

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.18 + phase: 0 /pseudo
(433 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
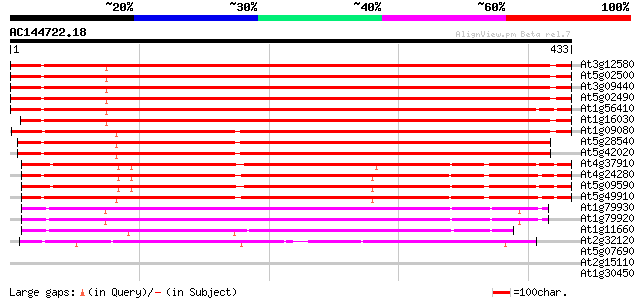
Score E
Sequences producing significant alignments: (bits) Value
At3g12580 putative protein 578 e-165
At5g02500 dnaK-type molecular chaperone hsc70.1 568 e-162
At3g09440 heat-shock protein (At-hsc70-3) 566 e-161
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 564 e-161
At1g56410 hypothetical protein 563 e-161
At1g16030 unknown protein 551 e-157
At1g09080 putative luminal binding protein 465 e-131
At5g28540 luminal binding protein 443 e-125
At5g42020 luminal binding protein (dbj|BAA13948.1) 441 e-124
At4g37910 heat shock protein 70 like protein 338 3e-93
At4g24280 hsp 70-like protein 332 2e-91
At5g09590 heat shock protein 70 (Hsc70-5) 331 5e-91
At5g49910 heat shock protein 70 (Hsc70-7) 327 1e-89
At1g79930 putative heat-shock protein 224 9e-59
At1g79920 putative heat-shock protein (At1g79920) 222 3e-58
At1g11660 heat-shock protein like 222 3e-58
At2g32120 70kD heat shock protein 185 5e-47
At5g07690 transcription factor-like protein 32 0.89
At2g15110 unknown protein 31 1.2
At1g30450 cation-chloride co-transporter, putative 30 2.0
>At3g12580 putative protein
Length = 650
Score = 578 bits (1490), Expect = e-165
Identities = 299/456 (65%), Positives = 365/456 (79%), Gaps = 29/456 (6%)
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP+NT+F FKV++G +KP IVVN+KGEEK+F
Sbjct: 59 KNQVAMNPTNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQF 118
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KMREIAEAFL +PVKNAV++VPAYFNDSQR+ATKDAG I+GLNVM+IIN
Sbjct: 119 SAEEISSMVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A+ V ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 179 EPTAAAIAYGLDKKASSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DI+ N +ALRRLRTACE+AKRTLS TIE+D++++GID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID 298
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F ++ITRA+FEE+NM+LF KCM V CL DAKMDK+SV DVVLVGGS+RIPKV+QLLQ+
Sbjct: 299 FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQLLQD 358
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ G +M+V
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 418
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+IPRNT+IP E+ + Y N+ V ++VY+
Sbjct: 419 LIPRNTTIPTKKEQIFSTYS----DNQPGVLIQVYE 450
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 568 bits (1464), Expect = e-162
Identities = 295/456 (64%), Positives = 355/456 (77%), Gaps = 29/456 (6%)
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MSGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP NT+F FK+ AG DKP I V YKGEEK F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDSSVQSDMKLWPFKIQAGPADKPMIYVEYKGEEKEF 118
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KMREIAEA+L +KNAV++VPAYFNDSQR+ATKDAG IAGLNVM+IIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DI+ N +ALRRLRT+CE+AKRTLS TIE+D++Y+GID
Sbjct: 239 FDNRMVNHFVQEFKRKSKKDITGNPRALRRLRTSCERAKRTLSSTAQTTIEIDSLYEGID 298
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F S+ITRA+FEE+NM+LF KCM V CL DAKMDK++V DVVLVGGS+RIPKV+QLLQ+
Sbjct: 299 FYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 358
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQ A+LS EG++ V +L+L DVTPLS G+ G +M+
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQGAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTT 418
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+IPRNT+IP E+ + Y N+ V ++VY+
Sbjct: 419 LIPRNTTIPTKKEQVFSTYS----DNQPGVLIQVYE 450
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 566 bits (1458), Expect = e-161
Identities = 296/456 (64%), Positives = 356/456 (77%), Gaps = 29/456 (6%)
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP NT+F F + +G +KP IVVNYKGE+K F
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKSGPAEKPMIVVNYKGEDKEF 118
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS+IL KMREIAEA+L T +KNAV++VPAYFNDSQR+ATKDAG IAGLNVM+IIN
Sbjct: 119 SAEEISSMILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DIS N +ALRRLRTACE+AKRTLS TIE+D+++ GID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFDGID 298
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F + ITRA+FEE+N++LF KCM V CL DAKMDKNS+DDVVLVGGS+RIPKV+QLL +
Sbjct: 299 FYAPITRARFEELNIDLFRKCMEPVEKCLRDAKMDKNSIDDVVLVGGSTRIPKVQQLLVD 358
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ G +M+V
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 418
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+I RNT+IP E+ + Y N+ V ++VY+
Sbjct: 419 LIQRNTTIPTKKEQVFSTYS----DNQPGVLIQVYE 450
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 564 bits (1453), Expect = e-161
Identities = 294/456 (64%), Positives = 355/456 (77%), Gaps = 29/456 (6%)
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP NT+F F +I+GT +KP IVV YKGEEK+F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDASVQSDRQLWPFTIISGTAEKPMIVVEYKGEEKQF 118
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KMREIAEAFL T VKNAV++VPAYFNDSQR+ATKDAG IAGLNV++IIN
Sbjct: 119 AAEEISSMVLIKMREIAEAFLGTTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A V ++N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DI+ +ALRRLRTACE+AKRTLS TIE+D++Y G D
Sbjct: 239 FDNRMVNHFVQEFKRKNKQDITGQPRALRRLRTACERAKRTLSSTAQTTIEIDSLYGGAD 298
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F S ITRA+FEEMNM+LF KCM V CL DAKMDK++V ++VLVGGS+RIPKV+QLLQ+
Sbjct: 299 FYSPITRARFEEMNMDLFRKCMEPVEKCLRDAKMDKSTVHEIVLVGGSTRIPKVQQLLQD 358
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ G +M+
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTT 418
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+I RNT+IP E+ + Y N+ V ++V++
Sbjct: 419 LIQRNTTIPTKKEQVFSTYS----DNQPGVLIQVFE 450
>At1g56410 hypothetical protein
Length = 617
Score = 563 bits (1452), Expect = e-161
Identities = 295/456 (64%), Positives = 357/456 (77%), Gaps = 29/456 (6%)
Query: 1 MEEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAA 60
M K EG AIGIDLGTTYSCVGVW Q+DRVEII NDQGNRTTPS VAFT+S+RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVW--QHDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAATNPSNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRF 98
KNQ A NP NT+F FKV G DKP I VNYKGEEK+F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDASVQSDMKFWPFKVTPGQADKPMIFVNYKGEEKQF 118
Query: 99 VAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIIN 158
AEEISS++L KMREIAEA+L + +KNAV++VPAYFNDSQR+ATKDAG IAGLNV++IIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 159 EPTAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQD 218
EPTAAA+AYGL K+A V +N+ IFDLGGGTFDVS+LT++ +FEVKATAGDTHLGG+D
Sbjct: 179 EPTAAAIAYGLDKKATSVGIKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 219 FDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID 278
FDN MV+HFV+EF+ K K+DIS +++ALRRLRTACE+AKRTLS T+E+D++++GID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDISGDARALRRLRTACERAKRTLSSTAQTTVEVDSLFEGID 298
Query: 279 FCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQE 338
F S ITRAKFEEMNM+LF KCM V CL D+KMDK+ V DVVLVGGS+RIPKV+QLLQ+
Sbjct: 299 FYSPITRAKFEEMNMDLFRKCMEPVMKCLRDSKMDKSMVHDVVLVGGSTRIPKVQQLLQD 358
Query: 339 VFKGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSV 397
F GKELCKSINPDEAVAYGA VQAA+LS EG++ V +L+L DVTPLS G+ +G +M+
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGIETIGGVMTT 418
Query: 398 MIPRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+I RNT+IP +K + + D N+ V ++VY+
Sbjct: 419 LIQRNTTIP--AKKEQEFTTTVD--NQPDVLIQVYE 450
>At1g16030 unknown protein
Length = 646
Score = 551 bits (1421), Expect = e-157
Identities = 283/448 (63%), Positives = 353/448 (78%), Gaps = 29/448 (6%)
Query: 9 AIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNP 68
AIGIDLGTTYSCVGVW NDRVEII NDQGNRTTPS VAFT+++RLIGDAAKNQ A NP
Sbjct: 8 AIGIDLGTTYSCVGVW--MNDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVALNP 65
Query: 69 SNTIF----------------------AFKVIAGTNDKPTIVVNYKGEEKRFVAEEISSV 106
NT+F FKV++G +KP IVV+YK EEK+F EEISS+
Sbjct: 66 QNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQFSPEEISSM 125
Query: 107 ILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALA 166
+L KM+E+AEAFL VKNAV++VPAYFNDSQR+ATKDAGAI+GLNV++IINEPTAAA+A
Sbjct: 126 VLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAIA 185
Query: 167 YGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDH 226
YGL K+ ++N+ IFDLGGGTFDVS+LT++ VFEVKATAGDTHLGG+DFDN +V+H
Sbjct: 186 YGLDKKGTKAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGEDFDNRLVNH 245
Query: 227 FVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRA 286
FV EFR K+K+DI+ N++ALRRLRTACE+AKRTLS TIE+D++++GIDF ++I+RA
Sbjct: 246 FVAEFRRKHKKDIAGNARALRRLRTACERAKRTLSSTAQTTIEIDSLHEGIDFYATISRA 305
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
+FEEMNM+LF KCM+ V L DAK+DK+SV DVVLVGGS+RIPK++QLLQ+ F GKELC
Sbjct: 306 RFEEMNMDLFRKCMDPVEKVLKDAKLDKSSVHDVVLVGGSTRIPKIQQLLQDFFNGKELC 365
Query: 347 KSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSI 405
KSINPDEAVAYGA VQAA+L+ EGS+ V +L+L DV PLS G+ G +M+V+IPRNT++
Sbjct: 366 KSINPDEAVAYGAAVQAAILTGEGSEKVQDLLLLDVAPLSLGLETAGGVMTVLIPRNTTV 425
Query: 406 PVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
P E+ + Y N+ V ++VY+
Sbjct: 426 PCKKEQVFSTYA----DNQPGVLIQVYE 449
>At1g09080 putative luminal binding protein
Length = 678
Score = 465 bits (1196), Expect = e-131
Identities = 247/454 (54%), Positives = 323/454 (70%), Gaps = 31/454 (6%)
Query: 2 EEKYEGVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAK 61
EE+ G IGIDLGTTYSCVGV+H N VEII NDQGNR TPS VAFT+++RLIG+AAK
Sbjct: 45 EEQKLGTVIGIDLGTTYSCVGVYH--NKHVEIIANDQGNRITPSWVAFTDTERLIGEAAK 102
Query: 62 NQAATNPSNTIFAFKVIAGT---------------------NDKPTIVVNYKGEEKRFVA 100
NQAA NP TIF K + G + KP I V KGEEK F
Sbjct: 103 NQAAKNPERTIFDPKRLIGRKFDDPDVQRDIKFLPYKVVNKDGKPYIQVKVKGEEKLFSP 162
Query: 101 EEISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEP 160
EEIS++IL+KM+E AEAFL +K+AVI+VPAYFND+QR+ATKDAGAIAGLNV++IINEP
Sbjct: 163 EEISAMILTKMKETAEAFLGKKIKDAVITVPAYFNDAQRQATKDAGAIAGLNVVRIINEP 222
Query: 161 TAAALAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFD 220
T AA+AYGL K+ + NI ++DLGGGTFDVSILT+ N VFEV +T+GDTHLGG+DFD
Sbjct: 223 TGAAIAYGLDKKGG---ESNILVYDLGGGTFDVSILTIDNGVFEVLSTSGDTHLGGEDFD 279
Query: 221 NIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFC 280
+ ++D+F+K + KY +DIS++ KAL +LR CE AKR+LS +E+++++ G+DF
Sbjct: 280 HRVMDYFIKLVKKKYNKDISKDHKALGKLRRECELAKRSLSNQHQVRVEIESLFDGVDFS 339
Query: 281 SSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVF 340
+TRA+FEE+NM+LF+K M V L DA + K+ +D++VLVGGS+RIPKV+Q+L++ F
Sbjct: 340 EPLTRARFEELNMDLFKKTMEPVKKALKDAGLKKSDIDEIVLVGGSTRIPKVQQMLKDFF 399
Query: 341 KGKELCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMI 399
GKE K NPDEAVAYGA VQ +LS EG + N++L DV PLS G+ +G +M+ +I
Sbjct: 400 DGKEPSKGTNPDEAVAYGAAVQGGVLSGEGGEETQNILLLDVAPLSLGIETVGGVMTNII 459
Query: 400 PRNTSIPVTVEKTKTYYECYDYYNKYSVTVKVYQ 433
PRNT IP + T Y+ + +VT+ VY+
Sbjct: 460 PRNTVIPTKKSQVFTTYQ----DQQTTVTINVYE 489
>At5g28540 luminal binding protein
Length = 669
Score = 443 bits (1140), Expect = e-125
Identities = 231/434 (53%), Positives = 310/434 (71%), Gaps = 28/434 (6%)
Query: 7 GVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV+ +N VEII NDQGNR TPS V FT+S+RLIG+AAKNQAA
Sbjct: 35 GSVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAV 92
Query: 67 NPSNTIFAFKVIAGT---------------------NDKPTIVVNYK-GEEKRFVAEEIS 104
NP T+F K + G + KP I V K GE K F EEIS
Sbjct: 93 NPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVNKDGKPYIQVKIKDGETKVFSPEEIS 152
Query: 105 SVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAA 164
++IL+KM+E AEA+L +K+AV++VPAYFND+QR+ATKDAG IAGLNV +IINEPTAAA
Sbjct: 153 AMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAA 212
Query: 165 LAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMV 224
+AYGL K+ ++NI +FDLGGGTFDVS+LT+ N VFEV +T GDTHLGG+DFD+ ++
Sbjct: 213 IAYGLDKKGG---EKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRVM 269
Query: 225 DHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSIT 284
++F+K + K+++DIS+++KAL +LR CE+AKR LS +E+++++ G+DF +T
Sbjct: 270 EYFIKLIKKKHQKDISKDNKALGKLRRECERAKRALSSQHQVRVEIESLFDGVDFSEPLT 329
Query: 285 RAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKE 344
RA+FEE+N +LF K M V + DA + K+ +D++VLVGGS+RIPKV+QLL++ F+GKE
Sbjct: 330 RARFEELNNDLFRKTMGPVKKAMDDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDFFEGKE 389
Query: 345 LCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNT 403
K +NPDEAVAYGA VQ +LS EG +++L DV PL+ G+ +G +M+ +IPRNT
Sbjct: 390 PNKGVNPDEAVAYGAAVQGGILSGEGGDETKDILLLDVAPLTLGIETVGGVMTKLIPRNT 449
Query: 404 SIPVTVEKTKTYYE 417
IP + T Y+
Sbjct: 450 VIPTKKSQVFTTYQ 463
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 441 bits (1134), Expect = e-124
Identities = 230/434 (52%), Positives = 309/434 (70%), Gaps = 28/434 (6%)
Query: 7 GVAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAAT 66
G IGIDLGTTYSCVGV+ +N VEII NDQGNR TPS V FT+S+RLIG+AAKNQAA
Sbjct: 35 GSVIGIDLGTTYSCVGVY--KNGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAV 92
Query: 67 NPSNTIFAFKVIAGT---------------------NDKPTIVVNYK-GEEKRFVAEEIS 104
NP T+F K + G + KP I V K GE K F EEIS
Sbjct: 93 NPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIVNKDGKPYIQVKIKDGETKVFSPEEIS 152
Query: 105 SVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAA 164
++IL+KM+E AEA+L +K+AV++VPAYFND+QR+ATKDAG IAGLNV +IINEPTAAA
Sbjct: 153 AMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAAA 212
Query: 165 LAYGLQKRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMV 224
+AYGL K+ ++NI +FDLGGGTFDVS+LT+ N VFEV +T GDTHLGG+DFD+ ++
Sbjct: 213 IAYGLDKKGG---EKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRIM 269
Query: 225 DHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSIT 284
++F+K + K+++DIS+++KAL +LR CE+AKR LS +E+++++ G+D +T
Sbjct: 270 EYFIKLIKKKHQKDISKDNKALGKLRRECERAKRALSSQHQVRVEIESLFDGVDLSEPLT 329
Query: 285 RAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKE 344
RA+FEE+N +LF K M V + DA + K+ +D++VLVGGS+RIPKV+QLL++ F+GKE
Sbjct: 330 RARFEELNNDLFRKTMGPVKKAMDDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDFFEGKE 389
Query: 345 LCKSINPDEAVAYGAVVQAALLS-EGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNT 403
K +NPDEAVAYGA VQ +LS EG +++L DV PL+ G+ +G +M+ +IPRNT
Sbjct: 390 PNKGVNPDEAVAYGAAVQGGILSGEGGDETKDILLLDVAPLTLGIETVGGVMTKLIPRNT 449
Query: 404 SIPVTVEKTKTYYE 417
IP + T Y+
Sbjct: 450 VIPTKKSQVFTTYQ 463
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 338 bits (867), Expect = 3e-93
Identities = 202/447 (45%), Positives = 276/447 (61%), Gaps = 38/447 (8%)
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
IGIDLGTT SCV V + RV I N +G+RTTPS VA + L+G AK QA TNP
Sbjct: 55 IGIDLGTTNSCVSVMEGKTARV--IENAEGSRTTPSVVAMNQKGELLVGTPAKRQAVTNP 112
Query: 69 SNTIFAFKVIAGTN------DKPTIVVNYK------------GEEKRFVAEEISSVILSK 110
+NTIF K + G K +V YK ++F +I + +L+K
Sbjct: 113 TNTIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEANGQKFSPSQIGANVLTK 172
Query: 111 MREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQ 170
M+E AEA+L + AV++VPAYFND+QR+ATKDAG IAGL+V +IINEPTAAAL+YG+
Sbjct: 173 MKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIINEPTAAALSYGMN 232
Query: 171 KRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKE 230
+ I +FDLGGGTFDVSIL + + VFEVKAT GDT LGG+DFDN ++++ V E
Sbjct: 233 NKEGV-----IAVFDLGGGTFDVSILEISSGVFEVKATNGDTFLGGEDFDNTLLEYLVNE 287
Query: 231 FRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCS----SITRA 286
F+ D+++++ AL+RLR A EKAK LS T I L I ++TR+
Sbjct: 288 FKRSDNIDLTKDNLALQRLREAAEKAKIELSSTTQTEINLPFITADASGAKHLNITLTRS 347
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
KFE + L E+ + CL DA + VD+V+LVGG +R+PKV++++ E+F GK C
Sbjct: 348 KFEGLVGKLIERTRSPCQNCLKDAGVTIKEVDEVLLVGGMTRVPKVQEIVSEIF-GKSPC 406
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIP 406
K +NPDEAVA GA +Q +L +V +L+L DV PLS G+ LG + + +IPRNT+IP
Sbjct: 407 KGVNPDEAVAMGAAIQGGIL---RGDVKDLLLLDVVPLSLGIETLGAVFTKLIPRNTTIP 463
Query: 407 VTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+K++ + D N+ V +KV Q
Sbjct: 464 --TKKSQVFSTAAD--NQMQVGIKVLQ 486
>At4g24280 hsp 70-like protein
Length = 718
Score = 332 bits (851), Expect = 2e-91
Identities = 204/447 (45%), Positives = 276/447 (61%), Gaps = 37/447 (8%)
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNS-QRLIGDAAKNQAATNP 68
+GIDLGTT S V + + I+ N +G RTTPS VA+T S RL+G AK QA NP
Sbjct: 81 VGIDLGTTNSAVAAM--EGGKPTIVTNAEGQRTTPSVVAYTKSGDRLVGQIAKRQAVVNP 138
Query: 69 SNTIFAFKVIAGTN----DKPTIVVNYK--------------GEEKRFVAEEISSVILSK 110
NT F+ K G D+ + V+Y+ K+F AEEIS+ +L K
Sbjct: 139 ENTFFSVKRFIGRKMNEVDEESKQVSYRVVRDENNNVKLECPAINKQFAAEEISAQVLRK 198
Query: 111 MREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQ 170
+ + A FL V AVI+VPAYFNDSQR ATKDAG IAGL V++IINEPTAA+LAYG
Sbjct: 199 LVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLAYGFD 258
Query: 171 KRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKE 230
++AN I +FDLGGGTFDVS+L V + VFEV +T+GDTHLGG DFD +VD E
Sbjct: 259 RKAN----ETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDWLAAE 314
Query: 231 FRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID----FCSSITRA 286
F+ D+ ++ +AL+RL A EKAK LS T + L I D +++TRA
Sbjct: 315 FKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSLPFITATADGPKHIETTLTRA 374
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
KFEE+ +L ++ V L DAK+ +D+V+LVGGS+RIP V++L+++V GKE
Sbjct: 375 KFEELCSDLLDRVRTPVENSLRDAKLSFKDIDEVILVGGSTRIPAVQELVRKV-TGKEPN 433
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIP 406
++NPDE VA GA VQA +L + +V ++VL DVTPLS G+ LG +M+ +IPRNT++P
Sbjct: 434 VTVNPDEVVALGAAVQAGVL---AGDVSDIVLLDVTPLSIGLETLGGVMTKIIPRNTTLP 490
Query: 407 VTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+ K++ + D + SV + V Q
Sbjct: 491 TS--KSEVFSTAAD--GQTSVEINVLQ 513
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 331 bits (848), Expect = 5e-91
Identities = 201/447 (44%), Positives = 274/447 (60%), Gaps = 38/447 (8%)
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAF-TNSQRLIGDAAKNQAATNP 68
IGIDLGTT SCV V +N +V I N +G RTTPS VAF T + L+G AK QA TNP
Sbjct: 60 IGIDLGTTNSCVAVMEGKNPKV--IENAEGARTTPSVVAFNTKGELLVGTPAKRQAVTNP 117
Query: 69 SNTIFAFKVIAGTN------DKPTIVVNYK------------GEEKRFVAEEISSVILSK 110
+NT+ K + G K +V YK +++ +I + IL+K
Sbjct: 118 TNTVSGTKRLIGRKFDDPQTQKEMKMVPYKIVRAPNGDAWVEANGQQYSPSQIGAFILTK 177
Query: 111 MREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQ 170
M+E AEA+L V AV++VPAYFND+QR+ATKDAG IAGL+V +IINEPTAAAL+YG+
Sbjct: 178 MKETAEAYLGKSVTKAVVTVPAYFNDAQRQATKDAGRIAGLDVERIINEPTAAALSYGMT 237
Query: 171 KRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKE 230
+ I +FDLGGGTFDVS+L + N VFEVKAT GDT LGG+DFDN ++D V E
Sbjct: 238 NKEGL-----IAVFDLGGGTFDVSVLEISNGVFEVKATNGDTFLGGEDFDNALLDFLVNE 292
Query: 231 FRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID----FCSSITRA 286
F+ D++++ AL+RLR A EKAK LS + I L I F ++TR+
Sbjct: 293 FKTTEGIDLAKDRLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHFNITLTRS 352
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
+FE + +L E+ + CL DA + VD+V+LVGG +R+PKV+ ++ E+F GK
Sbjct: 353 RFETLVNHLIERTRDPCKNCLKDAGISAKEVDEVLLVGGMTRVPKVQSIVAEIF-GKSPS 411
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIP 406
K +NPDEAVA GA +Q +L +V L+L DVTPLS G+ LG + + +I RNT+IP
Sbjct: 412 KGVNPDEAVAMGAALQGGIL---RGDVKELLLLDVTPLSLGIETLGGVFTRLITRNTTIP 468
Query: 407 VTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+K++ + D N+ V ++V Q
Sbjct: 469 --TKKSQVFSTAAD--NQTQVGIRVLQ 491
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 327 bits (837), Expect = 1e-89
Identities = 199/447 (44%), Positives = 273/447 (60%), Gaps = 37/447 (8%)
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQ-RLIGDAAKNQAATNP 68
+GIDLGTT S V + + I+ N +G RTTPS VA+T S+ RL+G AK QA NP
Sbjct: 81 VGIDLGTTNSAVAAM--EGGKPTIVTNAEGQRTTPSVVAYTKSKDRLVGQIAKRQAVVNP 138
Query: 69 SNTIFAFKVIAGT------------------NDKPTIVVNYKGEEKRFVAEEISSVILSK 110
NT F+ K G ++ + ++ K+F AEEIS+ +L K
Sbjct: 139 ENTFFSVKRFIGRRMNEVAEESKQVSYRVIKDENGNVKLDCPAIGKQFAAEEISAQVLRK 198
Query: 111 MREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQ 170
+ + A FL V AVI+VPAYFNDSQR ATKDAG IAGL V++IINEPTAA+LAYG +
Sbjct: 199 LVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLAYGFE 258
Query: 171 KRANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDHFVKE 230
+++N I +FDLGGGTFDVS+L V + VFEV +T+GDTHLGG DFD +VD
Sbjct: 259 RKSN----ETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDWLAST 314
Query: 231 FRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGID----FCSSITRA 286
F+ D+ ++ +AL+RL A EKAK LS T + L I D +++TR
Sbjct: 315 FKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSLPFITATADGPKHIETTLTRG 374
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
KFEE+ +L ++ V L DAK+ +D+V+LVGGS+RIP V+ L++++ GKE
Sbjct: 375 KFEELCSDLLDRVRTPVENSLRDAKLSFKDIDEVILVGGSTRIPAVQDLVRKL-TGKEPN 433
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELGNLMSVMIPRNTSIP 406
S+NPDE VA GA VQA +L S +V ++VL DVTPLS G+ LG +M+ +IPRNT++P
Sbjct: 434 VSVNPDEVVALGAAVQAGVL---SGDVSDIVLLDVTPLSLGLETLGGVMTKIIPRNTTLP 490
Query: 407 VTVEKTKTYYECYDYYNKYSVTVKVYQ 433
+ K++ + D + SV + V Q
Sbjct: 491 TS--KSEVFSTAAD--GQTSVEINVLQ 513
>At1g79930 putative heat-shock protein
Length = 831
Score = 224 bits (570), Expect = 9e-59
Identities = 141/442 (31%), Positives = 223/442 (49%), Gaps = 41/442 (9%)
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPS 69
+G D G V V ++ ++++ ND+ NR TP+ V F + QR IG A NP
Sbjct: 4 VGFDFGNENCLVAVARQRG--IDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPK 61
Query: 70 NTI----------------------FAFKVIAGTNDKPTIVVNYKGEEKRFVAEEISSVI 107
N+I F V G + P I NY GE++ F ++ ++
Sbjct: 62 NSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEKRAFTPTQVMGMM 121
Query: 108 LSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAY 167
LS ++ IAE L T V + I +P YF D QRRA DA IAGL+ +++I+E TA ALAY
Sbjct: 122 LSNLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLRLIHETTATALAY 181
Query: 168 GLQKR-ANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDH 226
G+ K D+ N+ D+G + V I K ++ + A D LGG+DFD ++ +H
Sbjct: 182 GIYKTDLPESDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNH 241
Query: 227 FVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRA 286
F +F+++YK D+S+N+KA RLR CEK K+ LS + A + ++ + D I R
Sbjct: 242 FAAKFKDEYKIDVSQNAKASLRLRATCEKLKKVLSANPLAPLNIECLMDEKDVRGVIKRE 301
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
+FEE+++ + E+ + L+DA + V V ++G SR+P + ++L E F GKE
Sbjct: 302 EFEEISIPILERVKRPLEKALSDAGLTVEDVHMVEVIGSGSRVPAMIKILTEFF-GKEPR 360
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELG------------NL 394
+++N E V+ G +Q A+LS K V + + P S ++ G
Sbjct: 361 RTMNASECVSRGCALQCAILSPTFK-VREFQVHESFPFSISLAWKGAASEAQNGGAENQQ 419
Query: 395 MSVMIPRNTSIPVTVEKTKTYY 416
+++ P+ IP K T+Y
Sbjct: 420 STIVFPKGNPIPSV--KALTFY 439
>At1g79920 putative heat-shock protein (At1g79920)
Length = 736
Score = 222 bits (566), Expect = 3e-58
Identities = 142/442 (32%), Positives = 221/442 (49%), Gaps = 41/442 (9%)
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPS 69
+G D G V V ++ ++++ ND+ NR TP+ V F + QR IG A NP
Sbjct: 4 VGFDFGNENCLVAVARQRG--IDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPK 61
Query: 70 NTI----------------------FAFKVIAGTNDKPTIVVNYKGEEKRFVAEEISSVI 107
N+I F V G + P I NY GE + F ++ ++
Sbjct: 62 NSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEIRAFTPTQVMGMM 121
Query: 108 LSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAY 167
LS ++ IAE L T V + I +P YF D QRRA DA IAGL+ + +I+E TA ALAY
Sbjct: 122 LSNLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLHLIHETTATALAY 181
Query: 168 GLQKRANCV-DKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMVDH 226
G+ K D+ N+ D+G + V I K ++ + A D LGG+DFD ++ +H
Sbjct: 182 GIYKTDLPENDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNH 241
Query: 227 FVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRA 286
F +F+++YK D+S+N+KA RLR CEK K+ LS + A + ++ + D I R
Sbjct: 242 FAAKFKDEYKIDVSQNAKASLRLRATCEKLKKVLSANPMAPLNIECLMAEKDVRGVIKRE 301
Query: 287 KFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKELC 346
+FEE+++ + E+ + L+DA + V V +VG SR+P + ++L E F GKE
Sbjct: 302 EFEEISIPILERVKRPLEKALSDAGLTVEDVHMVEVVGSGSRVPAMIKILTEFF-GKEPR 360
Query: 347 KSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVSELG------------NL 394
+++N E V+ G +Q A+LS K V + + P S ++ G
Sbjct: 361 RTMNASECVSRGCALQCAILSPTFK-VREFQVHESFPFSISLAWKGAATDAQNGGTENQQ 419
Query: 395 MSVMIPRNTSIPVTVEKTKTYY 416
+++ P+ IP K T+Y
Sbjct: 420 STIVFPKGNPIPSV--KALTFY 439
>At1g11660 heat-shock protein like
Length = 773
Score = 222 bits (566), Expect = 3e-58
Identities = 135/405 (33%), Positives = 221/405 (54%), Gaps = 30/405 (7%)
Query: 10 IGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFTNSQRLIGDAAKNQAATNPS 69
+G D+G + V ++ ++++ ND+ NR P+ V+F QR +G AA A +P
Sbjct: 4 VGFDVGNENCVIAVAKQRG--IDVLLNDESNRENPAMVSFGEKQRFMGAAAAASATMHPK 61
Query: 70 NTIFAFKVIAGTNDKPTIVVN----------------------YKGEEKRFVAEEISSVI 107
+TI K + G + V N Y GE + F +I ++
Sbjct: 62 STISQLKRLIGRKFREPDVQNDLRLFPFETSEDSDGGIQIRLRYMGEIQSFSPVQILGML 121
Query: 108 LSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAY 167
LS +++IAE L+TPV + VI +P+YF +SQR A DA AIAGL ++++++ TA AL Y
Sbjct: 122 LSHLKQIAEKSLKTPVSDCVIGIPSYFTNSQRLAYLDAAAIAGLRPLRLMHDSTATALGY 181
Query: 168 GLQKR---ANCVDKRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGGQDFDNIMV 224
G+ K AN +FI D+G V + + ++ V++ A D +LGG+DFD ++
Sbjct: 182 GIYKTDLVANSSPTYIVFI-DIGHCDTQVCVASFESGSMRVRSHAFDRNLGGRDFDEVLF 240
Query: 225 DHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSIT 284
+HF EF+ KY D+ N+KA RLR +CEK K+ LS + +A + ++ + + D S I
Sbjct: 241 NHFALEFKEKYNIDVYTNTKACVRLRASCEKVKKVLSANAEAQLNIECLMEEKDVRSFIK 300
Query: 285 RAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKE 344
R +FE+++ L E+ + LAD+ + + + V LVG SRIP + ++L +FK +E
Sbjct: 301 REEFEQLSAGLLERLIVPCQKALADSGLSLDQIHSVELVGSGSRIPAISKMLSSLFK-RE 359
Query: 345 LCKSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTPLSFGVS 389
L +++N E VA G +Q A+LS + V + ++D P + G S
Sbjct: 360 LGRTVNASECVARGCALQCAMLSPVFR-VRDYEVQDSYPFAIGFS 403
>At2g32120 70kD heat shock protein
Length = 563
Score = 185 bits (469), Expect = 5e-47
Identities = 131/432 (30%), Positives = 210/432 (48%), Gaps = 49/432 (11%)
Query: 8 VAIGIDLGTTYSCVGVWHEQNDRVEIIHNDQGNRTTPSCVAFT----------------- 50
+A+GID+GT+ + VW+ +V I+ N + + S V F
Sbjct: 29 IALGIDIGTSQCSIAVWN--GSQVHILRNTRNQKLIKSFVTFKDEVPAGGVSNQLAHEQE 86
Query: 51 --------NSQRLIGDAAKNQAATNPSNTIFAFKVIAGTNDKPTIVVNYKGEEKRFVAEE 102
N +RL+G + N F + + +P I + EE
Sbjct: 87 MLTGAAIFNMKRLVGRVDTDPVVHASKNLPFLVQTL-DIGVRPFIAALVNNAWRSTTPEE 145
Query: 103 ISSVILSKMREIAEAFLQTPVKNAVISVPAYFNDSQRRATKDAGAIAGLNVMQIINEPTA 162
+ ++ L ++R +AEA L+ PV+N V++VP F+ Q + A A+AGL+V++++ EPTA
Sbjct: 146 VLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQLTRFERACAMAGLHVLRLMPEPTA 205
Query: 163 AALAYGLQKRANCVD------KRNIFIFDLGGGTFDVSILTVKNNVFEVKATAGDTHLGG 216
AL Y Q++ D +R IF++G G DV++ V ++KA AG +GG
Sbjct: 206 IALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDVAVTATAGGVSQIKALAGSP-IGG 264
Query: 217 QDFDNIMVDHFVKEFRNKYKEDISENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKG 276
+D +N + N +A LR A + A L+ + IE+D + G
Sbjct: 265 EDI-----------LQNTIRHIAPPNEEASGLLRVAAQDAIHRLTDQENVQIEVD-LGNG 312
Query: 277 IDFCSSITRAKFEEMNMNLFEKCMNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLL 336
+ R +FEE+N +FE+C V CL DA+++ +DD+++VGG S IPKVR ++
Sbjct: 313 NKISKVLDRLEFEEVNQKVFEECERLVVQCLRDARVNGGDIDDLIMVGGCSYIPKVRTII 372
Query: 337 QEVFKGKELCKSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDV--TPLSFGVSELGNL 394
+ V K E+ K +NP EA GA ++ A+ S +L L + TPL+ GV GN
Sbjct: 373 KNVCKKDEIYKGVNPLEAAVRGAALEGAVTSGIHDPFGSLDLLTIQATPLAVGVRANGNK 432
Query: 395 MSVMIPRNTSIP 406
+IPRNT +P
Sbjct: 433 FIPVIPRNTMVP 444
>At5g07690 transcription factor-like protein
Length = 336
Score = 31.6 bits (70), Expect = 0.89
Identities = 16/41 (39%), Positives = 20/41 (48%)
Query: 343 KELCKSINPDEAVAYGAVVQAALLSEGSKNVPNLVLRDVTP 383
K L NPDE G++ +L SKNVP + D TP
Sbjct: 127 KPLAYDSNPDEQSQSGSISPKSLPPSSSKNVPEITSSDETP 167
>At2g15110 unknown protein
Length = 423
Score = 31.2 bits (69), Expect = 1.2
Identities = 48/230 (20%), Positives = 94/230 (40%), Gaps = 26/230 (11%)
Query: 131 PAYFNDSQRRATKDAGAIAGLNVMQIINEPTAAALAYGLQK------RANCVDKRNIFIF 184
P F D+ R AIA + M+ +N+ A L K C + +F
Sbjct: 130 PMTFADTMRTLVPPGSAIAPFDEMKEVNKENYLRFARKLGKLILEFNSVFCSHEDQLFDK 189
Query: 185 DLGGGTF----DVSILTV-KNNVFEVKATAGDTHLGGQDFDNIMVDHFVKEFRNKYKEDI 239
D+ +F D + V K N + A + + + +NI + +K +N Y + I
Sbjct: 190 DVEIESFKRSEDENAKAVEKANKVMNRMKAAELQVQKLEVNNIDLTAKLKAGKNAYLDAI 249
Query: 240 SENSKALRRLRTACEKAKRTLSYDTDATIELDAIYKGIDFCSSITRAKFEEMNM---NLF 296
+ ++A LRT EK K+ + + + RA+F + + N F
Sbjct: 250 EKETQARADLRTCKEKMKKMEEEQAEMIVAARTDER------RKVRAQFHDFSSKYGNFF 303
Query: 297 EKC--MNTVNICLADAKMDKNSVDDVVLVGGSSRIPKVRQLLQEVFKGKE 344
++ + T+ + +A+AK ++ ++++ IP + + L+ V +E
Sbjct: 304 KESEEVETLKVRVAEAKANRELLEEI----EKGEIPDLSKELESVRADEE 349
>At1g30450 cation-chloride co-transporter, putative
Length = 1321
Score = 30.4 bits (67), Expect = 2.0
Identities = 28/97 (28%), Positives = 42/97 (42%), Gaps = 10/97 (10%)
Query: 38 QGNRTTPSCVAFTNSQRLIGDAAKNQAATNPSNTIFAFKVIAGTNDKPTIVVNYKGEEKR 97
+GN S AF +QR I D + SN + A N KP +V + E+
Sbjct: 859 EGNSQEDSLEAFDAAQRRISDYL-GEIKRQGSNPLLA-------NGKPMVVNEQQVEKFL 910
Query: 98 FVAEEISSVILSKMREIAEAFLQTPVKNAVISVPAYF 134
+ +++S ILS R A + P ++ PAYF
Sbjct: 911 YTMLKLNSTILSYSRMAAVVLVSLP--PPPLNHPAYF 945
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.375
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,235,276
Number of Sequences: 26719
Number of extensions: 378496
Number of successful extensions: 1118
Number of sequences better than 10.0: 33
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1030
Number of HSP's gapped (non-prelim): 33
length of query: 433
length of database: 11,318,596
effective HSP length: 102
effective length of query: 331
effective length of database: 8,593,258
effective search space: 2844368398
effective search space used: 2844368398
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144722.18


