYGLD KG GE+NI VFDLGGGTFDVS+LTI VFEV +T G+THLGGEDFD+R+
Sbjct: 212 AIAYGLDKKG---GEKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRI 268
Query: 246 CWNKMCFTMNLIK 258
M + + LIK
Sbjct: 269 ----MEYFIKLIK 277
>At5g28540 luminal binding protein
Length = 669
Score = 297 bits (761), Expect = 3e-81
Identities = 160/253 (63%), Positives = 195/253 (76%), Gaps = 11/253 (4%)
Query: 7 GHAVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAI 66
G +GIDLGTTYSCVGV+ + VEII ND+GN+ITPS+V FTD +RL+G AAK+QAA+
Sbjct: 35 GSVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAV 92
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYK-GQEKNLCAEEI 125
NP+ TVFD KRLIGRKF D VQKD P+++++ + KP I VK K G+ K EEI
Sbjct: 93 NPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVNK-DGKPYIQVKIKDGETKVFSPEEI 151
Query: 126 SSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAA 185
S++IL MK+ AEAYL +K+AVVTVPAYFND+QR+AT +AG IAGLNV RIIN+PTAA
Sbjct: 152 SAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 211
Query: 186 AIAYGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
AIAYGLD KG GE+NI VFDLGGGTFDVS+LTI VFEV +T G+THLGGEDFD+R+
Sbjct: 212 AIAYGLDKKG---GEKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRV 268
Query: 246 CWNKMCFTMNLIK 258
M + + LIK
Sbjct: 269 ----MEYFIKLIK 277
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 228 bits (581), Expect = 2e-60
Identities = 134/237 (56%), Positives = 162/237 (67%), Gaps = 12/237 (5%)
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAF-TDDQRLVGGAAKDQAAINP 68
+GIDLGTT SCV V +E KN ++I N EG + TPS VAF T + LVG AK QA NP
Sbjct: 60 IGIDLGTTNSCVAV-MEGKNP-KVIENAEGARTTPSVVAFNTKGELLVGTPAKRQAVTNP 117
Query: 69 QNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSI 128
NTV KRLIGRKF DP QK++ P+K++ N V+ GQ+ + +I +
Sbjct: 118 TNTVSGTKRLIGRKFDDPQTQKEMKMVPYKIVRAPNGDAW--VEANGQQYS--PSQIGAF 173
Query: 129 ILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIA 188
IL MK+ AEAYL V AVVTVPAYFND+QR+AT +AG IAGL+V RIIN+PTAAA++
Sbjct: 174 ILTKMKETAEAYLGKSVTKAVVTVPAYFNDAQRQATKDAGRIAGLDVERIINEPTAAALS 233
Query: 189 YGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
YG+ NK E I VFDLGGGTFDVS+L I VFEVKAT G+T LGGEDFDN +
Sbjct: 234 YGMTNK-----EGLIAVFDLGGGTFDVSVLEISNGVFEVKATNGDTFLGGEDFDNAL 285
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 226 bits (577), Expect = 7e-60
Identities = 127/237 (53%), Positives = 159/237 (66%), Gaps = 12/237 (5%)
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDD-QRLVGGAAKDQAAINP 68
+GIDLGTT SCV V E +I N EG++ TPS VA + LVG AK QA NP
Sbjct: 55 IGIDLGTTNSCVSVM--EGKTARVIENAEGSRTTPSVVAMNQKGELLVGTPAKRQAVTNP 112
Query: 69 QNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSI 128
NT+F +KRLIGR+F DP QK++ P+K++ N V+ GQ+ +I +
Sbjct: 113 TNTIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAW--VEANGQK--FSPSQIGAN 168
Query: 129 ILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIA 188
+L MK+ AEAYL + AVVTVPAYFND+QR+AT +AG IAGL+V RIIN+PTAAA++
Sbjct: 169 VLTKMKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIINEPTAAALS 228
Query: 189 YGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
YG++NK E I VFDLGGGTFDVS+L I VFEVKAT G+T LGGEDFDN +
Sbjct: 229 YGMNNK-----EGVIAVFDLGGGTFDVSILEISSGVFEVKATNGDTFLGGEDFDNTL 280
>At4g24280 hsp 70-like protein
Length = 718
Score = 204 bits (518), Expect = 5e-53
Identities = 117/237 (49%), Positives = 151/237 (63%), Gaps = 11/237 (4%)
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDD-QRLVGGAAKDQAAINP 68
VGIDLGTT S V E + I+ N EG + TPS VA+T RLVG AK QA +NP
Sbjct: 81 VGIDLGTTNSAVAAM--EGGKPTIVTNAEGQRTTPSVVAYTKSGDRLVGQIAKRQAVVNP 138
Query: 69 QNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSI 128
+NT F KR IGRK ++ V ++ ++V+ N+ + ++ K AEEIS+
Sbjct: 139 ENTFFSVKRFIGRKMNE--VDEESKQVSYRVVRDENNN--VKLECPAINKQFAAEEISAQ 194
Query: 129 ILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIA 188
+L + A +L V AV+TVPAYFNDSQR AT +AG IAGL V+RIIN+PTAA++A
Sbjct: 195 VLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLA 254
Query: 189 YGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
YG D K I VFDLGGGTFDVS+L + VFEV +T+G+THLGG+DFD R+
Sbjct: 255 YGFDRK----ANETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRV 307
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 200 bits (509), Expect = 6e-52
Identities = 117/237 (49%), Positives = 150/237 (62%), Gaps = 11/237 (4%)
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQ-RLVGGAAKDQAAINP 68
VGIDLGTT S V E + I+ N EG + TPS VA+T + RLVG AK QA +NP
Sbjct: 81 VGIDLGTTNSAVAAM--EGGKPTIVTNAEGQRTTPSVVAYTKSKDRLVGQIAKRQAVVNP 138
Query: 69 QNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSI 128
+NT F KR IGR+ ++ V ++ ++VI N + G K AEEIS+
Sbjct: 139 ENTFFSVKRFIGRRMNE--VAEESKQVSYRVIKDENGNVKLDCPAIG--KQFAAEEISAQ 194
Query: 129 ILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIA 188
+L + A +L V AV+TVPAYFNDSQR AT +AG IAGL V+RIIN+PTAA++A
Sbjct: 195 VLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLA 254
Query: 189 YGLDNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFDNRM 245
YG + K I VFDLGGGTFDVS+L + VFEV +T+G+THLGG+DFD R+
Sbjct: 255 YGFERK----SNETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRV 307
>At1g79930 putative heat-shock protein
Length = 831
Score = 159 bits (402), Expect = 1e-39
Identities = 83/234 (35%), Positives = 134/234 (56%), Gaps = 3/234 (1%)
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQ 69
VG D G V V + ++++ NDE N+ TP+ V F D QR +G A +NP+
Sbjct: 4 VGFDFGNENCLVAV--ARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPK 61
Query: 70 NTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSII 129
N++ KRLIGR+FSDP +Q+DI PF V G + P+I Y G+++ ++ ++
Sbjct: 62 NSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEKRAFTPTQVMGMM 121
Query: 130 LANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIAY 189
L+N+K IAE L + V + + +P YF D QR+A ++A IAGL+ +R+I++ TA A+AY
Sbjct: 122 LSNLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLRLIHETTATALAY 181
Query: 190 GLDNKGRCDGER-NIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFD 242
G+ + ++ N+ D+G + V + K ++ + A + LGG DFD
Sbjct: 182 GIYKTDLPESDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFD 235
>At1g79920 putative heat-shock protein (At1g79920)
Length = 736
Score = 155 bits (393), Expect = 2e-38
Identities = 82/234 (35%), Positives = 132/234 (56%), Gaps = 3/234 (1%)
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQ 69
VG D G V V + ++++ NDE N+ TP+ V F D QR +G A +NP+
Sbjct: 4 VGFDFGNENCLVAV--ARQRGIDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPK 61
Query: 70 NTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSII 129
N++ KRLIGR+FSDP +Q+DI PF V G + P+I Y G+ + ++ ++
Sbjct: 62 NSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEIRAFTPTQVMGMM 121
Query: 130 LANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIAY 189
L+N+K IAE L + V + + +P YF D QR+A ++A IAGL+ + +I++ TA A+AY
Sbjct: 122 LSNLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLHLIHETTATALAY 181
Query: 190 GLDNKGRCDGER-NIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFD 242
G+ + ++ N+ D+G + V + K ++ + A + LGG DFD
Sbjct: 182 GIYKTDLPENDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFD 235
>At1g11660 heat-shock protein like
Length = 773
Score = 150 bits (380), Expect = 5e-37
Identities = 80/235 (34%), Positives = 141/235 (59%), Gaps = 4/235 (1%)
Query: 10 VGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQ 69
VG D+G +CV + + ++ ++++ NDE N+ P+ V+F + QR +G AA A ++P+
Sbjct: 4 VGFDVGNE-NCV-IAVAKQRGIDVLLNDESNRENPAMVSFGEKQRFMGAAAAASATMHPK 61
Query: 70 NTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPMIAVKYKGQEKNLCAEEISSII 129
+T+ KRLIGRKF +P VQ D+ +PF+ + I ++Y G+ ++ +I ++
Sbjct: 62 STISQLKRLIGRKFREPDVQNDLRLFPFETSEDSDGGIQIRLRYMGEIQSFSPVQILGML 121
Query: 130 LANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPTAAAIAY 189
L+++K+IAE LK+PV + V+ +P+YF +SQR A ++A AIAGL +R+++D TA A+ Y
Sbjct: 122 LSHLKQIAEKSLKTPVSDCVIGIPSYFTNSQRLAYLDAAAIAGLRPLRLMHDSTATALGY 181
Query: 190 GLDNKGRCDGERNIFV--FDLGGGTFDVSLLTIKGDVFEVKATAGNTHLGGEDFD 242
G+ ++ D+G V + + + V++ A + +LGG DFD
Sbjct: 182 GIYKTDLVANSSPTYIVFIDIGHCDTQVCVASFESGSMRVRSHAFDRNLGGRDFD 236
>At2g32120 70kD heat shock protein
Length = 563
Score = 125 bits (314), Expect = 2e-29
Identities = 86/244 (35%), Positives = 131/244 (53%), Gaps = 19/244 (7%)
Query: 9 AVGIDLGTTYSCVGVWLEEKNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINP 68
A+GID+GT+ + VW ++V I+ N K+ SFV F D+ + G +Q A
Sbjct: 30 ALGIDIGTSQCSIAVW--NGSQVHILRNTRNQKLIKSFVTFKDE--VPAGGVSNQLAHEQ 85
Query: 69 QN----TVFDAKRLIGRKFSDPVVQKDIMFWPFKVIS-GLNDKPMIAVKYKGQEKNLCAE 123
+ +F+ KRL+GR +DPVV PF V + + +P IA ++ E
Sbjct: 86 EMLTGAAIFNMKRLVGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNAWRSTTPE 144
Query: 124 EISSIILANMKKIAEAYLKSPVKNAVVTVPAYFNDSQRKATMNAGAIAGLNVMRIINDPT 183
E+ +I L ++ +AEA LK PV+N V+TVP F+ Q A A+AGL+V+R++ +PT
Sbjct: 145 EVLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQLTRFERACAMAGLHVLRLMPEPT 204
Query: 184 AAAIAYGL-------DNKGRCDGERNIFVFDLGGGTFDVSLLTIKGDVFEVKATAGNTHL 236
A A+ Y DN G ER +F++G G DV++ G V ++KA AG+ +
Sbjct: 205 AIALLYAQQQQMTTHDNMG-SGSERLAVIFNMGAGYCDVAVTATAGGVSQIKALAGSP-I 262
Query: 237 GGED 240
GGED
Sbjct: 263 GGED 266
>At3g61670 putative protein
Length = 790
Score = 29.6 bits (65), Expect = 1.7
Identities = 17/47 (36%), Positives = 22/47 (46%)
Query: 28 KNRVEIIHNDEGNKITPSFVAFTDDQRLVGGAAKDQAAINPQNTVFD 74
K+ + I NDEGNK S +RL+ A K I P N +D
Sbjct: 634 KDLTKSIQNDEGNKSNVSINGHPLTERLLRKAEKQAGVIQPGNYWYD 680
>At3g63200 unknown protein
Length = 384
Score = 28.1 bits (61), Expect = 4.9
Identities = 24/85 (28%), Positives = 38/85 (44%), Gaps = 14/85 (16%)
Query: 178 IINDPTAAAIAYGLDNK---GRCDGERNIFVFDLGGGTFDVS-----LLTIKGD-----V 224
++N+PTAAA+ + L NK +G ++ V LG G +S L GD V
Sbjct: 225 VMNNPTAAAVTHVLHNKRDFPSVNGVDDLLVLSLGNGPSTMSSSPGRKLRRNGDYSTSSV 284
Query: 225 FEVKATAGNTHLGGEDFDNRMCWNK 249
++ G + + N CWN+
Sbjct: 285 VDI-VVDGVSDTVDQMLGNAFCWNR 308
>At1g27750 unknown protein
Length = 1975
Score = 28.1 bits (61), Expect = 4.9
Identities = 11/42 (26%), Positives = 25/42 (59%)
Query: 67 NPQNTVFDAKRLIGRKFSDPVVQKDIMFWPFKVISGLNDKPM 108
N + ++ ++ + DP+V+ DI+ F+ ISGL+++ +
Sbjct: 1512 NHETSIPAVTKMFPSRVEDPMVRADILIQTFREISGLSEQSL 1553
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.409
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 5,871,938
Number of Sequences: 26719
Number of extensions: 246858
Number of successful extensions: 539
Number of sequences better than 10.0: 21
Number of HSP's better than 10.0 without gapping: 19
Number of HSP's successfully gapped in prelim test: 2
Number of HSP's that attempted gapping in prelim test: 482
Number of HSP's gapped (non-prelim): 22
length of query: 259
length of database: 11,318,596
effective HSP length: 97
effective length of query: 162
effective length of database: 8,726,853
effective search space: 1413750186
effective search space used: 1413750186
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 59 (27.3 bits)
Medicago: description of AC144722.9


