

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144722.10 + phase: 0 /pseudo
(1595 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
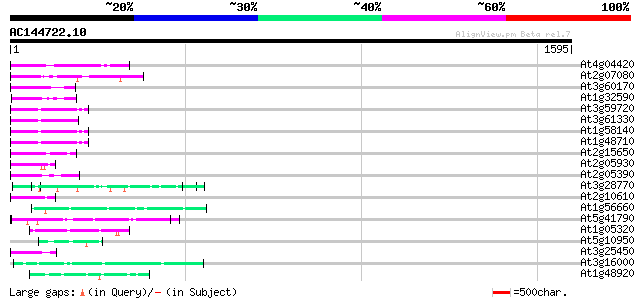
Score E
Sequences producing significant alignments: (bits) Value
At4g04420 putative transposon protein 88 5e-17
At2g07080 putative gag-protease polyprotein 87 1e-16
At3g60170 putative protein 70 7e-12
At1g32590 hypothetical protein, 5' partial 69 2e-11
At3g59720 copia-type reverse transcriptase-like protein 63 1e-09
At3g61330 copia-type polyprotein 62 2e-09
At1g58140 hypothetical protein 62 3e-09
At1g48710 hypothetical protein 62 3e-09
At2g15650 putative retroelement pol polyprotein 59 2e-08
At2g05930 copia-like retroelement pol polyprotein 58 4e-08
At2g05390 putative retroelement pol polyprotein 57 7e-08
At3g28770 hypothetical protein 55 3e-07
At2g10610 pseudogene 54 6e-07
At1g56660 hypothetical protein 51 5e-06
At5g41790 myosin heavy chain-like protein 51 6e-06
At1g05320 unknown protein 50 1e-05
At5g10950 putative protein 48 5e-05
At3g25450 hypothetical protein 47 7e-05
At3g16000 myosin heavy chain like protein 47 9e-05
At1g48920 unknown protein 47 1e-04
>At4g04420 putative transposon protein
Length = 1008
Score = 87.8 bits (216), Expect = 5e-17
Identities = 91/350 (26%), Positives = 155/350 (44%), Gaps = 44/350 (12%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
Q+E M++ E+IEE + + S L K Y V K+LR LPSR+ K TA+
Sbjct: 131 QFENLSMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGT 190
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEESP 122
+ D +++ E++V L+ +E+ + + SK +AL + K
Sbjct: 191 SLDTDSIDFEEVVGMLQAYELEITSGK-GGYSKGLALAASAK------------------ 231
Query: 123 DGDSDEDQSVK--MAMLSNKLEYLARK--QKKFLSKRGSYK---NSKNEDQKGCFNCKKP 175
+E Q +K M+M++ R+ +K F +G+ + S D+ C C+
Sbjct: 232 ---KNEIQELKDTMSMMAKDFSRAMRRVEKKGFGRNQGTDRYRDRSSKRDEIQCHECQGY 288
Query: 176 GHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAK 235
GH A+CP L+++ K S+ + +KF KS +SES S+ +++D K
Sbjct: 289 GHIKAECPSLKRKDLK-CSECNGLGHTKFDCVGSKSKPDKSCSSESESDSNDGDSEDYIK 347
Query: 236 AAVGLVATV-SSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKE 294
V V + + S++E+D EDE+ E D K++ + E L +
Sbjct: 348 GFVSFVGIIEEKDESSDSEADGEDEDN-----SADEDSDIEKDV-----NINEEFRKLYD 397
Query: 295 KYVDLMKQQKSTLLE-LKASE--EELKGFNLISTTYEDRLKSLCQKLQEK 341
++ L K++ + L E LK E E+LKG + L C +EK
Sbjct: 398 SWLMLSKEKVAWLEEKLKVQELTEKLKGELTAANQKNSELIQKCSVAEEK 447
>At2g07080 putative gag-protease polyprotein
Length = 627
Score = 86.7 bits (213), Expect = 1e-16
Identities = 101/401 (25%), Positives = 174/401 (43%), Gaps = 56/401 (13%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
Q+E +M+ DE I + S+ L + +++ K+Y V K+LR LP ++ +
Sbjct: 112 QFEYLKMEPDEKIVKFSSKISALANEAEVMGKTYKDQKLVKKLLRCLPPKFAAHKAVMRV 171
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEESP 122
A + + +S DLV LK+ EM ++ + K SK+IA A + SE +E
Sbjct: 172 AGNTDKISFVDLVGMLKLEEMKADQDKV-KPSKNIAF----------NADQGSEQFQEIK 220
Query: 123 DGDSDEDQSVKMAMLSNKL-EYLARKQKKFLSKRGSYKNSKNEDQK-----GCFNCKKPG 176
DG MA+L+ + L R + RG + S+N+D + C+ C G
Sbjct: 221 DG---------MALLARNFGKALKRVEIDGERSRGRFSRSENDDLRKKKEIQCYECGGFG 271
Query: 177 HFIADCPDLQKEKFK-------GKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEE 229
H +CP ++++ K G +K N SK + +KSL+ S SD E
Sbjct: 272 HIKPECPITKRKEMKCLKCKGVGHTKFECPNKSKLK---EKSLI---------SFSD-SE 318
Query: 230 ADDDAKAAVGLVATVSSEAVSE--AESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTN 287
+DD+ + + VA ++S S+ +++DS+ + E+ K + L DS +L
Sbjct: 319 SDDEGEELLNFVAFMASSDSSKFMSDTDSDCDEELNPKDKYRVLYDSWVQLSKDKLKLVK 378
Query: 288 ELTDLKEKYVDLMKQQKSTLLELKAS-------EEELKGFNLISTTYEDRLKSLCQKLQE 340
E L+ K ++ + K L + +++L DR K L ++L +
Sbjct: 379 EKLTLEAKLANVSTEDKQKLSGITVDGNSQDYYQKKLDCLQKECHRERDRTKLLERELND 438
Query: 341 KCDK-GSGNKHEIALDDFIMAGIDRSKVASMIYSTYKNKGK 380
K + NK +LD + G S+ + Y Y K K
Sbjct: 439 KYKQIRMLNKGLESLDKILAMGRTDSQQRGLGYQGYTGKIK 479
>At3g60170 putative protein
Length = 1339
Score = 70.5 bits (171), Expect = 7e-12
Identities = 48/185 (25%), Positives = 79/185 (41%), Gaps = 35/185 (18%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
++EL MK+ E I+ R T+V+ ++ + S VSKILRSL ++ V +IEE
Sbjct: 121 EFELLAMKEGEKIDTFLGRTLTVVNKMKTNGEVMEQSTIVSKILRSLTPKFNYVVCSIEE 180
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEESP 122
+ DL+TLS+++L SL VHE LN H +++ + + + ++ S
Sbjct: 181 SNDLSTLSIDELHGSLLVHEQRLNGHVQEEQALKVTHEERPSQGRGRGVFRGSRGR---- 236
Query: 123 DGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFIADC 182
RG ++ N C+ C GHF +C
Sbjct: 237 -------------------------------GRGRGRSGTNRAIVECYKCHNLGHFQYEC 265
Query: 183 PDLQK 187
P+ +K
Sbjct: 266 PEWEK 270
>At1g32590 hypothetical protein, 5' partial
Length = 1263
Score = 68.9 bits (167), Expect = 2e-11
Identities = 52/189 (27%), Positives = 82/189 (42%), Gaps = 40/189 (21%)
Query: 4 YELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEA 63
+E+ MK E+I +SR + + ++ L + S V KILR+L ++ V AIEE+
Sbjct: 79 FEVLEMKIGETITGYFSRVMEITNDMRNLGEDMPDSKVVEKILRTLVEKFTYVVCAIEES 138
Query: 64 KDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEESPD 123
++ L+V+ L SSL VHE +L+ H+ + ++ K+ ++ PD
Sbjct: 139 NNIKELTVDGLQSSLMVHEQNLSRHDVEE-----------RVLKAETQWR--------PD 179
Query: 124 GDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSY----KNSKNEDQKGCFNCKKPGHFI 179
G S RG Y + N D CF C K GH+
Sbjct: 180 GGRGRGGSPSRG-----------------RGRGGYQGRGRGYVNRDTVECFKCHKMGHYK 222
Query: 180 ADCPDLQKE 188
A+CP +KE
Sbjct: 223 AECPSWEKE 231
>At3g59720 copia-type reverse transcriptase-like protein
Length = 1272
Score = 63.2 bits (152), Expect = 1e-09
Identities = 52/223 (23%), Positives = 102/223 (45%), Gaps = 14/223 (6%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
++E +MK+ E + + +SR T+ + L+ + + K+LRSL ++ VT IEE
Sbjct: 119 EFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEE 178
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSI---ALPSKGKISKSSKAYKASESEE 119
KDL +++E L+ SL+ +E E KK + I L + ++ ++Y+ +
Sbjct: 179 TKDLEAMTIEQLLGSLQAYE------EKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQ 232
Query: 120 ESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFI 179
G + + K G K+ ++ C+NC K GH+
Sbjct: 233 VRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGK-GHPKSRYDKSSVKCYNCGKFGHYA 291
Query: 180 ADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSE 222
++C +KFK +K+++ K +++ LMA+++ + E
Sbjct: 292 SECKAPSNKKFK---EKANYVEEKIQEE-DMLLMASYKKDEQE 330
>At3g61330 copia-type polyprotein
Length = 1352
Score = 62.4 bits (150), Expect = 2e-09
Identities = 47/195 (24%), Positives = 88/195 (45%), Gaps = 10/195 (5%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
++E +MK+ E + + +SR T+ + L+ + + K+LRSL ++ VT IEE
Sbjct: 119 EFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEE 178
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIA---LPSKGKISKSSKAYKASESEE 119
KDL +++E L+ SL+ +E E KK + IA L + ++ ++Y+ +
Sbjct: 179 TKDLEAMTIEQLLGSLQAYE------EKKKKKEDIAEQVLNMQITKEENGQSYQRRGGGQ 232
Query: 120 ESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFI 179
G + + K G K+ ++ C+NC K GH+
Sbjct: 233 VRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGK-GHPKSRYDKSSVKCYNCGKFGHYA 291
Query: 180 ADCPDLQKEKFKGKS 194
++C +KF+ K+
Sbjct: 292 SECKAPSNKKFEEKA 306
>At1g58140 hypothetical protein
Length = 1320
Score = 61.6 bits (148), Expect = 3e-09
Identities = 51/223 (22%), Positives = 102/223 (44%), Gaps = 14/223 (6%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
++E +MK+ E + + +SR T+ + L+ + + K+LRSL ++ VT IEE
Sbjct: 119 EFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEE 178
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSI---ALPSKGKISKSSKAYKASESEE 119
KDL +++E L+ SL+ +E E KK + I L + ++ ++Y+ +
Sbjct: 179 TKDLEAMTIEQLLGSLQAYE------EKKKKKEDIVEQVLNMQITKEENGQSYQRRGGGQ 232
Query: 120 ESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFI 179
G + + K G K+ ++ C+NC K GH+
Sbjct: 233 VRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGK-GHPKSRYDKSSVKCYNCGKFGHYA 291
Query: 180 ADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSE 222
++C +KF+ +K+++ K +++ LMA+++ + E
Sbjct: 292 SECKAPSNKKFE---EKANYVEEKIQEE-DMLLMASYKKDEQE 330
>At1g48710 hypothetical protein
Length = 1352
Score = 61.6 bits (148), Expect = 3e-09
Identities = 51/223 (22%), Positives = 102/223 (44%), Gaps = 14/223 (6%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
++E +MK+ E + + +SR T+ + L+ + + K+LRSL ++ VT IEE
Sbjct: 119 EFEALQMKEGELVSDYFSRVLTVTNNLKRNGEKLDDVRIMEKVLRSLDLKFEHIVTVIEE 178
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSI---ALPSKGKISKSSKAYKASESEE 119
KDL +++E L+ SL+ +E E KK + I L + ++ ++Y+ +
Sbjct: 179 TKDLEAMTIEQLLGSLQAYE------EKKKKKEDIIEQVLNMQITKEENGQSYQRRGGGQ 232
Query: 120 ESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFI 179
G + + K G K+ ++ C+NC K GH+
Sbjct: 233 VRGRGRGGYGNGRGWRPHEDNTNQRGENSSRGRGK-GHPKSRYDKSSVKCYNCGKFGHYA 291
Query: 180 ADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSE 222
++C +KF+ +K+++ K +++ LMA+++ + E
Sbjct: 292 SECKAPSNKKFE---EKANYVEEKIQEE-DMLLMASYKKDEQE 330
>At2g15650 putative retroelement pol polyprotein
Length = 1347
Score = 59.3 bits (142), Expect = 2e-08
Identities = 44/187 (23%), Positives = 86/187 (45%), Gaps = 13/187 (6%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
+YE +M D+++I+ + L L + + + KIL SLP+++ V+ +E+
Sbjct: 121 EYENLKMYDNDNIKTFTDKLIVLEIQLTYHGEKKTNTQLIQKILISLPAKFDSIVSVLEQ 180
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEESP 122
+DL+ L++ +L+ LK E + E S K + + SKG + S ++++
Sbjct: 181 TRDLDALTMSELLGILKAQEARVTAREESTKEGAFYVRSKG---------RESGFKQDNT 231
Query: 123 DGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFIADC 182
+ ++D+ S+K ++K G K S + C+ C K GH+ +C
Sbjct: 232 NNRVNQDKKWCGFHKSSKHTEEECREKPKNDDHGKNKRSNIK----CYKCGKIGHYANEC 287
Query: 183 PDLQKEK 189
KE+
Sbjct: 288 RSKNKER 294
>At2g05930 copia-like retroelement pol polyprotein
Length = 916
Score = 58.2 bits (139), Expect = 4e-08
Identities = 43/150 (28%), Positives = 69/150 (45%), Gaps = 28/150 (18%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEE 62
Q+E M++ E+IEE + + S L K Y V K+LR LPSR+ K TA+
Sbjct: 143 QFENLTMEETENIEEFSGKISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGT 202
Query: 63 AKDLNTLSVEDLVSSLKVHEMSLNEHE----------TSKKSKSI--------------A 98
+ D N++ E++V + +E+ + + S K K +
Sbjct: 203 SLDTNSIDFEEVVGMFQAYELEITSGKGGYGHIKAECPSLKRKDLKCSECKGLGHIKFDC 262
Query: 99 LPSKGKISKSSKAYKASESEEESPDGDSDE 128
+ SK K +S +SESE +S DGDS++
Sbjct: 263 VGSKSKPDRSC----SSESESDSNDGDSED 288
Score = 39.3 bits (90), Expect = 0.018
Identities = 76/345 (22%), Positives = 132/345 (38%), Gaps = 58/345 (16%)
Query: 1 VHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAI 60
V+Q + ++++ ES +E + + G +K+S + D ++ +L +
Sbjct: 101 VNQNQFKQIQNCESAKEAWDKLAKAYEGTSSVKRSRI--DMLASQFENL---------TM 149
Query: 61 EEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEE 120
EE +++ S K+ ++ H KK K L K S+ +
Sbjct: 150 EETENIEEFSG-------KISAIASEAHNLGKKYKDKKLVKKLLRCLPSRFESKRTAMGT 202
Query: 121 SPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFIA 180
S D +S + + V + +LE + K + + + K +D K C CK GH
Sbjct: 203 SLDTNSIDFEEVVGMFQAYELEITSGKGG-YGHIKAECPSLKRKDLK-CSECKGLGHIKF 260
Query: 181 DCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGL 240
DC KSK SS +SES S+ +++D K V
Sbjct: 261 DC-------VGSKSKPDRSCSS-----------------ESESDSNDGDSEDYIKGFVSF 296
Query: 241 VATV-SSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDL 299
V + + S++E+D EDE+ + E + E E L + ++ L
Sbjct: 297 VGIIEEKDESSDSEADGEDEDNSADEDSDIEKDVKINE----------EFRKLYDSWLML 346
Query: 300 MKQQKSTLLE-LKASE--EELKGFNLISTTYEDRLKSLCQKLQEK 341
K++ + L E LK E E+LKG + L C +EK
Sbjct: 347 SKEKVAWLEEKLKVQELTEKLKGELTAANQKNSELTQKCSVAEEK 391
>At2g05390 putative retroelement pol polyprotein
Length = 1307
Score = 57.4 bits (137), Expect = 7e-08
Identities = 44/197 (22%), Positives = 86/197 (43%), Gaps = 30/197 (15%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 61
+++ MKD+E+I+E R + + + L + S V K L+SLP ++ + A+E
Sbjct: 77 EFDRLNMKDNETIDEFVGRISEISTKSESLGEEIEESKIVKKFLKSLPRKKYIHIIAALE 136
Query: 62 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEES 121
+ DLNT ED+V +K +E + + + S + +GK+ Y SES ++
Sbjct: 137 QILDLNTTGFEDIVGRMKTYEDRVCDEDDSPE-------EQGKL-----MYANSESSYDT 184
Query: 122 PDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFIAD 181
G R + + G +++ + C+ C K GH+ ++
Sbjct: 185 RGGRG-----------------RGRGRSSGRGRGGYGYQQRDKSKVICYRCDKTGHYASE 227
Query: 182 CPDLQKEKFKGKSKKSS 198
C D + K + ++ +
Sbjct: 228 CLDRLLKLIKAQEQQQN 244
>At3g28770 hypothetical protein
Length = 2081
Score = 55.1 bits (131), Expect = 3e-07
Identities = 120/560 (21%), Positives = 213/560 (37%), Gaps = 82/560 (14%)
Query: 9 MKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRS----LPSRWRPKVTAIEEAK 64
+KDDES+ + +L + +K + S + SKI+ + S +V +
Sbjct: 611 LKDDESVGAKTNNETSLEEKREQTQKGHDNSIN-SKIVDNKGGNADSNKEKEVHVGDSTN 669
Query: 65 DLNTLSVEDLVSSLKVH---------EMSLNEHETSKKSKSIALPSKGKISKSSKAYKAS 115
D N S ED S ++V E ++ S + K + SK K+
Sbjct: 670 DNNMESKEDTKSEVEVKKNDGSSEKGEEGKENNKDSMEDKKLENKESQTDSKDDKSVDDK 729
Query: 116 ESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKP 175
+ E + G+S +D+SV+ K E +++ KK + +N K E+ +G N K
Sbjct: 730 QEEAQIYGGESKDDKSVEAK--GKKKE--SKENKKTKTNENRVRN-KEENVQG--NKK-- 780
Query: 176 GHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMAT--WEDLDSESGSDKEEADDD 233
+ EK + KK S ++ + K L +T ++ SG D +E ++
Sbjct: 781 ----------ESEKVEKGEKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEE 830
Query: 234 AKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLK 293
+K S EA + E+ D N V +K ++L D E + N+ +K
Sbjct: 831 SK------DYQSVEAKEKNENGGVDTN-VGNKEDSKDLKDDRS-----VEVKANKEESMK 878
Query: 294 EKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIA 353
+K ++ + KS+ E ++ + KGSG +
Sbjct: 879 KKREEVQRNDKSSTKE---------------------VRDFANNMDIDVQKGSGESVKYK 917
Query: 354 LDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEG 413
D+ + +K I ++ K KGK +++SK ++K K+ K +V
Sbjct: 918 KDEKKEGNKEENK--DTINTSSKQKGKDKKKKKKESKNSNMKK-----KEEDKKEYV--- 967
Query: 414 TNAITAVQSKPEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIKPES 473
N + + + + K EN K +S+ + K + K E ++ K E+
Sbjct: 968 NNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEE-KKSKTKEEA 1026
Query: 474 KIPKQKDQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVQGRKSKTSKINPKGPMKIW 533
K K+K Q K EK + K + + L K + K K N K K
Sbjct: 1027 KKEKKKSQDKK---REEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHKSKKKED 1083
Query: 534 VPKSELAKNAGVLKGKRETK 553
+ E K+ + K+E K
Sbjct: 1084 KKEHEDNKSMKKEEDKKEKK 1103
Score = 49.3 bits (116), Expect = 2e-05
Identities = 107/474 (22%), Positives = 190/474 (39%), Gaps = 76/474 (16%)
Query: 61 EEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEE 120
EE++DL E+ + E +E+ SKK + K K + K + EE
Sbjct: 1053 EESRDLKAKKKEE-----ETKEKKESENHKSKKKEDKKEHEDNKSMKKEEDKKEKKKHEE 1107
Query: 121 SPDGDSDEDQSVKMAML----SNKLEYLARKQKKF----LSKRGSYKNSKNEDQ-----K 167
S +ED+ M L SNK + ++KK L K+ S K K E++ K
Sbjct: 1108 SKSRKKEEDKK-DMEKLEDQNSNKKKEDKNEKKKSQHVKLVKKESDKKEKKENEEKSETK 1166
Query: 168 GCFNCKKPGHFI-----ADCPDLQKEKFK---------------GKSKKSSFNSSKFRKQ 207
+ K + + D QK+K K + K++S +K +K+
Sbjct: 1167 EIESSKSQKNEVDKKEKKSSKDQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKE 1226
Query: 208 IKKSLMATWED---LDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYS 264
KK +D +SG KE + ++K A S+A ++A+SD E +NE+
Sbjct: 1227 TKKEKNKPKDDKKNTTKQSGGKKESMESESKEAEN---QQKSQATTQADSD-ESKNEI-- 1280
Query: 265 KIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASEEELKGFNLIS 324
L + S + ++ D E +++ Q S + +EE+ K ++
Sbjct: 1281 ----------LMQADSQADSHSDSQADSDESKNEILMQADSQATTQRNNEEDRKKQTSVA 1330
Query: 325 ------TTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRSKVASMIYSTYKNK 378
T E++ K K + K SG K E + A + A+ + ++K
Sbjct: 1331 ENKKQKETKEEKNKPKDDK--KNTTKQSGGKKESMESESKEAENQQKSQATTQADSDESK 1388
Query: 379 GKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAITAVQSKPEASGSQAKITSKPE 438
+ + ++ ++ +S S D D K+ + + + T ++ E + K TS E
Sbjct: 1389 NEILMQADSQADSHS-DSQAD--SDESKNEILMQADSQATTQRNNEE---DRKKQTSVAE 1442
Query: 439 NLKIKVM--TKSDPKSQKIKILKRSEPVHQNLIKPESKIPKQKDQKNKAATASE 490
N K K K+ PK K ++S +++ + ESK + QK++A T E
Sbjct: 1443 NKKQKETKEEKNKPKDDKKNTTEQSGGKKESM-ESESK-EAENQQKSQATTQGE 1494
Score = 48.1 bits (113), Expect = 4e-05
Identities = 94/472 (19%), Positives = 174/472 (35%), Gaps = 48/472 (10%)
Query: 87 EHETSKKSKSIALPSKGKISKSSKAYKASESEEESPDGDSDED---QSVKM--------- 134
E + SK +KS+ K+S + +A E E D +E QSV+
Sbjct: 789 EKKESKDAKSVETKDNKKLSSTENRDEAKERSGEDNKEDKEESKDYQSVEAKEKNENGGV 848
Query: 135 -AMLSNKLEYLARKQKKFL----SKRGSYKNSKNEDQKGCFNCKKPGHFIADCPDLQKEK 189
+ NK + K + + +K S K + E Q+ + K A+ D+ +K
Sbjct: 849 DTNVGNKEDSKDLKDDRSVEVKANKEESMKKKREEVQRNDKSSTKEVRDFANNMDIDVQK 908
Query: 190 FKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAK-AAVGLVATVSSEA 248
G+S K + + ++ K+ T + G DK++ ++K + + E
Sbjct: 909 GSGESVK--YKKDEKKEGNKEENKDTINTSSKQKGKDKKKKKKESKNSNMKKKEEDKKEY 966
Query: 249 VSEAESDSEDENEVYSKIPRQELVDSLKELLSLFE--------HRTNELTDLKEKYVDLM 300
V+ ED + +K +L + K+ E E + K K +
Sbjct: 967 VNNELKKQEDNKKETTKSENSKLKEENKDNKEKKESEDSASKNREKKEYEEKKSKTKEEA 1026
Query: 301 KQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIMA 360
K++K + K E++ + E R +K +E +K H+
Sbjct: 1027 KKEKKKSQDKKREEKDSEERKSKKEKEESRDLKAKKKEEETKEKKESENHK--------- 1077
Query: 361 GIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGTNAITAV 420
+ K + K+ K E+K E S + K ++ E N+
Sbjct: 1078 --SKKKEDKKEHEDNKSMKKEEDKKEKKKHEESKSRKKEEDKKDMEKL---EDQNSNKKK 1132
Query: 421 QSKPEASGSQ-AKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIKPESKIPKQK 479
+ K E SQ K+ K + K K K + + + K ++ S+ + K E K K
Sbjct: 1133 EDKNEKKKSQHVKLVKKESDKKEK---KENEEKSETKEIESSKSQKNEVDKKEKK--SSK 1187
Query: 480 DQKNKAATASEKTIPKGVKPKVLNDQKPLSIHPKVQGRKSKTSKINPKGPMK 531
DQ+ K +++ K +K + +K S+ + +++K K PK K
Sbjct: 1188 DQQKKKEKEMKESEEKKLKKNEEDRKKQTSVEENKKQKETKKEKNKPKDDKK 1239
Score = 33.9 bits (76), Expect = 0.78
Identities = 49/233 (21%), Positives = 91/233 (39%), Gaps = 45/233 (19%)
Query: 86 NEHETSKKSKSIALPSKGKISKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLA 145
N E KK S+A K K +K K ++ + ++ E K + ++ +
Sbjct: 1429 NNEEDRKKQTSVAENKKQKETKEEK------NKPKDDKKNTTEQSGGKKESMESESKEAE 1482
Query: 146 RKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFR 205
+QK + +G SKNE +Q + S +S + +
Sbjct: 1483 NQQKSQATTQGESDESKNEIL------------------MQADSQADTHANSQGDSDESK 1524
Query: 206 KQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSK 265
+I LM DS++ SD+ + + ++ S+A S+ +SD E +NE+ +
Sbjct: 1525 NEI---LMQADSQADSQTDSDESKNE--------ILMQADSQADSQTDSD-ESKNEILMQ 1572
Query: 266 IPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASEEELK 318
Q + SL +++ KE D + ++ S +E+K EE K
Sbjct: 1573 ADSQAKIGE-----SLEDNKVKG----KEDNGDEVGKENSKTIEVKGRHEESK 1616
>At2g10610 pseudogene
Length = 531
Score = 54.3 bits (129), Expect = 6e-07
Identities = 37/128 (28%), Positives = 62/128 (47%), Gaps = 5/128 (3%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 61
++ +MKD I++ + L S V K L+S+P R+ V A+E
Sbjct: 3 EFHRLKMKDSHKIDDFAGMLSEISVKSAALGVIIEESRLVKKFLKSVPRKRYIHIVAALE 62
Query: 62 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEES 121
+ DLN ++ +D+V LKV+E + + + ++ +S L S S S ++ EEE
Sbjct: 63 QVLDLNAITFDDIVGRLKVYEERVYDEDDPEEDQSKLLYS----SMDSSKRESEGEEEED 118
Query: 122 PDGDSDED 129
P G+ DED
Sbjct: 119 PIGEEDED 126
>At1g56660 hypothetical protein
Length = 522
Score = 51.2 bits (121), Expect = 5e-06
Identities = 127/532 (23%), Positives = 198/532 (36%), Gaps = 56/532 (10%)
Query: 61 EEAKDLNTLSVEDLVSSLKVHEMSLN-EHETSKKSKSIAL-------PSKGKISKSSKAY 112
E AK+ L V+ L E N E E K+KSI S GK SK K
Sbjct: 6 ENAKE-EKLHVKIKTQELDPKEKGENVEVEMEVKAKSIEKVKAKKDEESSGK-SKKDKEK 63
Query: 113 KASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNC 172
K ++ + D D+D+ M+S K E + + + E +KG
Sbjct: 64 KKGKNVDSEVKEDKDDDKKKDGKMVSKKHEE-GHGDLEVKESDVKVEEHEKEHKKG---- 118
Query: 173 KKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIK----KSLMATWEDLDSESGSDKE 228
K+ H + K+K K K S K +K K + + E+L+ E G +
Sbjct: 119 KEKKHEELEEEKEGKKKKNKKEKDESGPEEKNKKADKEKKHEDVSQEKEELEEEDGKKNK 178
Query: 229 EADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNE 288
+ + D E + ES S ++ +V K + E D KE + + ++
Sbjct: 179 KKEKDESGTEEKKKKPKKEKKQKEESKSNEDKKVKGKKEKGEKGDLEKE--DEEKKKEHD 236
Query: 289 LTDLKEKYVDLMKQQK-----STLLELKASEEELKGFNLISTTYED-RLKSLCQKLQ--E 340
TD + K D K +K S E K ++ K ST ED +LK K + E
Sbjct: 237 ETDQEMKEKDSKKNKKKEKDESCAEEKKKKPDKEKKEKDESTEKEDKKLKGKKGKGEKPE 296
Query: 341 KCDKGSGNKHEIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDC 400
K D+G K A + +D KNK K K KE + C+
Sbjct: 297 KEDEGKKTKEHDATEQ----EMDDEAADHKEGKKKKNKDKA------KKKETVIDEVCEK 346
Query: 401 IKDGLKSTFVPEGTNAITAVQSKPEASGSQAKITSKPEN-LKIKVMTK----SDPKSQKI 455
+ K E K K EN L+ +VM++ +P+++K
Sbjct: 347 ETKDKDDDEGETKQKKNKKKEKKSEKGEKDVKEDKKKENPLETEVMSRDIKLEEPEAEKK 406
Query: 456 KILKRSEPVHQNLIKPESKIPKQKDQKNKAATASEKTIPKGVKPKVLNDQ---KPLSIHP 512
+ E + ES+ K+K +K+K + T +PK+ D+ K S
Sbjct: 407 EEDDTEEKKKSKVEGGESEEGKKKKKKDKKKNKKKDT----KEPKMTEDEEEKKDDSKDV 462
Query: 513 KVQGRKSKTSKIN-----PKGPMKIWVPKSELAKNAGVLKGKRETKVMVPRQ 559
K++G K+K K + KG I K++LAK + E K + Q
Sbjct: 463 KIEGSKAKEEKKDKDVKKKKGGNDIGKLKTKLAKIDEKIGALMEEKAEIENQ 514
Score = 32.0 bits (71), Expect = 2.9
Identities = 47/211 (22%), Positives = 81/211 (38%), Gaps = 17/211 (8%)
Query: 38 ASDHVSKILRSLPSRWRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSI 97
A+DH + + + K T I+E + T +D K + E ++ K K +
Sbjct: 318 AADHKEGKKKKNKDKAKKKETVIDEVCEKETKDKDDDEGETKQKKNKKKEKKSEKGEKDV 377
Query: 98 ALPSKG----KISKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLS 153
K + S+ K E E E + D E++ K + + +K+KK
Sbjct: 378 KEDKKKENPLETEVMSRDIKLEEPEAEKKEEDDTEEK--KKSKVEGGESEEGKKKKKKDK 435
Query: 154 KRGSYKNSKNEDQKGCFNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFR--KQIKKS 211
K+ K++K KK D D++ E K K +K + K + I K
Sbjct: 436 KKNKKKDTKEPKMTEDEEEKKD-----DSKDVKIEGSKAKEEKKDKDVKKKKGGNDIGK- 489
Query: 212 LMATWEDLDSESGS---DKEEADDDAKAAVG 239
L +D + G+ +K E ++ K A G
Sbjct: 490 LKTKLAKIDEKIGALMEEKAEIENQIKDAEG 520
>At5g41790 myosin heavy chain-like protein
Length = 1305
Score = 50.8 bits (120), Expect = 6e-06
Identities = 101/484 (20%), Positives = 209/484 (42%), Gaps = 52/484 (10%)
Query: 4 YELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSL-----------PSR 52
+E + + + E+ ++ + L + L S A++ K L S+ S+
Sbjct: 476 HETHQRESSTRLSELETQLKLLEQRVVDLSASLNAAEEEKKSLSSMILEITDELKQAQSK 535
Query: 53 WRPKVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAY 112
+ VT + E+KD T +L S ++VHE + + K + S + K
Sbjct: 536 VQELVTELAESKDTLTQKENELSSFVEVHEAHKRDSSSQVKELEARVESAEEQVKELNQN 595
Query: 113 KASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNC 172
S EE+ + S+K+ + ++ L+ + ++ +GS+ NE F+
Sbjct: 596 LNSSEEEKKILSQQISEMSIKIKRAESTIQELSSESERL---KGSHAEKDNE----LFS- 647
Query: 173 KKPGHFIADCPDLQKEKFKGKSK--KSSFNSSKFR-KQIKKSLMATWEDLDSESGSDKEE 229
+ D + + + + + ++ SS+ R ++ +SL A E+ + S E
Sbjct: 648 ------LRDIHETHQRELSTQLRGLEAQLESSEHRVLELSESLKAAEEESRTMSTKISET 701
Query: 230 ADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVD-SLKELLSLFEHRTNE 288
+D+ + + +V +++++ E +E E++++ + +KEL + E
Sbjct: 702 SDELERTQI-MVQELTADSSKLKEQLAEKESKLFLLTEKDSKSQVQIKELEATVATLELE 760
Query: 289 LTDLKEKYVDLMKQ--QKSTLLE-LKASEEELKG-FNLISTTYEDR---LKSLCQKLQEK 341
L ++ + +DL + K+T++E L+A E+ + + T E+R L +L QKL++
Sbjct: 761 LESVRARIIDLETEIASKTTVVEQLEAQNREMVARISELEKTMEERGTELSALTQKLEDN 820
Query: 342 CDKGSGNKHEIALDDFIMAGID--RSKVASMIYSTYKNKGKGIGYSEEKS-KEYSLKSYC 398
DK S + E + A ID R+++ SM + + + + SEE S K L
Sbjct: 821 -DKQSSSSIET-----LTAEIDGLRAELDSMSVQKEEVEKQMVCKSEEASVKIKRLDDEV 874
Query: 399 DCIKDGLKSTFVPEGTNAITAVQSKPEASGSQAKITSKPENL--KIKV----MTKSDPKS 452
+ ++ + S I + E S ++IT+ E + K+KV + + + S
Sbjct: 875 NGLRQQVASLDSQRAELEIQLEKKSEEISEYLSQITNLKEEIINKVKVHESILEEINGLS 934
Query: 453 QKIK 456
+KIK
Sbjct: 935 EKIK 938
Score = 45.1 bits (105), Expect = 3e-04
Identities = 97/510 (19%), Positives = 207/510 (40%), Gaps = 67/510 (13%)
Query: 1 VHQYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRP-KVTA 59
V + ELF ++D I + S T S L+ +S + VS + SL + K +
Sbjct: 133 VKERELFSLRDIHEIHQRDS--STRASELEAQLES--SKQQVSDLSASLKAAEEENKAIS 188
Query: 60 IEEAKDLNTL-----SVEDLVSSL---------KVHEMS--LNEHETSKKSKSIALPSKG 103
+ + +N L ++++L++ L K E+S + HET ++ SI +
Sbjct: 189 SKNVETMNKLEQTQNTIQELMAELGKLKDSHREKESELSSLVEVHETHQRDSSIHVKELE 248
Query: 104 KISKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKN 163
+ +SSK A ++ + + + S K+A LSN+++ ++ +S+ G K S +
Sbjct: 249 EQVESSKKLVAELNQTLNNAEEEKKVLSQKIAELSNEIKEAQNTIQELVSESGQLKESHS 308
Query: 164 EDQKGCFNCKKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSES 223
+ F+ ++ + ++SS S+ Q++ S E S+
Sbjct: 309 VKDRDLFSL--------------RDIHETHQRESSTRVSELEAQLESS-----EQRISDL 349
Query: 224 GSDKEEADDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFE 283
D ++A+++ KA +SS+ + + + +N + +EL+D L EL +
Sbjct: 350 TVDLKDAEEENKA-------ISSKNLEIMDKLEQAQNTI------KELMDELGELKDRHK 396
Query: 284 HRTNELTDLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCD 343
+ +EL+ L + + K + L +EEE K + + ++ + +QE
Sbjct: 397 EKESELSSLVKSADQQVADMKQS---LDNAEEEKKMLSQRILDISNEIQEAQKTIQEHMS 453
Query: 344 KGSGNKHEIALDDFIMAGIDRSKVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKD 403
+ K + + + G+ S+ + + + L + + ++
Sbjct: 454 ESEQLKESHGVKERELTGLRDIHETHQRESSTRLSELETQLKLLEQRVVDLSASLNAAEE 513
Query: 404 GLK--STFVPEGTNAITAVQSK-----PEASGSQAKITSKPENL----KIKVMTKSDPKS 452
K S+ + E T+ + QSK E + S+ +T K L ++ K D S
Sbjct: 514 EKKSLSSMILEITDELKQAQSKVQELVTELAESKDTLTQKENELSSFVEVHEAHKRDSSS 573
Query: 453 QKIKILKRSEPVHQNLIKPESKIPKQKDQK 482
Q ++ R E + + + + +++K
Sbjct: 574 QVKELEARVESAEEQVKELNQNLNSSEEEK 603
Score = 40.4 bits (93), Expect = 0.008
Identities = 78/360 (21%), Positives = 146/360 (39%), Gaps = 65/360 (18%)
Query: 5 ELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDH----VSKILRSLPSRWRPKVTAI 60
ELF ++D I E + R L + L+ L+ +S+H +S+ L++ R T I
Sbjct: 644 ELFSLRD---IHETHQR--ELSTQLRGLEAQLESSEHRVLELSESLKAAEEESRTMSTKI 698
Query: 61 EEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEE 120
E D + +V L L E K+SK L K SKS K E+
Sbjct: 699 SETSD-ELERTQIMVQELTADSSKLKEQLAEKESKLFLLTEKD--SKSQVQIKELEATVA 755
Query: 121 SPDGDSDE------DQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKK 174
+ + + + D ++A + +E L + ++ +++ + K +++G
Sbjct: 756 TLELELESVRARIIDLETEIASKTTVVEQLEAQNREMVARISELE--KTMEERG------ 807
Query: 175 PGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIK------KSLMATWEDLDSESGSDKE 228
+ L +K + K+SS + +I S+ E+++ + E
Sbjct: 808 -----TELSALT-QKLEDNDKQSSSSIETLTAEIDGLRAELDSMSVQKEEVEKQMVCKSE 861
Query: 229 EA-------DDDAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSL 281
EA DD+ VA++ S+ +E E E ++E + E LS
Sbjct: 862 EASVKIKRLDDEVNGLRQQVASLDSQR-AELEIQLEKKSE------------EISEYLS- 907
Query: 282 FEHRTNELTDLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEK 341
++T+LKE+ ++ +K +S L E+ E++KG L T + L ++L+ K
Sbjct: 908 ------QITNLKEEIINKVKVHESILEEINGLSEKIKGRELELETLGKQRSELDEELRTK 961
>At1g05320 unknown protein
Length = 841
Score = 49.7 bits (117), Expect = 1e-05
Identities = 73/305 (23%), Positives = 130/305 (41%), Gaps = 33/305 (10%)
Query: 56 KVTAIEEAKDLNTLSVEDLVSSLKVHEMSLNEHETS-KKSKSIALPSKGKISKSSKAYKA 114
K TAIE+ +T +DL++ LK HE + EH+ ++ +A K ++ ++
Sbjct: 427 KETAIEKLNQKDT-EAKDLITKLKSHENVIEEHKRQVLEASGVADTRKVEVEEALLKLNT 485
Query: 115 SESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKK 174
ES E + ++ + V + L+ KL + F +K + K + K+
Sbjct: 486 LESTIEELEKENGDLAEVNI-KLNQKLANQGSETDDFQAKLSVLEAEKYQ------QAKE 538
Query: 175 PGHFIAD-CPDLQKEKFKGKSKKSSFNSSKFR-KQIKKSLMATWEDLDSESGSDKEEADD 232
I D L E+ + +S+ SS K + +I +S L ++ DK ++DD
Sbjct: 539 LQITIEDLTKQLTSERERLRSQISSLEEEKNQVNEIYQSTKNELVKLQAQLQVDKSKSDD 598
Query: 233 DAKAAVGLVATVSSEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDL 292
L A V+ ++V E++ ++ E++ K E V L L +H+ ++ L
Sbjct: 599 MVSQIEKLSALVAEKSVLESKF---EQVEIHLK-EEVEKVAELTSKLQEHKHKASDRDVL 654
Query: 293 KEKYVDLMK----------QQKSTL--------LELKASEEELKGFNLISTTYEDRLKSL 334
+EK + L K +QK L LK S+EEL + E +L L
Sbjct: 655 EEKAIQLHKELQASHTAISEQKEALSHKHSELEATLKKSQEELDAKKSVIVHLESKLNEL 714
Query: 335 CQKLQ 339
QK++
Sbjct: 715 EQKVK 719
Score = 35.8 bits (81), Expect = 0.20
Identities = 33/145 (22%), Positives = 66/145 (44%), Gaps = 10/145 (6%)
Query: 197 SSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESDS 256
S +KF ++IK E ++ +KEE D G V E +++ D
Sbjct: 9 SDVPQAKFLEKIKYCDDLLQEVTKEDTVMEKEEEDTIFD---GGFVKVEKEGINKKYDDD 65
Query: 257 EDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKSTLLELKASEEE 316
+DE ++ + SL++ L L + + ELT++KE + L + +++ ++ E+
Sbjct: 66 DDEKA-------EKQLKSLEDALQLHDVKHKELTEVKEAFDGLGLELENSRKKMIELEDR 118
Query: 317 LKGFNLISTTYEDRLKSLCQKLQEK 341
++ L + E+ K +L+EK
Sbjct: 119 IRISALEAEKLEELQKQSASELEEK 143
>At5g10950 putative protein
Length = 395
Score = 47.8 bits (112), Expect = 5e-05
Identities = 50/187 (26%), Positives = 76/187 (39%), Gaps = 29/187 (15%)
Query: 82 EMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEESPDGDSDEDQSVKMAMLSNKL 141
E+SL E + S+ P KGK S K G +DE K + +
Sbjct: 100 EISLQEESAGESSEGTPEPPKGKGKASLK-------------GRTDEAMPKKKQKIDSSS 146
Query: 142 EYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFIADCPDLQKEKFKGKSKKSSFNS 201
+ A++ +K SK + KN K D G I E+F+ K +
Sbjct: 147 KSKAKEVEKKASKPETEKNGKMGDIGGK---------IVAAASRMSERFRSKGNVDQKET 197
Query: 202 SKFRKQIKKSLMATW------EDLDSESGSDKEEADDDAKAAVGLVATVSSEAVSEAESD 255
SK K+ K S T +D D E+G D + + ++AK V +++ V+E SD
Sbjct: 198 SKASKKPKMSSKLTKRKHTDDQDEDEEAGDDIDTSSEEAKPKVLKSCNSNADEVAENSSD 257
Query: 256 SEDENEV 262
EDE +V
Sbjct: 258 -EDEPKV 263
Score = 30.8 bits (68), Expect = 6.6
Identities = 38/158 (24%), Positives = 60/158 (37%), Gaps = 28/158 (17%)
Query: 61 EEAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEE 120
E D++T S E LK + N E ++ S P K + S KA + E+E
Sbjct: 224 EAGDDIDTSSEEAKPKVLK--SCNSNADEVAENSSDEDEPKVLKTNNS----KADKDEDE 277
Query: 121 SPDGDSDEDQSVKMAMLSN---------------KLEYLARKQKKFLSKRGSYKNSKNED 165
+ SD++ K LSN ++ KK S +N NED
Sbjct: 278 EENETSDDEAEPKALKLSNSNSDNGENNSSDDEKEITISKITSKKIKSNTADEENGDNED 337
Query: 166 QKGCFNCKKPGH----FIADCP---DLQKEKFKGKSKK 196
+ + G F+ P D +++K KGK ++
Sbjct: 338 GEKAVDEMSDGEPLVSFLKKSPEGIDAKRKKMKGKKEE 375
>At3g25450 hypothetical protein
Length = 1343
Score = 47.4 bits (111), Expect = 7e-05
Identities = 34/131 (25%), Positives = 63/131 (47%), Gaps = 16/131 (12%)
Query: 3 QYELFRMKDDESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLP-SRWRPKVTAIE 61
+++ +MKD E+I++ R + + L + S V K L+SLP ++ V A+E
Sbjct: 113 EFDKLKMKDSETIDDYVGRISEITTKAAALGEDIEESKIVKKFLKSLPRKKYIHIVAALE 172
Query: 62 EAKDLNTLSVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEES 121
+ DL T + ED+ +K +E + + + S + +GK+ +E EEE
Sbjct: 173 QVLDLKTTTFEDIAGRIKTYEDRVWDDDDSHE-------DQGKL--------MTEVEEEV 217
Query: 122 PDGDSDEDQSV 132
D +E++ V
Sbjct: 218 VDDLEEEEEEV 228
>At3g16000 myosin heavy chain like protein
Length = 727
Score = 47.0 bits (110), Expect = 9e-05
Identities = 115/546 (21%), Positives = 207/546 (37%), Gaps = 38/546 (6%)
Query: 12 DESIEEMYSRFQTLVSGLQILKKSYVASDHVSKILRSLPSRWRPKVTAIEEAKDL-NTL- 69
+E+IE + ++ + L + +K + A K+ R + A EE L N L
Sbjct: 156 EETIESLKNQLKDRERALVLKEKDFEA-----KLQHEQEERKKEVEKAKEEQLSLINQLN 210
Query: 70 SVEDLVSSLKVHEMSLNEHETSKKSKSIALPSKGKISKSSKAYKASESEEESPDGDSDED 129
S +DLV+ L E+S +E + +K K + +SK+ + +A E++ D E
Sbjct: 211 SAKDLVTELG-RELS-SEKKLCEKLKDQIESLENSLSKAGEDKEALETKLREKL-DLVEG 267
Query: 130 QSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNCKKPGHFIADCPDLQKEK 189
++ +LS +L+ K ++F + +K E + N + DL + K
Sbjct: 268 LQDRINLLSLELKDSEEKAQRFNASL-----AKKEAELKELN----SIYTQTSRDLAEAK 318
Query: 190 FKGKSKKSSFNSSKFRKQIKKS----LMATWEDLDSESGSDKEEADDDAKAAVGLVATVS 245
+ K +K ++ K S L L +E S ++ D +K L T
Sbjct: 319 LEIKQQKEELIRTQSELDSKNSAIEELNTRITTLVAEKESYIQKLDSISKDYSALKLTSE 378
Query: 246 SEAVSEAESDSEDENEVYSKIPRQELVDSLKELLSLFEHRTNELTDLKEKYVDLMKQQKS 305
++A ++AE S E E+ Q+L ++L L +++ DL EKY D +
Sbjct: 379 TQAAADAELISRKEQEI------QQLNENLDRALDDVNKSKDKVADLTEKYEDSKRMLDI 432
Query: 306 TLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKGSGNKHEIALDDFIMAGIDRS 365
L +K EL+G DR+ L L E S + E+A+
Sbjct: 433 ELTTVKNLRHELEGTKKTLQASRDRVSDLETMLDESRALCSKLESELAIVHEEWKEAKER 492
Query: 366 KVASMIYSTYKNKGKGIGYSEEKSKEYSLKSYCDCIKDGLKSTFVPEGT--NAITAVQSK 423
++ KN+ + EK +K + + LK + V + + + K
Sbjct: 493 YERNLDAEKQKNEISASELALEKDLRRRVKDELEGVTHELKESSVKNQSLQKELVEIYKK 552
Query: 424 PEASGSQAKITSKPENLKIKVMTKSDPKSQKIKILKRSEPVHQNLIKPESKIPKQKDQKN 483
E S + + E K + + K + +IL E ++L + K D+ N
Sbjct: 553 VETSNKELE-----EEKKTVLSLNKEVKGMEKQILMERE-ARKSLETDLEEAVKSLDEMN 606
Query: 484 KAATASEKTIPKGVKPKVLNDQKPLSIHPKVQGRKSKTSKINPKGPMKIWVPKSELAKNA 543
K + + + K V N + + + G SK + + L K
Sbjct: 607 KNTSILSRELEK-VNTHASNLEDEKEVLQRSLGEAKNASKEAKENVEDAHILVMSLGKER 665
Query: 544 GVLKGK 549
VL+ K
Sbjct: 666 EVLEKK 671
>At1g48920 unknown protein
Length = 557
Score = 46.6 bits (109), Expect = 1e-04
Identities = 81/353 (22%), Positives = 143/353 (39%), Gaps = 44/353 (12%)
Query: 56 KVTAIEEAKD-LNTLSVEDLVSSL--KVHEMSLNEHETSKKSKSIALPSKGKISKSSKAY 112
KV+ ++ KD + + E V + KV ++ E+ ++ K+ +P+K KA
Sbjct: 34 KVSLKKQKKDVIAAVQKEKAVKKVPKKVESSDDSDSESEEEEKAKKVPAK-------KA- 85
Query: 113 KASESEEESPDGDSDEDQSVKMAMLSNKLEYLARKQKKFLSKRGSYKNSKNEDQKGCFNC 172
AS S+E S D SD++ + K A+ + +A+K SK S + + +
Sbjct: 86 -ASSSDESSDDSSSDDEPAPKKAVAATN-GTVAKK-----SKDDSSSSDDDSSDEEVAVT 138
Query: 173 KKPGHFIADCPDLQKEKFKGKSKKSSFNSSKFRKQIKKSLMATWEDLDSESGSDKEEADD 232
KKP A ++ +K SS +K K +D S S D +E +
Sbjct: 139 KKPAA-AAKNGSVKAKKESSSEDDSSSEDEPAKKPAAKIAKPAAKD-SSSSDDDSDEDSE 196
Query: 233 DAKAAVGLVATVSSEAVSEAES-------DSEDENEVYSKIPRQELVDSLKELLSLFEHR 285
D K A A +++A S ++S +SEDE P Q+ D+ S +
Sbjct: 197 DEKPATKKAAPAAAKAASSSDSSDEDSDEESEDEK------PAQKKADTKASKKSSSDES 250
Query: 286 TNELTDLKEKYVDLMKQQKSTLLELKASEEELKGFNLISTTYEDRLKSLCQKLQEKCDKG 345
+ D E + K++ S + + A + K ST K+L
Sbjct: 251 SESEEDESEDEEETPKKKSSDVEMVDAEKSSAKQPKTPSTPAAGGSKTLF------AANL 304
Query: 346 SGNKHEIALDDFIMAGIDRSKVASMIYSTYKNKG--KGIGYSEEKSKEYSLKS 396
S N +++F + +V + +ST ++ G +G G+ E S E + K+
Sbjct: 305 SFNIERADVENFFK---EAGEVVDVRFSTNRDDGSFRGFGHVEFASSEEAQKA 354
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.356 0.157 0.537
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,981,387
Number of Sequences: 26719
Number of extensions: 1111976
Number of successful extensions: 11937
Number of sequences better than 10.0: 265
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 217
Number of HSP's that attempted gapping in prelim test: 11052
Number of HSP's gapped (non-prelim): 839
length of query: 1595
length of database: 11,318,596
effective HSP length: 113
effective length of query: 1482
effective length of database: 8,299,349
effective search space: 12299635218
effective search space used: 12299635218
T: 11
A: 40
X1: 14 ( 7.2 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 37 (21.7 bits)
S2: 67 (30.4 bits)
Medicago: description of AC144722.10


