

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.13 + phase: 0
(600 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
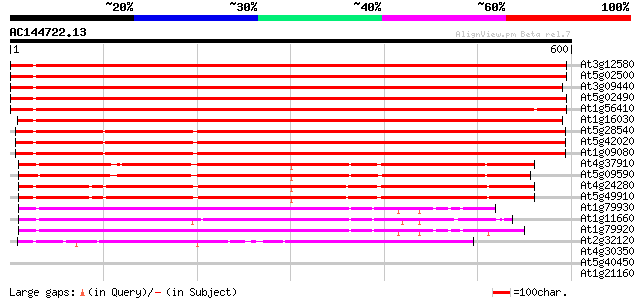
Score E
Sequences producing significant alignments: (bits) Value
At3g12580 putative protein 781 0.0
At5g02500 dnaK-type molecular chaperone hsc70.1 771 0.0
At3g09440 heat-shock protein (At-hsc70-3) 767 0.0
At5g02490 dnaK-type molecular chaperone hsc70.1 - like 761 0.0
At1g56410 hypothetical protein 749 0.0
At1g16030 unknown protein 738 0.0
At5g28540 luminal binding protein 617 e-177
At5g42020 luminal binding protein (dbj|BAA13948.1) 614 e-176
At1g09080 putative luminal binding protein 612 e-175
At4g37910 heat shock protein 70 like protein 434 e-122
At5g09590 heat shock protein 70 (Hsc70-5) 428 e-120
At4g24280 hsp 70-like protein 423 e-118
At5g49910 heat shock protein 70 (Hsc70-7) 419 e-117
At1g79930 putative heat-shock protein 253 2e-67
At1g11660 heat-shock protein like 253 2e-67
At1g79920 putative heat-shock protein (At1g79920) 250 1e-66
At2g32120 70kD heat shock protein 235 5e-62
At4g30350 unknown protein 35 0.16
At5g40450 unknown protein 31 2.3
At1g21160 transcription factor, putative 30 2.9
>At3g12580 putative protein
Length = 650
Score = 781 bits (2018), Expect = 0.0
Identities = 401/597 (67%), Positives = 478/597 (79%), Gaps = 4/597 (0%)
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NP NT+FDAKRLIGR+YSD +Q D WPFKVV G +KP I+V +KG EK F
Sbjct: 59 KNQVAMNPTNTVFDAKRLIGRRYSDPSVQADKSHWPFKVVSGPGEKPMIVVNHKGEEKQF 118
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KMREIAE FL SPVKNAVVTVPAYFNDSQR+ATKDAG I+GLNVMRIIN
Sbjct: 119 SAEEISSMVLIKMREIAEAFLGSPVKNAVVTVPAYFNDSQRQATKDAGVISGLNVMRIIN 178
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A+ V E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKASSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID 300
FDNRMVNHFV EF RK KDI+G+PR LRRLRTACER KRTLS TI+IDSLFEGID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDITGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFEGID 298
Query: 301 FHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKE 360
F+++ITRA+FE+LN+DLF+KC+E VE CL DA+++KSSV D VLVGGS+RIPKVQQLL++
Sbjct: 299 FYTTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSSVHDVVLVGGSTRIPKVQQLLQD 358
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G +++V
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 418
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQI 478
+IPRNT IP KK + + T D+ V ++VYEGER + +NNLLGKF S IPPAPRG
Sbjct: 419 LIPRNTTIPTKKEQIFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVP 478
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
I VCF ID +GILNVSA + T G + +ITITN+KGRLS EEIE+M+QEAE +K ED +
Sbjct: 479 QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKEEIEKMVQEAEKYKAEDEEH 538
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHEA 595
KKKV A N L++Y Y MR +KD+ ++S L + ++ KI A+ + ++ +D Q EA
Sbjct: 539 KKKVDAKNALENYAYNMRNTIKDEKIASKLDAADKKKIEDAIDQAIEWLDGNQLAEA 595
>At5g02500 dnaK-type molecular chaperone hsc70.1
Length = 651
Score = 771 bits (1991), Expect = 0.0
Identities = 390/597 (65%), Positives = 477/597 (79%), Gaps = 4/597 (0%)
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
M+ K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 1 MSGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NPVNT+FDAKRLIGR++SDS +Q+D++LWPFK+ G DKP I V+YKG EK F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDSSVQSDMKLWPFKIQAGPADKPMIYVEYKGEEKEF 118
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KMREIAE +L +KNAVVTVPAYFNDSQR+ATKDAG IAGLNVMRIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGVTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A V E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID 300
FDNRMVNHFV EF RK KDI+G+PR LRRLRT+CER KRTLS TI+IDSL+EGID
Sbjct: 239 FDNRMVNHFVQEFKRKSKKDITGNPRALRRLRTSCERAKRTLSSTAQTTIEIDSLYEGID 298
Query: 301 FHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKE 360
F+S+ITRA+FE+LN+DLF+KC+E VE CL DA+++KS+V D VLVGGS+RIPKVQQLL++
Sbjct: 299 FYSTITRARFEELNMDLFRKCMEPVEKCLRDAKMDKSTVHDVVLVGGSTRIPKVQQLLQD 358
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQ A+L E EK +L+L DVTPLSLG G +++
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQGAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTT 418
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQI 478
+IPRNT IP KK + + T D+ V ++VYEGER + +NNLLGKF S IPPAPRG
Sbjct: 419 LIPRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVP 478
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
I VCF ID +GILNVSA + T G + +ITITN+KGRLS +EIE+M+QEAE +K ED +
Sbjct: 479 QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEKYKSEDEEH 538
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHEA 595
KKKV+A N L++Y Y MR ++D+ + L + ++ KI ++ + ++ ++ Q EA
Sbjct: 539 KKKVEAKNALENYAYNMRNTIQDEKIGEKLPAADKKKIEDSIEQAIQWLEGNQLAEA 595
>At3g09440 heat-shock protein (At-hsc70-3)
Length = 649
Score = 767 bits (1980), Expect = 0.0
Identities = 386/593 (65%), Positives = 472/593 (79%), Gaps = 4/593 (0%)
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NP+NT+FDAKRLIGR+++DS +Q+DI+LWPF + G +KP I+V YKG +K F
Sbjct: 59 KNQVAMNPINTVFDAKRLIGRRFTDSSVQSDIKLWPFTLKSGPAEKPMIVVNYKGEDKEF 118
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSM+L KMREIAE +L + +KNAVVTVPAYFNDSQR+ATKDAG IAGLNVMRIIN
Sbjct: 119 SAEEISSMILIKMREIAEAYLGTTIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVMRIIN 178
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A V E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID 300
FDNRMVNHFV EF RK KDISG+PR LRRLRTACER KRTLS TI+IDSLF+GID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDISGNPRALRRLRTACERAKRTLSSTAQTTIEIDSLFDGID 298
Query: 301 FHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKE 360
F++ ITRA+FE+LN+DLF+KC+E VE CL DA+++K+S+DD VLVGGS+RIPKVQQLL +
Sbjct: 299 FYAPITRARFEELNIDLFRKCMEPVEKCLRDAKMDKNSIDDVVLVGGSTRIPKVQQLLVD 358
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G +++V
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTV 418
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQI 478
+I RNT IP KK + + T D+ V ++VYEGER + +NNLLGKF S IPPAPRG
Sbjct: 419 LIQRNTTIPTKKEQVFSTYSDNQPGVLIQVYEGERARTKDNNLLGKFELSGIPPAPRGVP 478
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
I VCF ID +GILNVSA + T G + +ITITN+KGRLS +EIE+M+QEAE +K ED +
Sbjct: 479 QITVCFDIDANGILNVSAEDKTTGQKNKITITNDKGRLSKDEIEKMVQEAEKYKSEDEEH 538
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQ 591
KKKV A N L++Y Y MR ++D+ + L ++ KI ++ ++ ++ Q
Sbjct: 539 KKKVDAKNALENYAYNMRNTIRDEKIGEKLAGDDKKKIEDSIEAAIEWLEANQ 591
>At5g02490 dnaK-type molecular chaperone hsc70.1 - like
Length = 653
Score = 761 bits (1964), Expect = 0.0
Identities = 384/597 (64%), Positives = 474/597 (79%), Gaps = 4/597 (0%)
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NPVNT+FDAKRLIGR++SD+ +Q+D +LWPF ++ G +KP I+V+YKG EK F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDASVQSDRQLWPFTIISGTAEKPMIVVEYKGEEKQF 118
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KMREIAE FL + VKNAVVTVPAYFNDSQR+ATKDAG IAGLNV+RIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAFLGTTVKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A V E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGEKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID 300
FDNRMVNHFV EF RK +DI+G PR LRRLRTACER KRTLS TI+IDSL+ G D
Sbjct: 239 FDNRMVNHFVQEFKRKNKQDITGQPRALRRLRTACERAKRTLSSTAQTTIEIDSLYGGAD 298
Query: 301 FHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKE 360
F+S ITRA+FE++N+DLF+KC+E VE CL DA+++KS+V + VLVGGS+RIPKVQQLL++
Sbjct: 299 FYSPITRARFEEMNMDLFRKCMEPVEKCLRDAKMDKSTVHEIVLVGGSTRIPKVQQLLQD 358
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G +++
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGLETAGGVMTT 418
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQI 478
+I RNT IP KK + + T D+ V ++V+EGER + +NNLLGKF S IPPAPRG
Sbjct: 419 LIQRNTTIPTKKEQVFSTYSDNQPGVLIQVFEGERARTKDNNLLGKFELSGIPPAPRGVP 478
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
I VCF ID +GILNVSA + T G + +ITITN+KGRLS E+IE+M+QEAE +K ED +
Sbjct: 479 QITVCFDIDANGILNVSAEDKTTGKKNKITITNDKGRLSKEDIEKMVQEAEKYKSEDEEH 538
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHEA 595
KKKV+A N L++Y Y MR ++D+ + L + ++ K+ ++ + ++ +D Q EA
Sbjct: 539 KKKVEAKNALENYAYNMRNTIRDEKIGEKLPAADKKKVEDSIEEAIQWLDGNQLGEA 595
>At1g56410 hypothetical protein
Length = 617
Score = 749 bits (1934), Expect = 0.0
Identities = 382/597 (63%), Positives = 468/597 (77%), Gaps = 6/597 (1%)
Query: 1 MAKKFKGPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAA 60
MA K +GPAIGIDLGTTYSCV VWQ ++R EII NDQGNRTTPS+VAFTD +RLIGDAA
Sbjct: 1 MAGKGEGPAIGIDLGTTYSCVGVWQ--HDRVEIIANDQGNRTTPSYVAFTDSERLIGDAA 58
Query: 61 KNQAASNPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHF 120
KNQ A NPVNT+FDAKRLIGR++SD+ +Q+D++ WPFKV PG DKP I V YKG EK F
Sbjct: 59 KNQVAMNPVNTVFDAKRLIGRRFSDASVQSDMKFWPFKVTPGQADKPMIFVNYKGEEKQF 118
Query: 121 CAEEISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIIN 180
AEEISSMVL KMREIAE +L S +KNAVVTVPAYFNDSQR+ATKDAG IAGLNV+RIIN
Sbjct: 119 AAEEISSMVLIKMREIAEAYLGSSIKNAVVTVPAYFNDSQRQATKDAGVIAGLNVLRIIN 178
Query: 181 EPTAAALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGED 240
EPTAAA+AYGL K+A V +N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGED
Sbjct: 179 EPTAAAIAYGLDKKATSVGIKNVLIFDLGGGTFDVSLLTIEEGIFEVKATAGDTHLGGED 238
Query: 241 FDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID 300
FDNRMVNHFV EF RK KDISG R LRRLRTACER KRTLS T+++DSLFEGID
Sbjct: 239 FDNRMVNHFVQEFKRKNKKDISGDARALRRLRTACERAKRTLSSTAQTTVEVDSLFEGID 298
Query: 301 FHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKE 360
F+S ITRAKFE++N+DLF+KC+E V CL D++++KS V D VLVGGS+RIPKVQQLL++
Sbjct: 299 FYSPITRAKFEEMNMDLFRKCMEPVMKCLRDSKMDKSMVHDVVLVGGSTRIPKVQQLLQD 358
Query: 361 FFNWKDICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSV 419
FFN K++C SINPDEAVAYGAAVQAA+L E EK +L+L DVTPLSLG G +++
Sbjct: 359 FFNGKELCKSINPDEAVAYGAAVQAAILSGEGNEKVQDLLLLDVTPLSLGIETIGGVMTT 418
Query: 420 VIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQI 478
+I RNT IP KK + + TT D+ V ++VYEGER + +NN+LG+F S IPPAPRG
Sbjct: 419 LIQRNTTIPAKKEQEFTTTVDNQPDVLIQVYEGERARTIDNNILGQFVLSGIPPAPRGIP 478
Query: 479 PIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKF 538
VCF ID +GILNVSA + G + +ITITN+KGRLS ++IE+M+QEAE +K ED +
Sbjct: 479 QFTVCFDIDSNGILNVSAEDKATGKKNKITITNDKGRLSKDDIEKMVQEAEKYKSEDEEH 538
Query: 539 KKKVKAMNDLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQHHEA 595
KKKV+A N L++Y Y + ++D + L + ++ K ++ + ++ +D+ Q EA
Sbjct: 539 KKKVEAKNGLENYAYNVGNTLRD--MGEKLPAADKKKFEDSIEEVIQWLDDNQLAEA 593
>At1g16030 unknown protein
Length = 646
Score = 738 bits (1906), Expect = 0.0
Identities = 375/585 (64%), Positives = 461/585 (78%), Gaps = 4/585 (0%)
Query: 9 AIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAASNP 68
AIGIDLGTTYSCV VW N+R EII NDQGNRTTPS+VAFTD +RLIGDAAKNQ A NP
Sbjct: 8 AIGIDLGTTYSCVGVWM--NDRVEIIPNDQGNRTTPSYVAFTDTERLIGDAAKNQVALNP 65
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSM 128
NT+FDAKRLIGRK+SD +Q+DI WPFKVV G +KP I+V YK EK F EEISSM
Sbjct: 66 QNTVFDAKRLIGRKFSDPSVQSDILHWPFKVVSGPGEKPMIVVSYKNEEKQFSPEEISSM 125
Query: 129 VLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALA 188
VL KM+E+AE FL VKNAVVTVPAYFNDSQR+ATKDAGAI+GLNV+RIINEPTAAA+A
Sbjct: 126 VLVKMKEVAEAFLGRTVKNAVVTVPAYFNDSQRQATKDAGAISGLNVLRIINEPTAAAIA 185
Query: 189 YGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNH 248
YGL K+ E+N+ IFDLGGGTFDVSLLTI+ +FEVKAT+GDTHLGGEDFDNR+VNH
Sbjct: 186 YGLDKKGTKAGEKNVLIFDLGGGTFDVSLLTIEEGVFEVKATAGDTHLGGEDFDNRLVNH 245
Query: 249 FVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSITRA 308
FV EF RK+ KDI+G+ R LRRLRTACER KRTLS TI+IDSL EGIDF+++I+RA
Sbjct: 246 FVAEFRRKHKKDIAGNARALRRLRTACERAKRTLSSTAQTTIEIDSLHEGIDFYATISRA 305
Query: 309 KFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKDIC 368
+FE++N+DLF+KC++ VE L DA+++KSSV D VLVGGS+RIPK+QQLL++FFN K++C
Sbjct: 306 RFEEMNMDLFRKCMDPVEKVLKDAKLDKSSVHDVVLVGGSTRIPKIQQLLQDFFNGKELC 365
Query: 369 VSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRNTPI 427
SINPDEAVAYGAAVQAA+L E +EK +L+L DV PLSLG G +++V+IPRNT +
Sbjct: 366 KSINPDEAVAYGAAVQAAILTGEGSEKVQDLLLLDVAPLSLGLETAGGVMTVLIPRNTTV 425
Query: 428 PVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF-SIPPAPRGQIPIKVCFSI 486
P KK + + T D+ V ++VYEGER + +NNLLG F IPPAPRG I VCF I
Sbjct: 426 PCKKEQVFSTYADNQPGVLIQVYEGERARTRDNNLLGTFELKGIPPAPRGVPQINVCFDI 485
Query: 487 DFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVKAMN 546
D +GILNVSA + T G + +ITITN+KGRLS EEIE+M+Q+AE +K ED + KKKV+A N
Sbjct: 486 DANGILNVSAEDKTAGVKNQITITNDKGRLSKEEIEKMVQDAEKYKAEDEQVKKKVEAKN 545
Query: 547 DLDDYLYTMRKVMKDDSVSSMLTSIERMKINSAMIKGMKLIDEKQ 591
L++Y Y MR +KD+ ++ LT ++ KI A+ + ++ I+ Q
Sbjct: 546 SLENYAYNMRNTIKDEKLAQKLTQEDKQKIEKAIDETIEWIEGNQ 590
>At5g28540 luminal binding protein
Length = 669
Score = 617 bits (1590), Expect = e-177
Identities = 324/592 (54%), Positives = 428/592 (71%), Gaps = 10/592 (1%)
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNR TPS+V FTD +RLIG+AAKNQAA
Sbjct: 35 GSVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAV 92
Query: 67 NPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGE-KHFCAEEI 125
NP T+FD KRLIGRK+ D +Q D +L P+++V D KP I VK K GE K F EEI
Sbjct: 93 NPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIV-NKDGKPYIQVKIKDGETKVFSPEEI 151
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAA 185
S+M+LTKM+E AE +L +K+AVVTVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 152 SAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 211
Query: 186 ALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRM 245
A+AYGL K+ E+NI +FDLGGGTFDVS+LTI N +FEV +T+GDTHLGGEDFD+R+
Sbjct: 212 AIAYGLDKKGG---EKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRV 268
Query: 246 VNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSI 305
+ +F+ +K+ KDIS + L +LR CER KR LS ++I+SLF+G+DF +
Sbjct: 269 MEYFIKLIKKKHQKDISKDNKALGKLRRECERAKRALSSQHQVRVEIESLFDGVDFSEPL 328
Query: 306 TRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWK 365
TRA+FE+LN DLF+K + V+ + DA ++KS +D+ VLVGGS+RIPKVQQLLK+FF K
Sbjct: 329 TRARFEELNNDLFRKTMGPVKKAMDDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDFFEGK 388
Query: 366 DICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRN 424
+ +NPDEAVAYGAAVQ +L E +++ +++L DV PL+LG G +++ +IPRN
Sbjct: 389 EPNKGVNPDEAVAYGAAVQGGILSGEGGDETKDILLLDVAPLTLGIETVGGVMTKLIPRN 448
Query: 425 TPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQIPIKVC 483
T IP KK++ + T +D + VS++V+EGER + LLGKF + IPPAPRG I+V
Sbjct: 449 TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLNGIPPAPRGTPQIEVT 508
Query: 484 FSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVK 543
F +D +GILNV A + G ++ITITNEKGRLS EEI+RM++EAE F +ED K K+K+
Sbjct: 509 FEVDANGILNVKAEDKASGKSEKITITNEKGRLSQEEIDRMVKEAEEFAEEDKKVKEKID 568
Query: 544 AMNDLDDYLYTMRKVMKD-DSVSSMLTSIERMKINSAMIKGMKLIDEKQHHE 594
A N L+ Y+Y M+ + D D ++ L E+ KI +A + ++ +DE Q+ E
Sbjct: 569 ARNALETYVYNMKNQVNDKDKLADKLEGDEKEKIEAATKEALEWLDENQNSE 620
>At5g42020 luminal binding protein (dbj|BAA13948.1)
Length = 668
Score = 614 bits (1583), Expect = e-176
Identities = 322/592 (54%), Positives = 427/592 (71%), Gaps = 10/592 (1%)
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V++ N EII NDQGNR TPS+V FTD +RLIG+AAKNQAA
Sbjct: 35 GSVIGIDLGTTYSCVGVYK--NGHVEIIANDQGNRITPSWVGFTDSERLIGEAAKNQAAV 92
Query: 67 NPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGE-KHFCAEEI 125
NP T+FD KRLIGRK+ D +Q D +L P+++V D KP I VK K GE K F EEI
Sbjct: 93 NPERTVFDVKRLIGRKFEDKEVQKDRKLVPYQIV-NKDGKPYIQVKIKDGETKVFSPEEI 151
Query: 126 SSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAA 185
S+M+LTKM+E AE +L +K+AVVTVPAYFND+QR+ATKDAG IAGLNV RIINEPTAA
Sbjct: 152 SAMILTKMKETAEAYLGKKIKDAVVTVPAYFNDAQRQATKDAGVIAGLNVARIINEPTAA 211
Query: 186 ALAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRM 245
A+AYGL K+ E+NI +FDLGGGTFDVS+LTI N +FEV +T+GDTHLGGEDFD+R+
Sbjct: 212 AIAYGLDKKGG---EKNILVFDLGGGTFDVSVLTIDNGVFEVLSTNGDTHLGGEDFDHRI 268
Query: 246 VNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSI 305
+ +F+ +K+ KDIS + L +LR CER KR LS ++I+SLF+G+D +
Sbjct: 269 MEYFIKLIKKKHQKDISKDNKALGKLRRECERAKRALSSQHQVRVEIESLFDGVDLSEPL 328
Query: 306 TRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWK 365
TRA+FE+LN DLF+K + V+ + DA ++KS +D+ VLVGGS+RIPKVQQLLK+FF K
Sbjct: 329 TRARFEELNNDLFRKTMGPVKKAMDDAGLQKSQIDEIVLVGGSTRIPKVQQLLKDFFEGK 388
Query: 366 DICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRN 424
+ +NPDEAVAYGAAVQ +L E +++ +++L DV PL+LG G +++ +IPRN
Sbjct: 389 EPNKGVNPDEAVAYGAAVQGGILSGEGGDETKDILLLDVAPLTLGIETVGGVMTKLIPRN 448
Query: 425 TPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQIPIKVC 483
T IP KK++ + T +D + VS++V+EGER + LLGKF + +PPAPRG I+V
Sbjct: 449 TVIPTKKSQVFTTYQDQQTTVSIQVFEGERSLTKDCRLLGKFDLTGVPPAPRGTPQIEVT 508
Query: 484 FSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVK 543
F +D +GILNV A + G ++ITITNEKGRLS EEI+RM++EAE F +ED K K+K+
Sbjct: 509 FEVDANGILNVKAEDKASGKSEKITITNEKGRLSQEEIDRMVKEAEEFAEEDKKVKEKID 568
Query: 544 AMNDLDDYLYTMRKVMKD-DSVSSMLTSIERMKINSAMIKGMKLIDEKQHHE 594
A N L+ Y+Y M+ + D D ++ L E+ KI +A + ++ +DE Q+ E
Sbjct: 569 ARNALETYVYNMKNQVSDKDKLADKLEGDEKEKIEAATKEALEWLDENQNSE 620
>At1g09080 putative luminal binding protein
Length = 678
Score = 612 bits (1578), Expect = e-175
Identities = 322/591 (54%), Positives = 423/591 (71%), Gaps = 9/591 (1%)
Query: 7 GPAIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAAS 66
G IGIDLGTTYSCV V+ N EII NDQGNR TPS+VAFTD +RLIG+AAKNQAA
Sbjct: 50 GTVIGIDLGTTYSCVGVYH--NKHVEIIANDQGNRITPSWVAFTDTERLIGEAAKNQAAK 107
Query: 67 NPVNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEIS 126
NP TIFD KRLIGRK+ D +Q DI+ P+KVV D KP I VK KG EK F EEIS
Sbjct: 108 NPERTIFDPKRLIGRKFDDPDVQRDIKFLPYKVV-NKDGKPYIQVKVKGEEKLFSPEEIS 166
Query: 127 SMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAA 186
+M+LTKM+E AE FL +K+AV+TVPAYFND+QR+ATKDAGAIAGLNV+RIINEPT AA
Sbjct: 167 AMILTKMKETAEAFLGKKIKDAVITVPAYFNDAQRQATKDAGAIAGLNVVRIINEPTGAA 226
Query: 187 LAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMV 246
+AYGL K+ E NI ++DLGGGTFDVS+LTI N +FEV +TSGDTHLGGEDFD+R++
Sbjct: 227 IAYGLDKKGG---ESNILVYDLGGGTFDVSILTIDNGVFEVLSTSGDTHLGGEDFDHRVM 283
Query: 247 NHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSIT 306
++F+ +KY KDIS + L +LR CE KR+LS ++I+SLF+G+DF +T
Sbjct: 284 DYFIKLVKKKYNKDISKDHKALGKLRRECELAKRSLSNQHQVRVEIESLFDGVDFSEPLT 343
Query: 307 RAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKD 366
RA+FE+LN+DLFKK +E V+ L DA ++KS +D+ VLVGGS+RIPKVQQ+LK+FF+ K+
Sbjct: 344 RARFEELNMDLFKKTMEPVKKALKDAGLKKSDIDEIVLVGGSTRIPKVQQMLKDFFDGKE 403
Query: 367 ICVSINPDEAVAYGAAVQAALLC-EATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRNT 425
NPDEAVAYGAAVQ +L E E++ N++L DV PLSLG G +++ +IPRNT
Sbjct: 404 PSKGTNPDEAVAYGAAVQGGVLSGEGGEETQNILLLDVAPLSLGIETVGGVMTNIIPRNT 463
Query: 426 PIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRFS-IPPAPRGQIPIKVCF 484
IP KK++ + T +D + V++ VYEGER +N LGKF + I PAPRG I+V F
Sbjct: 464 VIPTKKSQVFTTYQDQQTTVTINVYEGERSMTKDNRELGKFDLTGILPAPRGVPQIEVTF 523
Query: 485 SIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVKA 544
+D +GIL V A + Q ITITN+KGRL+ EEIE MI+EAE F +ED K+K+ A
Sbjct: 524 EVDANGILQVKAEDKVAKTSQSITITNDKGRLTEEEIEEMIREAEEFAEEDKIMKEKIDA 583
Query: 545 MNDLDDYLYTMRKVMKD-DSVSSMLTSIERMKINSAMIKGMKLIDEKQHHE 594
N L+ Y+Y M+ + D + ++ ++ ++ K+ + + ++ ++E + E
Sbjct: 584 RNKLETYVYNMKSTVADKEKLAKKISDEDKEKMEGVLKEALEWLEENVNAE 634
>At4g37910 heat shock protein 70 like protein
Length = 682
Score = 434 bits (1117), Expect = e-122
Identities = 254/560 (45%), Positives = 350/560 (62%), Gaps = 26/560 (4%)
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDD-KRLIGDAAKNQAASNP 68
IGIDLGTT SCV+V + + A +I N +G+RTTPS VA + L+G AK QA +NP
Sbjct: 55 IGIDLGTTNSCVSVMEGKT--ARVIENAEGSRTTPSVVAMNQKGELLVGTPAKRQAVTNP 112
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVV--PGNDDKPEIIVKYKGGEKHFCAEEIS 126
NTIF +KRLIGR++ D Q ++++ P+K+V P D E G+K F +I
Sbjct: 113 TNTIFGSKRLIGRRFDDPQTQKEMKMVPYKIVKAPNGDAWVEA-----NGQK-FSPSQIG 166
Query: 127 SMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAA 186
+ VLTKM+E AE +L + AVVTVPAYFND+QR+ATKDAG IAGL+V RIINEPTAAA
Sbjct: 167 ANVLTKMKETAEAYLGKSINKAVVTVPAYFNDAQRQATKDAGKIAGLDVQRIINEPTAAA 226
Query: 187 LAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMV 246
L+YG+ + E I +FDLGGGTFDVS+L I + +FEVKAT+GDT LGGEDFDN ++
Sbjct: 227 LSYGMNNK-----EGVIAVFDLGGGTFDVSILEISSGVFEVKATNGDTFLGGEDFDNTLL 281
Query: 247 NHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID----FH 302
+ V EF R D++ L+RLR A E+ K LS T I++ + +
Sbjct: 282 EYLVNEFKRSDNIDLTKDNLALQRLREAAEKAKIELSSTTQTEINLPFITADASGAKHLN 341
Query: 303 SSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFF 362
++TR+KFE L L ++ ++CL DA + VD+ +LVGG +R+PKVQ+++ E F
Sbjct: 342 ITLTRSKFEGLVGKLIERTRSPCQNCLKDAGVTIKEVDEVLLVGGMTRVPKVQEIVSEIF 401
Query: 363 NWKDICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIP 422
K C +NPDEAVA GAA+Q +L + +L+L DV PLSLG G + + +IP
Sbjct: 402 G-KSPCKGVNPDEAVAMGAAIQGGILRGDVK---DLLLLDVVPLSLGIETLGAVFTKLIP 457
Query: 423 RNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF-SIPPAPRGQIPIK 481
RNT IP KK++ + T D+ V ++V +GER A++N +LG+F IPPAPRG I+
Sbjct: 458 RNTTIPTKKSQVFSTAADNQMQVGIKVLQGEREMAADNKVLGEFDLVGIPPAPRGMPQIE 517
Query: 482 VCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKK 541
V F ID +GI VSA + G Q ITI G LS +EI RM++EAE +D + K+
Sbjct: 518 VTFDIDANGITTVSAKDKATGKEQNITI-RSSGGLSDDEINRMVKEAELNAQKDQEKKQL 576
Query: 542 VKAMNDLDDYLYTMRKVMKD 561
+ N D +Y++ K + +
Sbjct: 577 IDLRNSADTTIYSVEKSLSE 596
>At5g09590 heat shock protein 70 (Hsc70-5)
Length = 682
Score = 428 bits (1101), Expect = e-120
Identities = 252/556 (45%), Positives = 350/556 (62%), Gaps = 26/556 (4%)
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAF-TDDKRLIGDAAKNQAASNP 68
IGIDLGTT SCVAV + +N + +I N +G RTTPS VAF T + L+G AK QA +NP
Sbjct: 60 IGIDLGTTNSCVAVMEGKNPK--VIENAEGARTTPSVVAFNTKGELLVGTPAKRQAVTNP 117
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVV--PGNDDKPEIIVKYKGGEKHFCAEEIS 126
NT+ KRLIGRK+ D Q ++++ P+K+V P D E + + +I
Sbjct: 118 TNTVSGTKRLIGRKFDDPQTQKEMKMVPYKIVRAPNGDAWVE------ANGQQYSPSQIG 171
Query: 127 SMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAA 186
+ +LTKM+E AE +L V AVVTVPAYFND+QR+ATKDAG IAGL+V RIINEPTAAA
Sbjct: 172 AFILTKMKETAEAYLGKSVTKAVVTVPAYFNDAQRQATKDAGRIAGLDVERIINEPTAAA 231
Query: 187 LAYGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMV 246
L+YG+ + E I +FDLGGGTFDVS+L I N +FEVKAT+GDT LGGEDFDN ++
Sbjct: 232 LSYGMTNK-----EGLIAVFDLGGGTFDVSVLEISNGVFEVKATNGDTFLGGEDFDNALL 286
Query: 247 NHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID----FH 302
+ V EF D++ L+RLR A E+ K LS + I++ + F+
Sbjct: 287 DFLVNEFKTTEGIDLAKDRLALQRLREAAEKAKIELSSTSQTEINLPFITADASGAKHFN 346
Query: 303 SSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFF 362
++TR++FE L L ++ + ++CL DA I VD+ +LVGG +R+PKVQ ++ E F
Sbjct: 347 ITLTRSRFETLVNHLIERTRDPCKNCLKDAGISAKEVDEVLLVGGMTRVPKVQSIVAEIF 406
Query: 363 NWKDICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIP 422
K +NPDEAVA GAA+Q +L ++ L+L DVTPLSLG G + + +I
Sbjct: 407 G-KSPSKGVNPDEAVAMGAALQGGILRGDVKE---LLLLDVTPLSLGIETLGGVFTRLIT 462
Query: 423 RNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF-SIPPAPRGQIPIK 481
RNT IP KK++ + T D+ + V +RV +GER A++N LLG+F IPP+PRG I+
Sbjct: 463 RNTTIPTKKSQVFSTAADNQTQVGIRVLQGEREMATDNKLLGEFDLVGIPPSPRGVPQIE 522
Query: 482 VCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKK 541
V F ID +GI+ VSA + T G Q+ITI G LS ++I++M++EAE +D + K+
Sbjct: 523 VTFDIDANGIVTVSAKDKTTGKVQQITI-RSSGGLSEDDIQKMVREAELHAQKDKERKEL 581
Query: 542 VKAMNDLDDYLYTMRK 557
+ N D +Y++ K
Sbjct: 582 IDTKNTADTTIYSIEK 597
>At4g24280 hsp 70-like protein
Length = 718
Score = 423 bits (1087), Expect = e-118
Identities = 246/558 (44%), Positives = 343/558 (61%), Gaps = 21/558 (3%)
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDD-KRLIGDAAKNQAASNP 68
+GIDLGTT S VA + + I+ N +G RTTPS VA+T RL+G AK QA NP
Sbjct: 81 VGIDLGTTNSAVAAME--GGKPTIVTNAEGQRTTPSVVAYTKSGDRLVGQIAKRQAVVNP 138
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSM 128
NT F KR IGRK ++ + + + ++VV D+ + ++ K F AEEIS+
Sbjct: 139 ENTFFSVKRFIGRKMNE--VDEESKQVSYRVV--RDENNNVKLECPAINKQFAAEEISAQ 194
Query: 129 VLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALA 188
VL K+ + A +FL V AV+TVPAYFNDSQR ATKDAG IAGL V+RIINEPTAA+LA
Sbjct: 195 VLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLA 254
Query: 189 YGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNH 248
YG ++AN I +FDLGGGTFDVS+L + + +FEV +TSGDTHLGG+DFD R+V+
Sbjct: 255 YGFDRKAN----ETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDW 310
Query: 249 FVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID----FHSS 304
EF + D+ + L+RL A E+ K LS T + + + D ++
Sbjct: 311 LAAEFKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSLPFITATADGPKHIETT 370
Query: 305 ITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNW 364
+TRAKFE+L DL + VE+ L DA++ +D+ +LVGGS+RIP VQ+L+++
Sbjct: 371 LTRAKFEELCSDLLDRVRTPVENSLRDAKLSFKDIDEVILVGGSTRIPAVQELVRK-VTG 429
Query: 365 KDICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRN 424
K+ V++NPDE VA GAAVQA +L ++VL DVTPLS+G G +++ +IPRN
Sbjct: 430 KEPNVTVNPDEVVALGAAVQAGVLAGDVS---DIVLLDVTPLSIGLETLGGVMTKIIPRN 486
Query: 425 TPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF-SIPPAPRGQIPIKVC 483
T +P K++ + T D + V + V +GER +N LG FR IPPAPRG I+V
Sbjct: 487 TTLPTSKSEVFSTAADGQTSVEINVLQGEREFVRDNKSLGSFRLDGIPPAPRGVPQIEVK 546
Query: 484 FSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVK 543
F ID +GIL+VSA + G +Q+ITIT L +E+++M+QEAE F +D + + +
Sbjct: 547 FDIDANGILSVSAVDKGTGKKQDITITG-ASTLPKDEVDQMVQEAERFAKDDKEKRDAID 605
Query: 544 AMNDLDDYLYTMRKVMKD 561
N D +Y K +K+
Sbjct: 606 TKNQADSVVYQTEKQLKE 623
>At5g49910 heat shock protein 70 (Hsc70-7)
Length = 718
Score = 419 bits (1076), Expect = e-117
Identities = 244/558 (43%), Positives = 341/558 (60%), Gaps = 21/558 (3%)
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDK-RLIGDAAKNQAASNP 68
+GIDLGTT S VA + + I+ N +G RTTPS VA+T K RL+G AK QA NP
Sbjct: 81 VGIDLGTTNSAVAAME--GGKPTIVTNAEGQRTTPSVVAYTKSKDRLVGQIAKRQAVVNP 138
Query: 69 VNTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSM 128
NT F KR IGR+ ++ + + + ++V+ D+ + + K F AEEIS+
Sbjct: 139 ENTFFSVKRFIGRRMNE--VAEESKQVSYRVI--KDENGNVKLDCPAIGKQFAAEEISAQ 194
Query: 129 VLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALA 188
VL K+ + A +FL V AV+TVPAYFNDSQR ATKDAG IAGL V+RIINEPTAA+LA
Sbjct: 195 VLRKLVDDASRFLNDKVTKAVITVPAYFNDSQRTATKDAGRIAGLEVLRIINEPTAASLA 254
Query: 189 YGLQKRANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNH 248
YG ++++N I +FDLGGGTFDVS+L + + +FEV +TSGDTHLGG+DFD R+V+
Sbjct: 255 YGFERKSN----ETILVFDLGGGTFDVSVLEVGDGVFEVLSTSGDTHLGGDDFDKRVVDW 310
Query: 249 FVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGID----FHSS 304
F + D+ + L+RL A E+ K LS T + + + D ++
Sbjct: 311 LASTFKKDEGIDLLKDKQALQRLTEAAEKAKIELSSLTQTNMSLPFITATADGPKHIETT 370
Query: 305 ITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNW 364
+TR KFE+L DL + VE+ L DA++ +D+ +LVGGS+RIP VQ L+++
Sbjct: 371 LTRGKFEELCSDLLDRVRTPVENSLRDAKLSFKDIDEVILVGGSTRIPAVQDLVRK-LTG 429
Query: 365 KDICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRGDLLSVVIPRN 424
K+ VS+NPDE VA GAAVQA +L ++VL DVTPLSLG G +++ +IPRN
Sbjct: 430 KEPNVSVNPDEVVALGAAVQAGVLSGDVS---DIVLLDVTPLSLGLETLGGVMTKIIPRN 486
Query: 425 TPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF-SIPPAPRGQIPIKVC 483
T +P K++ + T D + V + V +GER +N +G FR IPPAPRG I+V
Sbjct: 487 TTLPTSKSEVFSTAADGQTSVEINVLQGEREFVRDNKSIGSFRLDGIPPAPRGVPQIEVK 546
Query: 484 FSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMKFKKKVK 543
F ID +GIL+VSA++ G +Q+ITIT L +E++ M+QEAE F ED + + +
Sbjct: 547 FDIDANGILSVSASDKGTGKKQDITITG-ASTLPKDEVDTMVQEAERFAKEDKEKRDAID 605
Query: 544 AMNDLDDYLYTMRKVMKD 561
N D +Y K +K+
Sbjct: 606 TKNQADSVVYQTEKQLKE 623
>At1g79930 putative heat-shock protein
Length = 831
Score = 253 bits (647), Expect = 2e-67
Identities = 163/526 (30%), Positives = 264/526 (49%), Gaps = 26/526 (4%)
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAASNPV 69
+G D G VAV + + +++ ND+ NR TP+ V F D +R IG A NP
Sbjct: 4 VGFDFGNENCLVAVARQRG--IDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPK 61
Query: 70 NTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSMV 129
N+I KRLIGR++SD +Q DI+ PF V G D P I Y G ++ F ++ M+
Sbjct: 62 NSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEKRAFTPTQVMGMM 121
Query: 130 LTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALAY 189
L+ ++ IAEK L + V + + +P YF D QR+A DA IAGL+ +R+I+E TA ALAY
Sbjct: 122 LSNLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLRLIHETTATALAY 181
Query: 190 GLQKR-ANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNH 248
G+ K ++ N+ D+G + V + K ++ + + D LGG DFD + NH
Sbjct: 182 GIYKTDLPESDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNH 241
Query: 249 FVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSITRA 308
F +F +Y D+S + + RLR CE+ K+ LS + A ++I+ L + D I R
Sbjct: 242 FAAKFKDEYKIDVSQNAKASLRLRATCEKLKKVLSANPLAPLNIECLMDEKDVRGVIKRE 301
Query: 309 KFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKDIC 368
+FE++++ + ++ +E LSDA + V ++G SR+P + ++L EFF K+
Sbjct: 302 EFEEISIPILERVKRPLEKALSDAGLTVEDVHMVEVIGSGSRVPAMIKILTEFFG-KEPR 360
Query: 369 VSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRG------------DL 416
++N E V+ G A+Q A+L T K + + P S+ +G
Sbjct: 361 RTMNASECVSRGCALQCAIL-SPTFKVREFQVHESFPFSISLAWKGAASEAQNGGAENQQ 419
Query: 417 LSVVIPRNTPIPVKKTKNYY---TTKDDLSYVSVRVYEGERLKASENNLLGKFRFSIPPA 473
++V P+ PIP K +Y T D+ Y V + +G F+ S
Sbjct: 420 STIVFPKGNPIPSVKALTFYRSGTFSVDVQYSDVNDLQAP--PKISTYTIGPFQSS--KG 475
Query: 474 PRGQIPIKVCFSIDFDGILNVSATEDTCGNRQEITITNEKGRLSTE 519
R ++ +KV ++ GI++V + E+ +T E +T+
Sbjct: 476 ERAKLKVKV--RLNLHGIVSVESATLLEEEEVEVPVTKEHSEETTK 519
>At1g11660 heat-shock protein like
Length = 773
Score = 253 bits (646), Expect = 2e-67
Identities = 170/540 (31%), Positives = 290/540 (53%), Gaps = 25/540 (4%)
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAASNPV 69
+G D+G +AV + + +++ ND+ NR P+ V+F + +R +G AA A +P
Sbjct: 4 VGFDVGNENCVIAVAKQRG--IDVLLNDESNRENPAMVSFGEKQRFMGAAAAASATMHPK 61
Query: 70 NTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSMV 129
+TI KRLIGRK+ + +QND+RL+PF+ +D +I ++Y G + F +I M+
Sbjct: 62 STISQLKRLIGRKFREPDVQNDLRLFPFETSEDSDGGIQIRLRYMGEIQSFSPVQILGML 121
Query: 130 LTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALAY 189
L+ +++IAEK L++PV + V+ +P+YF +SQR A DA AIAGL +R++++ TA AL Y
Sbjct: 122 LSHLKQIAEKSLKTPVSDCVIGIPSYFTNSQRLAYLDAAAIAGLRPLRLMHDSTATALGY 181
Query: 190 GLQKR---ANCVEERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMV 246
G+ K AN +FI D+G V + + ++ V++ + D +LGG DFD +
Sbjct: 182 GIYKTDLVANSSPTYIVFI-DIGHCDTQVCVASFESGSMRVRSHAFDRNLGGRDFDEVLF 240
Query: 247 NHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSIT 306
NHF +EF KY D+ + + RLR +CE+ K+ LS + A ++I+ L E D S I
Sbjct: 241 NHFALEFKEKYNIDVYTNTKACVRLRASCEKVKKVLSANAEAQLNIECLMEEKDVRSFIK 300
Query: 307 RAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKD 366
R +FEQL+ L ++ I + L+D+ + + LVG SRIP + ++L F ++
Sbjct: 301 REEFEQLSAGLLERLIVPCQKALADSGLSLDQIHSVELVGSGSRIPAISKMLSSLFK-RE 359
Query: 367 ICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLG-TLVRGDLLS----VVI 421
+ ++N E VA G A+Q A+L + + ++D P ++G + +G + + ++
Sbjct: 360 LGRTVNASECVARGCALQCAML-SPVFRVRDYEVQDSYPFAIGFSSDKGPINTPSNELLF 418
Query: 422 PRNTPIPVKKTKNYY---TTKDDLSYVSVRVYEGERLKASENNLLGKFRFSIPPAPRGQI 478
P+ P K + T + + Y + + + ++G F S A R
Sbjct: 419 PKGQIFPSVKVLTLHRENTFQLEAFYANHNELSPDIPTQISSFMIGPFHISHGEAAR--- 475
Query: 479 PIKVCFSIDFDGILNV-SATEDTCGNRQEITITNEKGRLSTEEIERMIQEAENFKDEDMK 537
+KV ++ GI+ + SAT ++ + E I K +++EE MI E EN + MK
Sbjct: 476 -VKVRVQLNLHGIVTIDSATLESKLSVSEQLIEYHKENITSEE---MISE-ENHQSSAMK 530
>At1g79920 putative heat-shock protein (At1g79920)
Length = 736
Score = 250 bits (639), Expect = 1e-66
Identities = 168/561 (29%), Positives = 272/561 (47%), Gaps = 30/561 (5%)
Query: 10 IGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAASNPV 69
+G D G VAV + + +++ ND+ NR TP+ V F D +R IG A NP
Sbjct: 4 VGFDFGNENCLVAVARQRG--IDVVLNDESNRETPAIVCFGDKQRFIGTAGAASTMMNPK 61
Query: 70 NTIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDDKPEIIVKYKGGEKHFCAEEISSMV 129
N+I KRLIGR++SD +Q DI+ PF V G D P I Y G + F ++ M+
Sbjct: 62 NSISQIKRLIGRQFSDPELQRDIKSLPFSVTEGPDGYPLIHANYLGEIRAFTPTQVMGMM 121
Query: 130 LTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPTAAALAY 189
L+ ++ IAEK L + V + + +P YF D QR+A DA IAGL+ + +I+E TA ALAY
Sbjct: 122 LSNLKGIAEKNLNTAVVDCCIGIPVYFTDLQRRAVLDAATIAGLHPLHLIHETTATALAY 181
Query: 190 GLQKRANCVEER-NIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLGGEDFDNRMVNH 248
G+ K ++ N+ D+G + V + K ++ + + D LGG DFD + NH
Sbjct: 182 GIYKTDLPENDQLNVAFIDIGHASMQVCIAGFKKGQLKILSHAFDRSLGGRDFDEVLFNH 241
Query: 249 FVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFEGIDFHSSITRA 308
F +F +Y D+S + + RLR CE+ K+ LS + A ++I+ L D I R
Sbjct: 242 FAAKFKDEYKIDVSQNAKASLRLRATCEKLKKVLSANPMAPLNIECLMAEKDVRGVIKRE 301
Query: 309 KFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQLLKEFFNWKDIC 368
+FE++++ + ++ +E LSDA + V +VG SR+P + ++L EFF K+
Sbjct: 302 EFEEISIPILERVKRPLEKALSDAGLTVEDVHMVEVVGSGSRVPAMIKILTEFFG-KEPR 360
Query: 369 VSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGTLVRG------------DL 416
++N E V+ G A+Q A+L T K + + P S+ +G
Sbjct: 361 RTMNASECVSRGCALQCAIL-SPTFKVREFQVHESFPFSISLAWKGAATDAQNGGTENQQ 419
Query: 417 LSVVIPRNTPIPVKKTKNYY---TTKDDLSYVSVRVYEGERLKASENNLLGKFRFSIPPA 473
++V P+ PIP K +Y T D+ Y V + +G F+ S
Sbjct: 420 STIVFPKGNPIPSVKALTFYRSGTFSIDVQYSDVNDLQAP--PKISTYTIGPFQSS--KG 475
Query: 474 PRGQIPIKVCFSIDFDGILNVSATEDTCGNRQEITIT----NEKGRLSTEEIERMIQEAE 529
R ++ +KV ++ GI++V + E+++T E ++ T++ A
Sbjct: 476 ERAKLKVKV--RLNLHGIVSVESATLLEEEEVEVSVTKDQSEETAKMDTDKASAEAAPAS 533
Query: 530 NFKDEDMKFKKKVKAMNDLDD 550
D +M+ K D+
Sbjct: 534 GDSDVNMQDAKDTSDATGTDN 554
Score = 28.9 bits (63), Expect = 8.6
Identities = 14/56 (25%), Positives = 27/56 (48%)
Query: 514 GRLSTEEIERMIQEAENFKDEDMKFKKKVKAMNDLDDYLYTMRKVMKDDSVSSMLT 569
G L T E+E+ +++ +D ++ N ++ Y+Y MR + D + LT
Sbjct: 592 GALKTVEVEKAVEKEFEMALQDRVMEETKDRKNAVESYVYDMRNKLSDKYQNISLT 647
>At2g32120 70kD heat shock protein
Length = 563
Score = 235 bits (600), Expect = 5e-62
Identities = 166/502 (33%), Positives = 254/502 (50%), Gaps = 32/502 (6%)
Query: 9 AIGIDLGTTYSCVAVWQDQNNRAEIIHNDQGNRTTPSFVAFTDDKRLIGDAAKNQAASNP 68
A+GID+GT+ +AVW ++ I+ N + + SFV F D+ G NQ A
Sbjct: 30 ALGIDIGTSQCSIAVWN--GSQVHILRNTRNQKLIKSFVTFKDEVPAGG--VSNQLAHEQ 85
Query: 69 VN----TIFDAKRLIGRKYSDSVIQNDIRLWPFKVVPGNDD-KPEIIVKYKGGEKHFCAE 123
IF+ KRL+GR +D V+ L PF V + +P I + E
Sbjct: 86 EMLTGAAIFNMKRLVGRVDTDPVVHASKNL-PFLVQTLDIGVRPFIAALVNNAWRSTTPE 144
Query: 124 EISSMVLTKMREIAEKFLESPVKNAVVTVPAYFNDSQRKATKDAGAIAGLNVMRIINEPT 183
E+ ++ L ++R +AE L+ PV+N V+TVP F+ Q + A A+AGL+V+R++ EPT
Sbjct: 145 EVLAIFLVELRLMAEAQLKRPVRNVVLTVPVSFSRFQLTRFERACAMAGLHVLRLMPEPT 204
Query: 184 AAALAYGLQKRANCVE------ERNIFIFDLGGGTFDVSLLTIKNNLFEVKATSGDTHLG 237
A AL Y Q++ + ER IF++G G DV++ + ++KA +G +G
Sbjct: 205 AIALLYAQQQQMTTHDNMGSGSERLAVIFNMGAGYCDVAVTATAGGVSQIKALAGSP-IG 263
Query: 238 GEDFDNRMVNHFVMEFNRKYMKDISGSPRVLRRLRTACERTKRTLSFDTNATIDIDSLFE 297
GED + H ++ SG LR A + L+ N I++D L
Sbjct: 264 GEDILQNTIRHIAPP-----NEEASGL------LRVAAQDAIHRLTDQENVQIEVD-LGN 311
Query: 298 GIDFHSSITRAKFEQLNVDLFKKCIETVESCLSDAQIEKSSVDDAVLVGGSSRIPKVQQL 357
G + R +FE++N +F++C V CL DA++ +DD ++VGG S IPKV+ +
Sbjct: 312 GNKISKVLDRLEFEEVNQKVFEECERLVVQCLRDARVNGGDIDDLIMVGGCSYIPKVRTI 371
Query: 358 LKEFFNWKDICVSINPDEAVAYGAAVQAALLCEATEK--SLNLVLRDVTPLSLGTLVRGD 415
+K +I +NP EA GAA++ A+ + SL+L+ TPL++G G+
Sbjct: 372 IKNVCKKDEIYKGVNPLEAAVRGAALEGAVTSGIHDPFGSLDLLTIQATPLAVGVRANGN 431
Query: 416 LLSVVIPRNTPIPVKKTKNYYTTKDDLSYVSVRVYEGERLKASENNLLGKFRF-SIPPAP 474
VIPRNT +P +K + T +D+ + +YEGE EN+LLG F+ IPPAP
Sbjct: 432 KFIPVIPRNTMVPARKDLFFTTVQDNQKEALIIIYEGEGETVEENHLLGYFKLVGIPPAP 491
Query: 475 RGQIPIKVCFSIDFDGILNVSA 496
+G I VC ID L V A
Sbjct: 492 KGVPEINVCMDIDASNALRVFA 513
>At4g30350 unknown protein
Length = 924
Score = 34.7 bits (78), Expect = 0.16
Identities = 34/143 (23%), Positives = 63/143 (43%), Gaps = 21/143 (14%)
Query: 304 SITRAKFEQLNVDLFKKCIETVESCLSD-AQIEKSSVDDAVLV------------GGSSR 350
SI+ + Q+ + C ++S +D A++EK D V G +
Sbjct: 420 SISPTRSFQIPMSKMSCCSRCLQSYENDVAKVEKDLTGDNRSVLPQWLQNAKANDDGDKK 479
Query: 351 IPKVQQLLKEFFNWKDICVSINPDEAVAYGAAVQAALLCEATEKSLNLVLRDVTPLSLGT 410
+ K QQ+++ W D+C+ ++P+++V+ A + + +S D+TP G+
Sbjct: 480 LTKDQQIVELQKKWNDLCLRLHPNQSVSERIAPSTLSMMKINTRS------DITP--PGS 531
Query: 411 LVRGDLLSVVIPRNTPIPVKKTK 433
V DL+ R P KKT+
Sbjct: 532 PVGTDLVLGRPNRGLSSPEKKTR 554
>At5g40450 unknown protein
Length = 2910
Score = 30.8 bits (68), Expect = 2.3
Identities = 26/92 (28%), Positives = 43/92 (46%), Gaps = 15/92 (16%)
Query: 487 DFDGILNVSATEDT------CGNRQEITITNEKGRLSTEEIERMIQ----EAENFKD--- 533
D + + +VS T+D N+Q+ ITNE L +IE +Q E+EN KD
Sbjct: 2748 DIESLSSVSKTQDKPEPEYEVPNQQKREITNEVPSLENSKIEEELQKKDEESENTKDLFS 2807
Query: 534 --EDMKFKKKVKAMNDLDDYLYTMRKVMKDDS 563
++ + K A L D++ K +D++
Sbjct: 2808 VVKETEPTLKEPARKSLSDHIQKEPKTEEDEN 2839
>At1g21160 transcription factor, putative
Length = 1088
Score = 30.4 bits (67), Expect = 2.9
Identities = 16/50 (32%), Positives = 27/50 (54%)
Query: 512 EKGRLSTEEIERMIQEAENFKDEDMKFKKKVKAMNDLDDYLYTMRKVMKD 561
E+ RL EE ER I+E + E+++ K+K++ M + L K +D
Sbjct: 243 EEERLRKEEEERRIEEEREREAEEIRQKRKIRKMEKKQEGLILTAKQKRD 292
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.382
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,019,915
Number of Sequences: 26719
Number of extensions: 561219
Number of successful extensions: 1780
Number of sequences better than 10.0: 34
Number of HSP's better than 10.0 without gapping: 21
Number of HSP's successfully gapped in prelim test: 13
Number of HSP's that attempted gapping in prelim test: 1671
Number of HSP's gapped (non-prelim): 44
length of query: 600
length of database: 11,318,596
effective HSP length: 105
effective length of query: 495
effective length of database: 8,513,101
effective search space: 4213984995
effective search space used: 4213984995
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144722.13


