

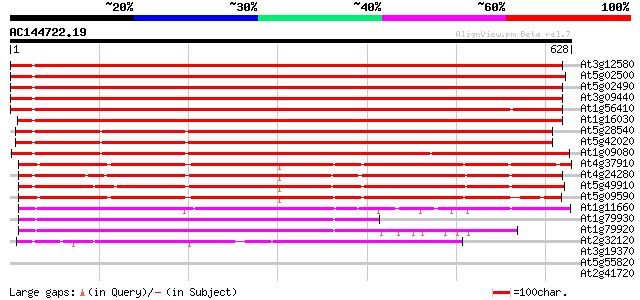
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.19 + phase: 0
(628 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At3g12580 putative protein 793 0.0
At5g02500 dnaK-type molecular chaperone hsc70.1 781 0.0
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 776 0.0
At3g09440 heat-shock protein (At-hsc70-3) 772 0.0
At1g56410 hypothetical protein 761 0.0
At1g16030 unknown protein 751 0.0
At5g28540 luminal binding protein 624 e-179
At5g42020 luminal binding protein (dbj|BAA13948.1) 621 e-178
At1g09080 putative luminal binding protein 617 e-177
At4g37910 heat shock protein 70 like protein 449 e-126
At4g24280 hsp 70-like protein 443 e-124
At5g49910 heat shock protein 70 (Hsc70-7) 440 e-123
At5g09590 heat shock protein 70 (Hsc70-5) 439 e-123
At1g11660 heat-shock protein like 259 3e-69
At1g79930 putative heat-shock protein 251 1e-66
At1g79920 putative heat-shock protein (At1g79920) 250 2e-66
At2g32120 70kD heat shock protein 241 9e-64
At3g19370 unknown protein 38 0.015
At5g55820 unknown protein 36 0.074
At2g41720 salt-inducible protein-like 35 0.097
>At3g12580 putative protein
Length = 650
Score = 793 bits (2049), Expect = 0.0
Identities = 402/621 (64%), Positives = 503/621 (80%), Gaps = 5/621 (0%)
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP+NT+FD KRLIGR+YSD VQ D+ WPFKV+SG KP I+V + G+EK+F
Sbjct: 59 KNQVAMNPTNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQF 118
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KMREIAEAFL SPVKNAV+TVPAYFNDSQR+ATKDAG I+GLNVMRIIN
Sbjct: 119 SAEEISSMVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A+ V ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKASSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+KN +DI+ NP+ALRRLRTACERAKRTLS + TIE+D++++GID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID 298
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F ++ITRA+FEE+NM+LF KCME V CL DAKM+K+SV DVVLVGGS+RIPKV+QLLQ+
Sbjct: 299 FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQLLQD 358
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+V
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 418
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+IPRNT+IP K+ + T D+Q VLI+VYEGER + DNNLLG F LS +P APRG P
Sbjct: 419 LIPRNTTIPTKKEQIFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVP 478
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+QEAE +KA+D +
Sbjct: 479 QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKAEDEEH 538
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
KKKV+A+NAL++Y Y +R +K++ +SKL DK+KI A+ + + +Q + F
Sbjct: 539 KKKVDAKNALENYAYNMRNTIKDEKIASKLDAADKKKIEDAIDQAIEWLDGNQLAEADEF 598
Query: 598 VDFLKELESIFESAMNKINKG 618
D +KELES+ + ++ +G
Sbjct: 599 EDKMKELESLCNPIIARMYQG 619
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 781 bits (2016), Expect = 0.0
Identities = 397/625 (63%), Positives = 498/625 (79%), Gaps = 5/625 (0%)
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M+ K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MSGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP NT+FD KRLIGR++SDS VQ D LWPFK+ +G +KP I V+Y G+EK F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDSSVQSDMKLWPFKIQAGPADKPMIYVEYKGEEKEF 118
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KMREIAEA+L +KNAV+TVPAYFNDSQR+ATKDAG IAGLNVMRIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+K+ +DI+ NP+ALRRLRT+CERAKRTLS + TIE+D++Y+GID
Sbjct: 239 FDNRMVNHFVQEFKRKSKKDITGNPRALRRLRTSCERAKRTLSSTAQTTIEIDSLYEGID 298
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F S+ITRA+FEE+NM+LF KCME V CL DAKM+K++V DVVLVGGS+RIPKV+QLLQ+
Sbjct: 299 FYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 358
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQGAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTT 418
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+IPRNT+IP K+ + T D+Q VLI+VYEGER + DNNLLG F LS +P APRG P
Sbjct: 419 LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVP 478
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+QEAE +K++D +
Sbjct: 479 QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEKYKSEDEEH 538
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
KKKVEA+NAL++Y Y +R ++++ KL DK+KI ++ + + +Q + F
Sbjct: 539 KKKVEAKNALENYAYNMRNTIQDEKIGEKLPAADKKKIEDSIEQAIQWLEGNQLAEADEF 598
Query: 598 VDFLKELESIFESAMNKINKGDSDE 622
D +KELESI + K+ +G E
Sbjct: 599 EDKMKELESICNPIIAKMYQGAGGE 623
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 776 bits (2004), Expect = 0.0
Identities = 393/621 (63%), Positives = 493/621 (79%), Gaps = 5/621 (0%)
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP NT+FD KRLIGR++SD+ VQ DR LWPF +ISG KP I+V+Y G+EK+F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDASVQSDRQLWPFTIISGTAEKPMIVVEYKGEEKQF 118
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KMREIAEAFL + VKNAV+TVPAYFNDSQR+ATKDAG IAGLNV+RIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAFLGTTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+KN +DI+ P+ALRRLRTACERAKRTLS + TIE+D++Y G D
Sbjct: 239 FDNRMVNHFVQEFKRKNKQDITGQPRALRRLRTACERAKRTLSSTAQTTIEIDSLYGGAD 298
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F S ITRA+FEEMNM+LF KCME V CL DAKM+K++V ++VLVGGS+RIPKV+QLLQ+
Sbjct: 299 FYSPITRARFEEMNMDLFRKCMEPVEKCLRDAKMDKSTVHEIVLVGGSTRIPKVQQLLQD 358
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTT 418
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+I RNT+IP K+ + T D+Q VLI+V+EGER + DNNLLG F LS +P APRG P
Sbjct: 419 LIQRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELSGIPPAPRGVP 478
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ IE+M+QEAE +K++D +
Sbjct: 479 QITVCFDIDANGILNVSAEDKTTGKKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEH 538
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
KKKVEA+NAL++Y Y +R ++++ KL DK+K+ ++ + + +Q + F
Sbjct: 539 KKKVEAKNALENYAYNMRNTIRDEKIGEKLPAADKKKVEDSIEEAIQWLDGNQLGEADEF 598
Query: 598 VDFLKELESIFESAMNKINKG 618
D +KELES+ + K+ +G
Sbjct: 599 EDKMKELESVCNPIIAKMYQG 619
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 772 bits (1994), Expect = 0.0
Identities = 395/621 (63%), Positives = 492/621 (78%), Gaps = 5/621 (0%)
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP NT+FD KRLIGR+++DS VQ D LWPF + SG KP I+V Y G++K F
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKSGPAEKPMIVVNYKGEDKEF 118
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS+IL KMREIAEA+L + +KNAV+TVPAYFNDSQR+ATKDAG IAGLNVMRIIN
Sbjct: 119 SAEEISSMILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+KN +DIS NP+ALRRLRTACERAKRTLS + TIE+D+++ GID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFDGID 298
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F + ITRA+FEE+N++LF KCME V CL DAKM+KNS+DDVVLVGGS+RIPKV+QLL +
Sbjct: 299 FYAPITRARFEELNIDLFRKCMEPVEKCLRDAKMDKNSIDDVVLVGGSTRIPKVQQLLVD 358
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+V
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 418
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+I RNT+IP K+ + T D+Q VLI+VYEGER + DNNLLG F LS +P APRG P
Sbjct: 419 LIQRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVP 478
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF IDA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+QEAE +K++D +
Sbjct: 479 QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEKYKSEDEEH 538
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
KKKV+A+NAL++Y Y +R ++++ KL DK+KI ++ + +Q + F
Sbjct: 539 KKKVDAKNALENYAYNMRNTIRDEKIGEKLAGDDKKKIEDSIEAAIEWLEANQLAECDEF 598
Query: 598 VDFLKELESIFESAMNKINKG 618
D +KELESI + K+ +G
Sbjct: 599 EDKMKELESICNPIIAKMYQG 619
>At1g56410 hypothetical protein
Length = 617
Score = 761 bits (1966), Expect = 0.0
Identities = 390/621 (62%), Positives = 488/621 (77%), Gaps = 7/621 (1%)
Query: 1 MTEKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVWQ +DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRF 120
KNQ A NP NT+FD KRLIGR++SD+ VQ D WPFKV G +KP I V Y G+EK+F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDASVQSDMKFWPFKVTPGQADKPMIFVNYKGEEKQF 118
Query: 121 VAEEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIIN 180
AEEISS++L KMREIAEA+L S +KNAV+TVPAYFNDSQR+ATKDAG IAGLNV+RIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 181 EPTAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGED 240
EPTAAA+AYGL K+A V +N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGIKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID 300
FDNRM+ HFV++FK+KN +DIS + +ALRRLRTACERAKRTLS + T+E+D++++GID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDISGDARALRRLRTACERAKRTLSSTAQTTVEVDSLFEGID 298
Query: 301 FSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQE 360
F S ITRAKFEEMNM+LF KCME V CL D+KM+K+ V DVVLVGGS+RIPKV+QLLQ+
Sbjct: 299 FYSPITRAKFEEMNMDLFRKCMEPVMKCLRDSKMDKSMVHDVVLVGGSTRIPKVQQLLQD 358
Query: 361 FFQGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSV 419
FF GKELC SINPDEAVAYGAA+Q A+L E + V +L+L DVTPLSLG+ G +M+
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGIETIGGVMTT 418
Query: 420 VIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHP 478
+I RNT+IP K+ E+ TT D+Q VLI+VYEGER + DNN+LG F LS +P APRG P
Sbjct: 419 LIQRNTTIPAKKEQEFTTTVDNQPDVLIQVYEGERARTIDNNILGQFVLSGIPPAPRGIP 478
Query: 479 H-KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKF 537
VCF ID++GILNVSA ++ +G KN+ITITN+KGRLS+ IE+M+QEAE +K++D +
Sbjct: 479 QFTVCFDIDSNGILNVSAEDKATGKKNKITITNDKGRLSKDDIEKMVQEAEKYKSEDEEH 538
Query: 538 KKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
KKKVEA+N L++Y Y V +++ KL DK+K ++ + + D+Q + F
Sbjct: 539 KKKVEAKNGLENYAYNVGNTLRD--MGEKLPAADKKKFEDSIEEVIQWLDDNQLAEADEF 596
Query: 598 VDFLKELESIFESAMNKINKG 618
+KELES++ + + K+ +G
Sbjct: 597 EHKMKELESVWSTIITKMYQG 617
>At1g16030 unknown protein
Length = 646
Score = 751 bits (1938), Expect = 0.0
Identities = 380/613 (61%), Positives = 484/613 (77%), Gaps = 5/613 (0%)
Query: 9 AIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNP 68
AIGIDLGTTYSCVGVW NDRVEII NDQGNRTTPS VAFT+++RLIGDAAKNQ A NP
Sbjct: 8 AIGIDLGTTYSCVGVWM--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVALNP 65
Query: 69 SNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSV 128
NT+FD KRLIGRK+SD VQ D L WPFKV+SG KP I+V Y +EK+F EEISS+
Sbjct: 66 QNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQFSPEEISSM 125
Query: 129 ILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALA 188
+L KM+E+AEAFL VKNAV+TVPAYFNDSQR+ATKDAG I+GLNV+RIINEPTAAA+A
Sbjct: 126 VLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAIA 185
Query: 189 YGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
YGL K+ ++N+ IFDLGGGTFDVS+LT+++ +EVKATAGDTHLGGEDFDNR++ H
Sbjct: 186 YGLDKKGTKAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGEDFDNRLVNH 245
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSITRA 308
FV +F++K+ +DI+ N +ALRRLRTACERAKRTLS + TIE+D++++GIDF ++I+RA
Sbjct: 246 FVAEFRRKHKKDIAGNARALRRLRTACERAKRTLSSTAQTTIEIDSLHEGIDFYATISRA 305
Query: 309 KFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELC 368
+FEEMNM+LF KCM+ V L DAK++K+SV DVVLVGGS+RIPK++QLLQ+FF GKELC
Sbjct: 306 RFEEMNMDLFRKCMDPVEKVLKDAKLDKSSVHDVVLVGGSTRIPKIQQLLQDFFNGKELC 365
Query: 369 NSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRNTSI 427
SINPDEAVAYGAA+Q A+L E + V +L+L DV PLSLG+ G +M+V+IPRNT++
Sbjct: 366 KSINPDEAVAYGAAVQAAILTGEGSEKVQDLLLLDVAPLSLGLETAGGVMTVLIPRNTTV 425
Query: 428 PVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KVCFTI 485
P K+ + T D+Q VLI+VYEGER + DNNLLG F L +P APRG P VCF I
Sbjct: 426 PCKKEQVFSTYADNQPGVLIQVYEGERARTRDNNLLGTFELKGIPPAPRGVPQINVCFDI 485
Query: 486 DADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARN 545
DA+GILNVSA ++T+G KN+ITITN+KGRLS+ +IE+M+Q+AE +KA+D + KKKVEA+N
Sbjct: 486 DANGILNVSAEDKTAGVKNQITITNDKGRLSKEEIEKMVQDAEKYKAEDEQVKKKVEAKN 545
Query: 546 ALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFLKELE 605
+L++Y Y +R +K++ + KLT DK+KI A+ + I +Q + F LKELE
Sbjct: 546 SLENYAYNMRNTIKDEKLAQKLTQEDKQKIEKAIDETIEWIEGNQLAEVDEFEYKLKELE 605
Query: 606 SIFESAMNKINKG 618
I ++K+ +G
Sbjct: 606 GICNPIISKMYQG 618
>At5g28540 luminal binding protein
Length = 669
Score = 624 bits (1608), Expect = e-179
Identities = 325/606 (53%), Positives = 446/606 (72%), Gaps = 11/606 (1%)
Query: 7 GVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV++ N VEII NDQGNR TPS V FT+S+RLIG+AAKNQAA
Sbjct: 35 GSVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAV 92
Query: 67 NPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYM-GKEKRFVAEEI 125
NP T+FDVKRLIGRK+ D VQ DR L P+++++ + KP I VK G+ K F EEI
Sbjct: 93 NPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVN-KDGKPYIQVKIKDGETKVFSPEEI 151
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
S++IL+KM+E AEA+L +K+AV+TVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 152 SAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 211
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
A+AYGL K+ ++NI +FDLGGGTFDVS+LT+ + +EV +T GDTHLGGEDFD+R+
Sbjct: 212 AIAYGLDKKGG---EKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRV 268
Query: 246 LTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSI 305
+ +F+K KKK+ +DIS++ KAL +LR CERAKR LS + +E+++++ G+DFS +
Sbjct: 269 MEYFIKLIKKKHQKDISKDNKALGKLRRECERAKRALSSQHQVRVEIESLFDGVDFSEPL 328
Query: 306 TRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGK 365
TRA+FEE+N +LF K M V + DA ++K+ +D++VLVGGS+RIPKV+QLL++FF+GK
Sbjct: 329 TRARFEELNNDLFRKTMGPVKKAMDDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDFFEGK 388
Query: 366 ELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
E +NPDEAVAYGAA+Q +L E +++L DV PL+LG+ G +M+ +IPRN
Sbjct: 389 EPNKGVNPDEAVAYGAAVQGGILSGEGGDETKDILLLDVAPLTLGIETVGGVMTKLIPRN 448
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVC 482
T IP K++ + T +D Q+TV I+V+EGER D LLG F L+ +P APRG P +V
Sbjct: 449 TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLNGIPPAPRGTPQIEVT 508
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F +DA+GILNV A ++ SG +ITITNEKGRLS+ +I+RM++EAE F +D K K+K++
Sbjct: 509 FEVDANGILNVKAEDKASGKSEKITITNEKGRLSQEEIDRMVKEAEEFAEEDKKVKEKID 568
Query: 543 ARNALDDYLYKVR-KVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFL 601
ARNAL+ Y+Y ++ +V D + KL +KEKI +A + + ++Q + + + L
Sbjct: 569 ARNALETYVYNMKNQVNDKDKLADKLEGDEKEKIEAATKEALEWLDENQNSEKEEYDEKL 628
Query: 602 KELESI 607
KE+E++
Sbjct: 629 KEVEAV 634
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 621 bits (1602), Expect = e-178
Identities = 324/606 (53%), Positives = 445/606 (72%), Gaps = 11/606 (1%)
Query: 7 GVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV++ N VEII NDQGNR TPS V FT+S+RLIG+AAKNQAA
Sbjct: 35 GSVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAV 92
Query: 67 NPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYM-GKEKRFVAEEI 125
NP T+FDVKRLIGRK+ D VQ DR L P+++++ + KP I VK G+ K F EEI
Sbjct: 93 NPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVN-KDGKPYIQVKIKDGETKVFSPEEI 151
Query: 126 SSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAA 185
S++IL+KM+E AEA+L +K+AV+TVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 152 SAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 211
Query: 186 ALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRM 245
A+AYGL K+ ++NI +FDLGGGTFDVS+LT+ + +EV +T GDTHLGGEDFD+R+
Sbjct: 212 AIAYGLDKKGG---EKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRI 268
Query: 246 LTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSI 305
+ +F+K KKK+ +DIS++ KAL +LR CERAKR LS + +E+++++ G+D S +
Sbjct: 269 MEYFIKLIKKKHQKDISKDNKALGKLRRECERAKRALSSQHQVRVEIESLFDGVDLSEPL 328
Query: 306 TRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGK 365
TRA+FEE+N +LF K M V + DA ++K+ +D++VLVGGS+RIPKV+QLL++FF+GK
Sbjct: 329 TRARFEELNNDLFRKTMGPVKKAMDDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDFFEGK 388
Query: 366 ELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
E +NPDEAVAYGAA+Q +L E +++L DV PL+LG+ G +M+ +IPRN
Sbjct: 389 EPNKGVNPDEAVAYGAAVQGGILSGEGGDETKDILLLDVAPLTLGIETVGGVMTKLIPRN 448
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS-LPLAPRGHPH-KVC 482
T IP K++ + T +D Q+TV I+V+EGER D LLG F L+ +P APRG P +V
Sbjct: 449 TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLTGVPPAPRGTPQIEVT 508
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F +DA+GILNV A ++ SG +ITITNEKGRLS+ +I+RM++EAE F +D K K+K++
Sbjct: 509 FEVDANGILNVKAEDKASGKSEKITITNEKGRLSQEEIDRMVKEAEEFAEEDKKVKEKID 568
Query: 543 ARNALDDYLYKVR-KVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFL 601
ARNAL+ Y+Y ++ +V D + KL +KEKI +A + + ++Q + + + L
Sbjct: 569 ARNALETYVYNMKNQVSDKDKLADKLEGDEKEKIEAATKEALEWLDENQNSEKEEYDEKL 628
Query: 602 KELESI 607
KE+E++
Sbjct: 629 KEVEAV 634
>At1g09080 putative luminal binding protein
Length = 678
Score = 617 bits (1591), Expect = e-177
Identities = 327/629 (51%), Positives = 444/629 (69%), Gaps = 12/629 (1%)
Query: 3 EKYEGVAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKN 62
E+ G IGIDLGTTYSCVGV+ N VEII NDQGNR TPS VAFT+++RLIG+AAKN
Sbjct: 46 EQKLGTVIGIDLGTTYSCVGVYH--NKHVEIIANDQGNRITPSWVAFTDTERLIGEAAKN 103
Query: 63 QAATNPSNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVA 122
QAA NP TIFD KRLIGRK+ D VQ D P+KV++ + KP I VK G+EK F
Sbjct: 104 QAAKNPERTIFDPKRLIGRKFDDPDVQRDIKFLPYKVVN-KDGKPYIQVKVKGEEKLFSP 162
Query: 123 EEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEP 182
EEIS++IL+KM+E AEAFL +K+AVITVPAYFND+QR+ATKDAG IAGLNV+RIINEP
Sbjct: 163 EEISAMILTKMKETAEAFLGKKIKDAVITVPAYFNDAQRQATKDAGAIAGLNVVRIINEP 222
Query: 183 TAAALAYGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFD 242
T AA+AYGL K+ + NI ++DLGGGTFDVSILT+ + +EV +T+GDTHLGGEDFD
Sbjct: 223 TGAAIAYGLDKKGG---ESNILVYDLGGGTFDVSILTIDNGVFEVLSTSGDTHLGGEDFD 279
Query: 243 NRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFS 302
+R++ +F+K KKK N+DIS++ KAL +LR CE AKR+LS + +E+++++ G+DFS
Sbjct: 280 HRVMDYFIKLVKKKYNKDISKDHKALGKLRRECELAKRSLSNQHQVRVEIESLFDGVDFS 339
Query: 303 SSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFF 362
+TRA+FEE+NM+LF+K ME V L DA ++K+ +D++VLVGGS+RIPKV+Q+L++FF
Sbjct: 340 EPLTRARFEELNMDLFKKTMEPVKKALKDAGLKKSDIDEIVLVGGSTRIPKVQQMLKDFF 399
Query: 363 QGKELCNSINPDEAVAYGAAIQ-AVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVI 421
GKE NPDEAVAYGAA+Q VL E + N++L DV PLSLG+ G +M+ +I
Sbjct: 400 DGKEPSKGTNPDEAVAYGAAVQGGVLSGEGGEETQNILLLDVAPLSLGIETVGGVMTNII 459
Query: 422 PRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSLS--LPLAPRGHPH 479
PRNT IP K++ + T +D Q+TV I VYEGER DN LG F L+ LP APRG P
Sbjct: 460 PRNTVIPTKKSQVFTTYQDQQTTVTINVYEGERSMTKDNRELGKFDLTGILP-APRGVPQ 518
Query: 480 -KVCFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFK 538
+V F +DA+GIL V A ++ + ITITN+KGRL+ +IE MI+EAE F +D K
Sbjct: 519 IEVTFEVDANGILQVKAEDKVAKTSQSITITNDKGRLTEEEIEEMIREAEEFAEEDKIMK 578
Query: 539 KKVEARNALDDYLYKVRK-VMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVF 597
+K++ARN L+ Y+Y ++ V + + K++ DKEK+ + + + ++ + +
Sbjct: 579 EKIDARNKLETYVYNMKSTVADKEKLAKKISDEDKEKMEGVLKEALEWLEENVNAEKEDY 638
Query: 598 VDFLKELESIFESAMNKINKGDSDEKSDS 626
+ LKE+E + + + + + E D+
Sbjct: 639 DEKLKEVELVCDPVIKSVYEKTEGENEDT 667
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 449 bits (1155), Expect = e-126
Identities = 270/626 (43%), Positives = 378/626 (60%), Gaps = 28/626 (4%)
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
IGIDLGTT SCV V + + RV I N +G+RTTPS VA + L+G AK QA TNP
Sbjct: 55 IGIDLGTTNSCVSVMEGKTARV--IENAEGSRTTPSVVAMNQKGELLVGTPAKRQAVTNP 112
Query: 69 SNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSV 128
+NTIF KRLIGR++ D Q + + P+K++ N + ++F +I +
Sbjct: 113 TNTIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWV----EANGQKFSPSQIGAN 168
Query: 129 ILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALA 188
+L+KM+E AEA+L + AV+TVPAYFND+QR+ATKDAG+IAGL+V RIINEPTAAAL+
Sbjct: 169 VLTKMKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIINEPTAAALS 228
Query: 189 YGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
YG+ + I +FDLGGGTFDVSIL + +EVKAT GDT LGGEDFDN +L +
Sbjct: 229 YGMNNKEGV-----IAVFDLGGGTFDVSILEISSGVFEVKATNGDTFLGGEDFDNTLLEY 283
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID----FSSS 304
V +FK+ +N D++++ AL+RLR A E+AK LS T+ I L I + +
Sbjct: 284 LVNEFKRSDNIDLTKDNLALQRLREAAEKAKIELSSTTQTEINLPFITADASGAKHLNIT 343
Query: 305 ITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQG 364
+TR+KFE + L E+ +CL DA + VD+V+LVGG +R+PKV++++ E F G
Sbjct: 344 LTRSKFEGLVGKLIERTRSPCQNCLKDAGVTIKEVDEVLLVGGMTRVPKVQEIVSEIF-G 402
Query: 365 KELCNSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
K C +NPDEAVA GAAIQ + VK +L+L DV PLSLG+ G + + +IPRN
Sbjct: 403 KSPCKGVNPDEAVAMGAAIQGGILRGDVK---DLLLLDVVPLSLGIETLGAVFTKLIPRN 459
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KVC 482
T+IP K++ + T D+Q V IKV +GER +DN +LG F L +P APRG P +V
Sbjct: 460 TTIPTKKSQVFSTAADNQMQVGIKVLQGEREMAADNKVLGEFDLVGIPPAPRGMPQIEVT 519
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F IDA+GI VSA ++ +G + ITI G LS +I RM++EAE +D + K+ ++
Sbjct: 520 FDIDANGITTVSAKDKATGKEQNITI-RSSGGLSDDEINRMVKEAELNAQKDQEKKQLID 578
Query: 543 ARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGDSQQEDTFVFVDFLK 602
RN+ D +Y V K + K+ +I +A++ ++ + ED V+
Sbjct: 579 LRNSADTTIYSVEKSL--SEYREKIPAEIASEIETAVSDLRTAMAGEDVEDIKAKVEAAN 636
Query: 603 ELESIFESAMNKINKGDSDEKSDSDS 628
+ S M +KG SD S
Sbjct: 637 KAVSKIGEHM---SKGSGSSGSDGSS 659
>At4g24280 hsp 70-like protein
Length = 718
Score = 443 bits (1139), Expect = e-124
Identities = 270/618 (43%), Positives = 380/618 (60%), Gaps = 27/618 (4%)
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
+GIDLGTT S V + + I+ N +G RTTPS VA+T S RL+G AK QA NP
Sbjct: 81 VGIDLGTTNSAVAAME--GGKPTIVTNAEGQRTTPSVVAYTKSGDRLVGQIAKRQAVVNP 138
Query: 69 SNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSV 128
NT F VKR IGRK ++ V + ++V+ NN + ++ K+F AEEIS+
Sbjct: 139 ENTFFSVKRFIGRKMNE--VDEESKQVSYRVVRDENNN--VKLECPAINKQFAAEEISAQ 194
Query: 129 ILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALA 188
+L K+ + A FL V AVITVPAYFNDSQR ATKDAG IAGL V+RIINEPTAA+LA
Sbjct: 195 VLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLA 254
Query: 189 YGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
YG ++AN I +FDLGGGTFDVS+L V D +EV +T+GDTHLGG+DFD R++
Sbjct: 255 YGFDRKAN----ETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDW 310
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID----FSSS 304
+FKK D+ ++ +AL+RL A E+AK LS T+ + L I D ++
Sbjct: 311 LAAEFKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSLPFITATADGPKHIETT 370
Query: 305 ITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQG 364
+TRAKFEE+ +L ++ V + L DAK+ +D+V+LVGGS+RIP V++L+++ G
Sbjct: 371 LTRAKFEELCSDLLDRVRTPVENSLRDAKLSFKDIDEVILVGGSTRIPAVQELVRK-VTG 429
Query: 365 KELCNSINPDEAVAYGAAIQA-VLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPR 423
KE ++NPDE VA GAA+QA VL + V ++VL DVTPLS+G+ G +M+ +IPR
Sbjct: 430 KEPNVTVNPDEVVALGAAVQAGVLAGD----VSDIVLLDVTPLSIGLETLGGVMTKIIPR 485
Query: 424 NTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KV 481
NT++P ++ + T D Q++V I V +GER DN LG F L +P APRG P +V
Sbjct: 486 NTTLPTSKSEVFSTAADGQTSVEINVLQGEREFVRDNKSLGSFRLDGIPPAPRGVPQIEV 545
Query: 482 CFTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKV 541
F IDA+GIL+VSAV++ +G K +ITIT L + ++++M+QEAE F D + + +
Sbjct: 546 KFDIDANGILSVSAVDKGTGKKQDITITG-ASTLPKDEVDQMVQEAERFAKDDKEKRDAI 604
Query: 542 EARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGD-SQQEDTFVFVDF 600
+ +N D +Y+ K +K K+ KEK+ + + + K IG S QE
Sbjct: 605 DTKNQADSVVYQTEKQLK--ELGEKIPGEVKEKVEAKLQELKDKIGSGSTQEIKDAMAAL 662
Query: 601 LKELESIFESAMNKINKG 618
+E+ I +S N+ G
Sbjct: 663 NQEVMQIGQSLYNQPGAG 680
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 440 bits (1132), Expect = e-123
Identities = 266/620 (42%), Positives = 378/620 (60%), Gaps = 25/620 (4%)
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQ-RLIGDAAKNQAATNP 68
+GIDLGTT S V + + I+ N +G RTTPS VA+T S+ RL+G AK QA NP
Sbjct: 81 VGIDLGTTNSAVAAME--GGKPTIVTNAEGQRTTPSVVAYTKSKDRLVGQIAKRQAVVNP 138
Query: 69 SNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSV 128
NT F VKR IGR+ ++ + ++ ++VI N + +GK+ F AEEIS+
Sbjct: 139 ENTFFSVKRFIGRRMNEVAEESKQV--SYRVIKDENGNVKLDCPAIGKQ--FAAEEISAQ 194
Query: 129 ILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALA 188
+L K+ + A FL V AVITVPAYFNDSQR ATKDAG IAGL V+RIINEPTAA+LA
Sbjct: 195 VLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLA 254
Query: 189 YGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
YG ++++N I +FDLGGGTFDVS+L V D +EV +T+GDTHLGG+DFD R++
Sbjct: 255 YGFERKSN----ETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDW 310
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID----FSSS 304
FKK D+ ++ +AL+RL A E+AK LS T+ + L I D ++
Sbjct: 311 LASTFKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSLPFITATADGPKHIETT 370
Query: 305 ITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQG 364
+TR KFEE+ +L ++ V + L DAK+ +D+V+LVGGS+RIP V+ L+++ G
Sbjct: 371 LTRGKFEELCSDLLDRVRTPVENSLRDAKLSFKDIDEVILVGGSTRIPAVQDLVRK-LTG 429
Query: 365 KELCNSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
KE S+NPDE VA GAA+QA + S V ++VL DVTPLSLG+ G +M+ +IPRN
Sbjct: 430 KEPNVSVNPDEVVALGAAVQAGVLSGDVS---DIVLLDVTPLSLGLETLGGVMTKIIPRN 486
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KVC 482
T++P ++ + T D Q++V I V +GER DN +G F L +P APRG P +V
Sbjct: 487 TTLPTSKSEVFSTAADGQTSVEINVLQGEREFVRDNKSIGSFRLDGIPPAPRGVPQIEVK 546
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F IDA+GIL+VSA ++ +G K +ITIT L + +++ M+QEAE F +D + + ++
Sbjct: 547 FDIDANGILSVSASDKGTGKKQDITITG-ASTLPKDEVDTMVQEAERFAKEDKEKRDAID 605
Query: 543 ARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGD-SQQEDTFVFVDFL 601
+N D +Y+ K +K K+ KEK+ + + + K I S QE
Sbjct: 606 TKNQADSVVYQTEKQLK--ELGEKIPGPVKEKVEAKLQELKEKIASGSTQEIKDTMAALN 663
Query: 602 KELESIFESAMNKINKGDSD 621
+E+ I +S N+ G +D
Sbjct: 664 QEVMQIGQSLYNQPQPGGAD 683
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 439 bits (1128), Expect = e-123
Identities = 263/616 (42%), Positives = 377/616 (60%), Gaps = 37/616 (6%)
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAF-TNSQRLIGDAAKNQAATNP 68
IGIDLGTT SCV V + +N +V I N +G RTTPS VAF T + L+G AK QA TNP
Sbjct: 60 IGIDLGTTNSCVAVMEGKNPKV--IENAEGARTTPSVVAFNTKGELLVGTPAKRQAVTNP 117
Query: 69 SNTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSV 128
+NT+ KRLIGRK+ D Q + + P+K++ N + +++ +I +
Sbjct: 118 TNTVSGTKRLIGRKFDDPQTQKEMKMVPYKIVRAPNGDAWV----EANGQQYSPSQIGAF 173
Query: 129 ILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALA 188
IL+KM+E AEA+L V AV+TVPAYFND+QR+ATKDAG IAGL+V RIINEPTAAAL+
Sbjct: 174 ILTKMKETAEAYLGKSVTKAVVTVPAYFNDAQRQATKDAGRIAGLDVERIINEPTAAALS 233
Query: 189 YGLQKRANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
YG+ + I +FDLGGGTFDVS+L + + +EVKAT GDT LGGEDFDN +L
Sbjct: 234 YGMTNKEGL-----IAVFDLGGGTFDVSVLEISNGVFEVKATNGDTFLGGEDFDNALLDF 288
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGID----FSSS 304
V +FK D++++ AL+RLR A E+AK LS ++ I L I F+ +
Sbjct: 289 LVNEFKTTEGIDLAKDRLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHFNIT 348
Query: 305 ITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQG 364
+TR++FE + +L E+ + +CL DA + VD+V+LVGG +R+PKV+ ++ E F G
Sbjct: 349 LTRSRFETLVNHLIERTRDPCKNCLKDAGISAKEVDEVLLVGGMTRVPKVQSIVAEIF-G 407
Query: 365 KELCNSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRGDIMSVVIPRN 424
K +NPDEAVA GAA+Q + VK L+L DVTPLSLG+ G + + +I RN
Sbjct: 408 KSPSKGVNPDEAVAMGAALQGGILRGDVK---ELLLLDVTPLSLGIETLGGVFTRLITRN 464
Query: 425 TSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSL-SLPLAPRGHPH-KVC 482
T+IP K++ + T D+Q+ V I+V +GER +DN LLG F L +P +PRG P +V
Sbjct: 465 TTIPTKKSQVFSTAADNQTQVGIRVLQGEREMATDNKLLGEFDLVGIPPSPRGVPQIEVT 524
Query: 483 FTIDADGILNVSAVEETSGNKNEITITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVE 542
F IDA+GI+ VSA ++T+G +ITI G LS I++M++EAE +D + K+ ++
Sbjct: 525 FDIDANGIVTVSAKDKTTGKVQQITI-RSSGGLSEDDIQKMVREAELHAQKDKERKELID 583
Query: 543 ARNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKG-KSLIGDSQQEDTFVFVDFL 601
+N D +Y + K + +EKI S + K + + D + + D L
Sbjct: 584 TKNTADTTIYSIEKSLGE----------YREKIPSEIAKEIEDAVADLRSASS---GDDL 630
Query: 602 KELESIFESAMNKINK 617
E+++ E+A ++K
Sbjct: 631 NEIKAKIEAANKAVSK 646
>At1g11660 heat-shock protein like
Length = 773
Score = 259 bits (662), Expect = 3e-69
Identities = 188/685 (27%), Positives = 336/685 (48%), Gaps = 80/685 (11%)
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPS 69
+G D+G + V +++ ++++ ND+ NR P+ V+F QR +G AA A +P
Sbjct: 4 VGFDVGNENCVIAVAKQRG--IDVLLNDESNRENPAMVSFGEKQRFMGAAAAASATMHPK 61
Query: 70 NTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSVI 129
+TI +KRLIGRK+ + VQ D L+PF+ ++ I ++YMG+ + F +I ++
Sbjct: 62 STISQLKRLIGRKFREPDVQNDLRLFPFETSEDSDGGIQIRLRYMGEIQSFSPVQILGML 121
Query: 130 LSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALAY 189
LS +++IAE L++PV + VI +P+YF +SQR A DA IAGL +R++++ TA AL Y
Sbjct: 122 LSHLKQIAEKSLKTPVSDCVIGIPSYFTNSQRLAYLDAAAIAGLRPLRLMHDSTATALGY 181
Query: 190 GLQKR---ANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRML 246
G+ K AN +FI D+G V + + + + V++ A D +LGG DFD +
Sbjct: 182 GIYKTDLVANSSPTYIVFI-DIGHCDTQVCVASFESGSMRVRSHAFDRNLGGRDFDEVLF 240
Query: 247 THFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSIT 306
HF +FK+K N D+ N KA RLR +CE+ K+ LS + EA + ++ + + D S I
Sbjct: 241 NHFALEFKEKYNIDVYTNTKACVRLRASCEKVKKVLSANAEAQLNIECLMEEKDVRSFIK 300
Query: 307 RAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKE 366
R +FE+++ L E+ + LAD+ + + + V LVG SRIP + ++L F+ +E
Sbjct: 301 REEFEQLSAGLLERLIVPCQKALADSGLSLDQIHSVELVGSGSRIPAISKMLSSLFK-RE 359
Query: 367 LCNSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVS-----IRGDIMSVVI 421
L ++N E VA G A+Q + S V V + ++D P ++G S I ++
Sbjct: 360 LGRTVNASECVARGCALQCAMLS-PVFRVRDYEVQDSYPFAIGFSSDKGPINTPSNELLF 418
Query: 422 PRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDS-------DNNLLGLFSLSLPLAP 474
P+ P + V T ++T ++ + + S + ++G F +S A
Sbjct: 419 PKGQIFPSVK----VLTLHRENTFQLEAFYANHNELSPDIPTQISSFMIGPFHISHGEAA 474
Query: 475 RGHPHKVCFTIDADGILNV------------------------------------SAVEE 498
R KV ++ GI+ + SA+++
Sbjct: 475 R---VKVRVQLNLHGIVTIDSATLESKLSVSEQLIEYHKENITSEEMISEENHQSSAMKD 531
Query: 499 TSGNKNEITITNE---------------KGRLSRGQIERMIQEAENFKAQDMKFKKKVEA 543
S + + +I NE G L++ ++ Q + QD+K + +
Sbjct: 532 GSLDPSSGSIGNEPKAIKRMEIPVVANVSGALTKDELSEAKQRENSLVEQDLKMESTKDK 591
Query: 544 RNALDDYLYKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKS-LIGDSQQEDTFVFVDFLK 602
+NAL+ ++Y++R M N + + T ++E I + + + L D E +++ L
Sbjct: 592 KNALESFVYEMRDKMLN-TYRNTATESERECIARNLQETEEWLYEDGDDESENAYIEKLN 650
Query: 603 ELESIFESAMNKINKGDSDEKSDSD 627
+++ + + N+ G+ ++ D
Sbjct: 651 DVKKLIDPIENRFKDGEERVQASKD 675
>At1g79930 putative heat-shock protein
Length = 831
Score = 251 bits (640), Expect = 1e-66
Identities = 143/406 (35%), Positives = 223/406 (54%), Gaps = 5/406 (1%)
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPS 69
+G D G V V +++ ++++ ND+ NR TP+ V F + QR IG A NP
Sbjct: 4 VGFDFGNENCLVAVARQRG--IDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPK 61
Query: 70 NTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSVI 129
N+I +KRLIGR++SD +Q D PF V G + P I Y+G+++ F ++ ++
Sbjct: 62 NSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEKRAFTPTQVMGMM 121
Query: 130 LSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALAY 189
LS ++ IAE L + V + I +P YF D QRRA DA IAGL+ +R+I+E TA ALAY
Sbjct: 122 LSNLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLRLIHETTATALAY 181
Query: 190 GLQKR-ANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
G+ K D+ N+ D+G + V I K ++ + A D LGG DFD + H
Sbjct: 182 GIYKTDLPESDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNH 241
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSITRA 308
F KFK + D+S+N KA RLR CE+ K+ LS + A + ++ + D I R
Sbjct: 242 FAAKFKDEYKIDVSQNAKASLRLRATCEKLKKVLSANPLAPLNIECLMDEKDVRGVIKRE 301
Query: 309 KFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELC 368
+FEE+++ + E+ + L+DA + V V ++G SR+P + ++L EFF GKE
Sbjct: 302 EFEEISIPILERVKRPLEKALSDAGLTVEDVHMVEVIGSGSRVPAMIKILTEFF-GKEPR 360
Query: 369 NSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRG 414
++N E V+ G A+Q + S + K V + + P S+ ++ +G
Sbjct: 361 RTMNASECVSRGCALQCAILSPTFK-VREFQVHESFPFSISLAWKG 405
Score = 35.0 bits (79), Expect = 0.13
Identities = 21/96 (21%), Positives = 49/96 (50%), Gaps = 2/96 (2%)
Query: 513 GRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVMKNDSTSSKLTPVDK 572
G L ++E+ +++ QD ++ + +NA++ Y+Y +R + +D +T ++
Sbjct: 592 GALKTVEVEKAVEKEFEMALQDRVMEETKDRKNAVESYVYDMRNKL-SDKYQEYITDSER 650
Query: 573 EKINSAMTKGKS-LIGDSQQEDTFVFVDFLKELESI 607
E + + + + L D + E V+V L+EL+ +
Sbjct: 651 EAFLANLQEVEDWLYEDGEDETKGVYVAKLEELKKV 686
>At1g79920 putative heat-shock protein (At1g79920)
Length = 736
Score = 250 bits (638), Expect = 2e-66
Identities = 185/648 (28%), Positives = 295/648 (44%), Gaps = 93/648 (14%)
Query: 10 IGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPS 69
+G D G V V +++ ++++ ND+ NR TP+ V F + QR IG A NP
Sbjct: 4 VGFDFGNENCLVAVARQRG--IDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPK 61
Query: 70 NTIFDVKRLIGRKYSDSIVQMDRLLWPFKVISGANNKPTIIVKYMGKEKRFVAEEISSVI 129
N+I +KRLIGR++SD +Q D PF V G + P I Y+G+ + F ++ ++
Sbjct: 62 NSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEIRAFTPTQVMGMM 121
Query: 130 LSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEPTAAALAY 189
LS ++ IAE L + V + I +P YF D QRRA DA IAGL+ + +I+E TA ALAY
Sbjct: 122 LSNLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLHLIHETTATALAY 181
Query: 190 GLQKR-ANFVDKRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHLGGEDFDNRMLTH 248
G+ K D+ N+ D+G + V I K ++ + A D LGG DFD + H
Sbjct: 182 GIYKTDLPENDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNH 241
Query: 249 FVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFSSSITRA 308
F KFK + D+S+N KA RLR CE+ K+ LS + A + ++ + D I R
Sbjct: 242 FAAKFKDEYKIDVSQNAKASLRLRATCEKLKKVLSANPMAPLNIECLMAEKDVRGVIKRE 301
Query: 309 KFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEFFQGKELC 368
+FEE+++ + E+ + L+DA + V V +VG SR+P + ++L EFF GKE
Sbjct: 302 EFEEISIPILERVKRPLEKALSDAGLTVEDVHMVEVVGSGSRVPAMIKILTEFF-GKEPR 360
Query: 369 NSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDVTPLSLGVSIRG------------DI 416
++N E V+ G A+Q + S + K V + + P S+ ++ +G
Sbjct: 361 RTMNASECVSRGCALQCAILSPTFK-VREFQVHESFPFSISLAWKGAATDAQNGGTENQQ 419
Query: 417 MSVVIPRNTSIP-VKRTN-----------EYVTTEDDQSTVLIKVY--------EGERLK 456
++V P+ IP VK +Y D Q+ I Y +GER K
Sbjct: 420 STIVFPKGNPIPSVKALTFYRSGTFSIDVQYSDVNDLQAPPKISTYTIGPFQSSKGERAK 479
Query: 457 DSDN---NLLGLFSLSLPLAPRGHPHKVCFTID---------------------ADGILN 492
NL G+ S+ +V T D D +N
Sbjct: 480 LKVKVRLNLHGIVSVESATLLEEEEVEVSVTKDQSEETAKMDTDKASAEAAPASGDSDVN 539
Query: 493 VSAVEETS---GNKNEITITNEK-----------------------------GRLSRGQI 520
+ ++TS G N + + EK G L ++
Sbjct: 540 MQDAKDTSDATGTDNGVPESAEKPVQMETDSKAEAPKKKVKKTNVPLSELVYGALKTVEV 599
Query: 521 ERMIQEAENFKAQDMKFKKKVEARNALDDYLYKVRKVMKNDSTSSKLT 568
E+ +++ QD ++ + +NA++ Y+Y +R + + + LT
Sbjct: 600 EKAVEKEFEMALQDRVMEETKDRKNAVESYVYDMRNKLSDKYQNISLT 647
>At2g32120 70kD heat shock protein
Length = 563
Score = 241 bits (615), Expect = 9e-64
Identities = 170/514 (33%), Positives = 260/514 (50%), Gaps = 33/514 (6%)
Query: 8 VAIGIDLGTTYSCVGVWQEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATN 67
+A+GID+GT+ + VW +V I+ N + + S V F + G NQ A
Sbjct: 29 IALGIDIGTSQCSIAVWN--GSQVHILRNTRNQKLIKSFVTFKDEVPAGG--VSNQLAHE 84
Query: 68 PSN----TIFDVKRLIGRKYSDSIVQMDRLLWPFKVIS-GANNKPTIIVKYMGKEKRFVA 122
IF++KRL+GR +D +V + L PF V + +P I +
Sbjct: 85 QEMLTGAAIFNMKRLVGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNAWRSTTP 143
Query: 123 EEISSVILSKMREIAEAFLESPVKNAVITVPAYFNDSQRRATKDAGEIAGLNVMRIINEP 182
EE+ ++ L ++R +AEA L+ PV+N V+TVP F+ Q + A +AGL+V+R++ EP
Sbjct: 144 EEVLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQLTRFERACAMAGLHVLRLMPEP 203
Query: 183 TAAALAYGLQKRANFVD------KRNIFIFDLGGGTFDVSILTVKDNNYEVKATAGDTHL 236
TA AL Y Q++ D +R IF++G G DV++ ++KA AG +
Sbjct: 204 TAIALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDVAVTATAGGVSQIKALAGSP-I 262
Query: 237 GGEDFDNRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIY 296
GGED + H N +A LR A + A L+ IE+D +
Sbjct: 263 GGEDILQNTIRHIAPP-----------NEEASGLLRVAAQDAIHRLTDQENVQIEVD-LG 310
Query: 297 KGIDFSSSITRAKFEEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQ 356
G S + R +FEE+N +FE+C V CL DA++ +DD+++VGG S IPKVR
Sbjct: 311 NGNKISKVLDRLEFEEVNQKVFEECERLVVQCLRDARVNGGDIDDLIMVGGCSYIPKVRT 370
Query: 357 LLQEFFQGKELCNSINPDEAVAYGAAIQAVLHSESVKSVPNLVLRDV--TPLSLGVSIRG 414
+++ + E+ +NP EA GAA++ + S +L L + TPL++GV G
Sbjct: 371 IIKNVCKKDEIYKGVNPLEAAVRGAALEGAVTSGIHDPFGSLDLLTIQATPLAVGVRANG 430
Query: 415 DIMSVVIPRNTSIPVKRTNEYVTTEDDQSTVLIKVYEGERLKDSDNNLLGLFSL-SLPLA 473
+ VIPRNT +P ++ + T +D+Q LI +YEGE +N+LLG F L +P A
Sbjct: 431 NKFIPVIPRNTMVPARKDLFFTTVQDNQKEALIIIYEGEGETVEENHLLGYFKLVGIPPA 490
Query: 474 PRGHPH-KVCFTIDADGILNVSAVEETSGNKNEI 506
P+G P VC IDA L V A G+ + +
Sbjct: 491 PKGVPEINVCMDIDASNALRVFAAVLMPGSSSPV 524
>At3g19370 unknown protein
Length = 704
Score = 38.1 bits (87), Expect = 0.015
Identities = 26/97 (26%), Positives = 49/97 (49%), Gaps = 8/97 (8%)
Query: 496 VEETSGNKNEITIT----NEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEARNALDDYL 551
VE G ++ + IT NE G +++ +I+R + ++ K + ++ +K E R L++ +
Sbjct: 435 VERKKGEEDPLCITWKSNNESGPMTKDEIKRHLGLTKSDKVEKIESDEKQELRKKLEESV 494
Query: 552 YKVRKVMKNDSTSSKLTPVDKEKINSAMTKGKSLIGD 588
K+R N K +KEK+ + M KS+ D
Sbjct: 495 EKIR----NLEAEMKTLRENKEKVEAEMETEKSMKED 527
>At5g55820 unknown protein
Length = 1826
Score = 35.8 bits (81), Expect = 0.074
Identities = 29/98 (29%), Positives = 47/98 (47%), Gaps = 7/98 (7%)
Query: 452 GERLKDSD---NNLLGLFSLSLPLAPRGHPHKVCFTIDADGILNVSAVEETSGNKN--EI 506
G RL++ + NN++ + +PL + P T D + V A+E +K E
Sbjct: 1477 GPRLQEKEPRHNNIVSNITSFVPLVQQQKPAPALITGKRD--VKVKALEAAEASKRIAEQ 1534
Query: 507 TITNEKGRLSRGQIERMIQEAENFKAQDMKFKKKVEAR 544
+ K + ++ER QE EN K Q+++ KKK E R
Sbjct: 1535 KENDRKLKKEAMKLERAKQEQENLKKQEIEKKKKEEDR 1572
>At2g41720 salt-inducible protein-like
Length = 822
Score = 35.4 bits (80), Expect = 0.097
Identities = 29/126 (23%), Positives = 60/126 (47%), Gaps = 5/126 (3%)
Query: 243 NRMLTHFVKKFKKKNNEDISRNPKALRRLRTACERAKRTLSYDTEATIELDAIYKGIDFS 302
N L V+ F++ + I N ++ L AC R+K+ ++ DT + A +GI+ +
Sbjct: 438 NGFLAEAVEIFRQMEQDGIKPNVVSVCTLLAACSRSKKKVNVDTVLSA---AQSRGINLN 494
Query: 303 SSITRAKF-EEMNMNLFEKCMETVNSCLADAKMEKNSVDDVVLVGGSSRIPKVRQLLQEF 361
++ + +N EK + S + K++ +SV +L+ GS R+ K + +
Sbjct: 495 TAAYNSAIGSYINAAELEKAIALYQS-MRKKKVKADSVTFTILISGSCRMSKYPEAISYL 553
Query: 362 FQGKEL 367
+ ++L
Sbjct: 554 KEMEDL 559
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.315 0.132 0.363
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,361,612
Number of Sequences: 26719
Number of extensions: 572233
Number of successful extensions: 1516
Number of sequences better than 10.0: 49
Number of HSP's better than 10.0 without gapping: 26
Number of HSP's successfully gapped in prelim test: 23
Number of HSP's that attempted gapping in prelim test: 1394
Number of HSP's gapped (non-prelim): 65
length of query: 628
length of database: 11,318,596
effective HSP length: 105
effective length of query: 523
effective length of database: 8,513,101
effective search space: 4452351823
effective search space used: 4452351823
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 42 (22.0 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144722.19


