

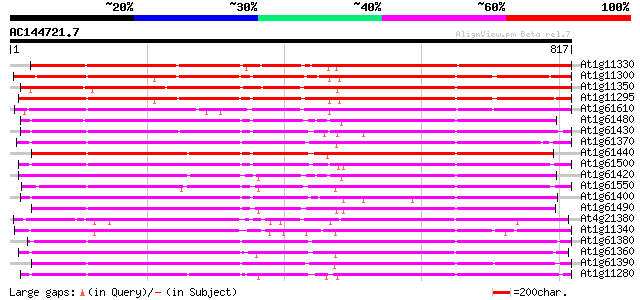
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.7 + phase: 0 /pseudo
(817 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g11330 receptor-like protein kinase, putative 743 0.0
At1g11300 putative s-locus protein kinase (PPC:1.7.2) 717 0.0
At1g11350 putative receptor-like protein kinase gi|4008010; simi... 703 0.0
At1g11295 putative s-locus protein kinase (PPC:1.7.2) 678 0.0
At1g61610 659 0.0
At1g61480 S-like receptor protein kinase; member of gene cluster... 628 e-180
At1g61430 S-like receptor protein kinase; member of gene cluster... 625 e-179
At1g61370 S-like receptor protein kinase; member of gene cluster... 623 e-178
At1g61440 S-like receptor protein kinase; member of gene cluster... 618 e-177
At1g61500 616 e-176
At1g61420 S-like receptor protein kinase; member of gene cluster... 615 e-176
At1g61550 613 e-175
At1g61400 S-like receptor protein kinase; member of gene cluster... 608 e-174
At1g61490 S-like receptor protein kinase; member of gene cluster... 605 e-173
At4g21380 receptor-like serine/threonine protein kinase ARK3 601 e-172
At1g11340 receptor kinase, putative 600 e-171
At1g61380 S-like receptor protein kinase 600 e-171
At1g61360 S-like receptor protein kinase; member of gene cluster... 596 e-170
At1g61390 S-like receptor protein kinase; member of gene cluster... 596 e-170
At1g11280 serine/threonine kinase like protein 590 e-168
>At1g11330 receptor-like protein kinase, putative
Length = 840
Score = 743 bits (1918), Expect = 0.0
Identities = 396/823 (48%), Positives = 543/823 (65%), Gaps = 50/823 (6%)
Query: 31 DTITSSKSLKDNE--TITSNNTNFKLGFFSPLNSTNR--YLGIWY--INKTNNIWIANRD 84
D IT S +KD+E T+ + F+ GFF+P+NST R Y+GIWY I +W+AN+D
Sbjct: 31 DRITFSSPIKDSESETLLCKSGIFRFGFFTPVNSTTRLRYVGIWYEKIPIQTVVWVANKD 90
Query: 85 QPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISS--STNST-AQLADSGNLILRDI-S 140
P+ D++G+++I++DGN + + N ++WSTN+S + N+T QL DSGNL+L+D +
Sbjct: 91 SPINDTSGVISIYQDGNLAVTDGRNR-LVWSTNVSVPVAPNATWVQLMDSGNLMLQDNRN 149
Query: 141 SGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLERLDAPEV 200
+G +W+SF HP D+ +P M + + TG + S S +DPS+G+Y+A + PE+
Sbjct: 150 NGEILWESFKHPYDSFMPRMTLGTDGRTGGNLKLTSWTSHDDPSTGNYTAGIAPFTFPEL 209
Query: 201 FIWKDKNIHWRTGPWNGRVFLGSPRMLTE-YLAGWRFDQDTDGTTYITYNFADKTMFGIL 259
IWK+ WR+GPWNG+VF+G P M + +L G+ + D GT I+ ++A+ +
Sbjct: 210 LIWKNNVPTWRSGPWNGQVFIGLPNMDSLLFLDGFNLNSDNQGT--ISMSYANDSFMYHF 267
Query: 260 SLTPHGTLKLIEYMNKKELFRLEVD--QNECDFYGKCGPFGNCDNSTVPICSCFDGFEPK 317
+L P G + ++ +R+ V +CD YG+CG FG+C P C C GF PK
Sbjct: 268 NLDPEGIIYQKDWSTSMRTWRIGVKFPYTDCDAYGRCGRFGSCHAGENPPCKCVKGFVPK 327
Query: 318 NSVEWSLGNWTNGCVRKEGMNLKCEM---VKNGSSIVKQDGFKVYHNMKPPDFNVRTNNA 374
N+ EW+ GNW+NGC+RK L+CE V NG K DGF MK P + + A
Sbjct: 328 NNTEWNGGNWSNGCMRKAP--LQCERQRNVSNGGGGGKADGFLKLQKMKVP-ISAERSEA 384
Query: 375 DQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAELVAVKKE 434
+ C CL NCSC AYAYD I CM W+G+L+D+Q F G+DLF+RV A +
Sbjct: 385 SEQVCPKVCLDNCSCTAYAYDRGIGCMLWSGDLVDMQSFLGSGIDLFIRV-----AHSEL 439
Query: 435 KGHNKSFLIIVIAGVIGALILVICAYLL----WRKCSARHKARE---------------- 474
K H+ + +++ A VIG +++ LL ++K A+ ++ E
Sbjct: 440 KTHS-NLAVMIAAPVIGVMLIAAVCVLLACRKYKKRPAKDRSAELMFKRMEALTSDNESA 498
Query: 475 HQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQ 534
Q+KL ELPL++F+ L T+T+ F N LG+GGFGPVYKG + +GQEIAVKRLS+ SGQ
Sbjct: 499 SNQIKLKELPLFEFQVLATSTDSFSLRNKLGQGGFGPVYKGKLPEGQEIAVKRLSRKSGQ 558
Query: 535 GIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDW 594
G+EE MNEVVVISKLQHRNLV+LLGCC+E E++LVYE+MP KSLDA+LFDP+++K LDW
Sbjct: 559 GLEELMNEVVVISKLQHRNLVKLLGCCIEGEERMLVYEYMPKKSLDAYLFDPMKQKILDW 618
Query: 595 RKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDD 654
+ R NI+EGI RG++YLHRDSRL+IIHRDLKASNILLD ++ PKISDFGLARI + E D
Sbjct: 619 KTRFNIMEGICRGLLYLHRDSRLKIIHRDLKASNILLDENLNPKISDFGLARIFRANE-D 677
Query: 655 EANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVG 714
EANT+RVVGTYGYM PEYAMEG FSEKSDV+S GV+ LEI+SGRRNSS E+ L+L+
Sbjct: 678 EANTRRVVGTYGYMSPEYAMEGFFSEKSDVFSLGVIFLEIISGRRNSSSHKEENNLNLLA 737
Query: 715 FAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEI 774
+AWKLW + SL DP V+D CFE + +C+HIGLLCVQE+ DRPN+S V+ ML +E
Sbjct: 738 YAWKLWNDGEAASLADPAVFDKCFEKEIEKCVHIGLLCVQEVANDRPNVSNVIWMLTTEN 797
Query: 775 THLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
L P + AF+ ++ + S ESS +S Q S N+V+L+ V G
Sbjct: 798 MSLADPKQPAFIVRRGA-SEAESSDQSSQKVSINDVSLTAVTG 839
>At1g11300 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 717 bits (1852), Expect = 0.0
Identities = 391/840 (46%), Positives = 540/840 (63%), Gaps = 52/840 (6%)
Query: 6 HNSNYFFIITFLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNR 65
H S+ F+ ++ C S A S K L D+ETI S+ F+ GFFSP+NST+R
Sbjct: 4 HESSSPFVCILVLSCFFLSVSLAQERAFFSGK-LNDSETIVSSFRTFRFGFFSPVNSTSR 62
Query: 66 YLGIWY--INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISS--S 121
Y GIWY ++ IW+AN+D+P+ DS+G++++ +DGN ++ + V+ WSTN+S+ S
Sbjct: 63 YAGIWYNSVSVQTVIWVANKDKPINDSSGVISVSQDGNLVVTDGQRRVL-WSTNVSTQAS 121
Query: 122 TNST-AQLADSGNLILRDISSGATIWDSFTHPADAAVPTMRIAAN-QVTGKKISFVSRKS 179
NST A+L DSGNL+L++ SS A +W+SF +P D+ +P M + N ++ G ++ S KS
Sbjct: 122 ANSTVAELLDSGNLVLKEASSDAYLWESFKYPTDSWLPNMLVGTNARIGGGNVTITSWKS 181
Query: 180 DNDPSSGHYSASLERLDAPEVFIWKDKNIH---WRTGPWNGRVFLGSPRMLTE-YLAGWR 235
+DPS G Y+A+L PE+FI + N + WR+GPWNG++F G P + +L +
Sbjct: 182 PSDPSPGSYTAALVLAAYPELFIMNNNNNNSTVWRSGPWNGQMFNGLPDVYAGVFLYRFI 241
Query: 236 FDQDTDGTTYITYNFADKTMFGILSLTPHGTLKLIEYMNKKELFR--LEVDQNECDFYGK 293
+ DT+G+ +T ++A+ + + G++ ++ + + L+V ECD Y +
Sbjct: 242 VNDDTNGS--VTMSYANDSTLRYFYMDYRGSVIRRDWSETRRNWTVGLQVPATECDNYRR 299
Query: 294 CGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQ 353
CG F C+ P+CSC GF P+N +EW+ GNW+ GC R+ + L+CE N S
Sbjct: 300 CGEFATCNPRKNPLCSCIRGFRPRNLIEWNNGNWSGGCTRR--VPLQCERQNNNGSA--- 354
Query: 354 DGFKVYHNMKPPDFNVRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKF 413
DGF MK PDF R + A + +C CL CSC+A A+ CM W G L+D Q+
Sbjct: 355 DGFLRLRRMKLPDF-ARRSEASEPECLRTCLQTCSCIAAAHGLGYGCMIWNGSLVDSQEL 413
Query: 414 PNGGVDLFVRVPAELVAVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWR---KCSARH 470
G+DL++R+ + K K + ++AG G ++ C L R K A+
Sbjct: 414 SASGLDLYIRLAHSEI---KTKDKRPILIGTILAG--GIFVVAACVLLARRIVMKKRAKK 468
Query: 471 KAREHQQM-------------KLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVM 517
K R+ +Q+ KL ELPL++F+ L ATN F N LG+GGFGPVYKG +
Sbjct: 469 KGRDAEQIFERVEALAGGNKGKLKELPLFEFQVLAAATNNFSLRNKLGQGGFGPVYKGKL 528
Query: 518 EDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNK 577
++GQEIAVKRLS+ASGQG+EE +NEVVVISKLQHRNLV+LLGCC+ E++LVYEFMP K
Sbjct: 529 QEGQEIAVKRLSRASGQGLEELVNEVVVISKLQHRNLVKLLGCCIAGEERMLVYEFMPKK 588
Query: 578 SLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIP 637
SLD +LFD + K LDW+ R NII GI RG++YLHRDSRLRIIHRDLKASNILLD ++IP
Sbjct: 589 SLDYYLFDSRRAKLLDWKTRFNIINGICRGLLYLHRDSRLRIIHRDLKASNILLDENLIP 648
Query: 638 KISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSG 697
KISDFGLARI G +DEANT+RVVGTYGYM PEYAM GLFSEKSDV+S GV+LLEI+SG
Sbjct: 649 KISDFGLARIFP-GNEDEANTRRVVGTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIISG 707
Query: 698 RRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELP 757
RRNS+ +L+ + W +W E I SL+DPE++D FE + +CIHIGLLCVQE
Sbjct: 708 RRNSN-------STLLAYVWSIWNEGEINSLVDPEIFDLLFEKEIHKCIHIGLLCVQEAA 760
Query: 758 RDRPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
DRP++STV ML SEI +P P + AF+ +++ ESS+ S +S NNVT+++V G
Sbjct: 761 NDRPSVSTVCSMLSSEIADIPEPKQPAFI-SRNNVPEAESSENSDLKDSINNVTITDVTG 819
>At1g11350 putative receptor-like protein kinase gi|4008010; similar
to EST gb|AI999419.1
Length = 830
Score = 703 bits (1815), Expect = 0.0
Identities = 379/838 (45%), Positives = 532/838 (63%), Gaps = 50/838 (5%)
Query: 17 LIFCTIYSCYSA----INDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYI 72
LI C+S D IT S +D+ET+ SN++ F+ GFFSP+NST RY GIW+
Sbjct: 5 LILLLTLICFSLRLCLATDVITFSSEFRDSETVVSNHSTFRFGFFSPVNSTGRYAGIWFN 64
Query: 73 N--KTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNIS---SSTNSTAQ 127
N +W+AN + P+ DS+G+V+I K+GN ++++ G + WSTN+ ++ A+
Sbjct: 65 NIPVQTVVWVANSNSPINDSSGMVSISKEGNLVVMDG-RGQVHWSTNVLVPVAANTFYAR 123
Query: 128 LADSGNLILRDISSGAT--IWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSS 185
L ++GNL+L ++ +W+SF HP + +PTM +A + TG+ + S KS DPS
Sbjct: 124 LLNTGNLVLLGTTNTGDEILWESFEHPQNIYLPTMSLATDTKTGRSLKLRSWKSPFDPSP 183
Query: 186 GHYSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAG-WRFDQDTDGTT 244
G YSA L L PE+ +WKD + WR+GPWNG+ F+G P M +Y + +D
Sbjct: 184 GRYSAGLIPLPFPELVVWKDDLLMWRSGPWNGQYFIGLPNM--DYRINLFELTLSSDNRG 241
Query: 245 YITYNFADKTMFGILSLTPHGTLKLIEYMNKKELFR--LEVDQNECDFYGKCGPFGNC-- 300
++ ++A T+ L G++ ++ + ++ L+V +CD Y CG F +C
Sbjct: 242 SVSMSYAGNTLLYHFLLDSEGSVFQRDWNVAIQEWKTWLKVPSTKCDTYATCGQFASCRF 301
Query: 301 DNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYH 360
+ + P C C GF+P++ EW+ GNWT GCVRK L+CE N K DGF
Sbjct: 302 NPGSTPPCMCIRGFKPQSYAEWNNGNWTQGCVRKAP--LQCESRDNNDGSRKSDGFVRVQ 359
Query: 361 NMKPPDFNVRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDL 420
MK P N + + A++ C CL NCSC AY++D I C+ W+G L+D+Q+F GV
Sbjct: 360 KMKVPH-NPQRSGANEQDCPESCLKNCSCTAYSFDRGIGCLLWSGNLMDMQEFSGTGVVF 418
Query: 421 FVRVPAELVAVKKEKGHNKSFLIIVIAGVIGALILVICAYL-LWRKCSARHKAREHQ--- 476
++R L + +K N+S ++I + ++GA + L LW+ R K R +
Sbjct: 419 YIR----LADSEFKKRTNRS-IVITVTLLVGAFLFAGTVVLALWKIAKHREKNRNTRLLN 473
Query: 477 -----------------QMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMED 519
Q KL ELPL++F+ L ATN F N LG+GGFG VYKG +++
Sbjct: 474 ERMEALSSNDVGAILVNQYKLKELPLFEFQVLAVATNNFSITNKLGQGGFGAVYKGRLQE 533
Query: 520 GQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSL 579
G +IAVKRLS+ SGQG+EEF+NEVVVISKLQHRNLVRLLG C+E E++LVYEFMP L
Sbjct: 534 GLDIAVKRLSRTSGQGVEEFVNEVVVISKLQHRNLVRLLGFCIEGEERMLVYEFMPENCL 593
Query: 580 DAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKI 639
DA+LFDP++++ LDW+ R NII+GI RG+MYLHRDSRL+IIHRDLKASNILLD ++ PKI
Sbjct: 594 DAYLFDPVKQRLLDWKTRFNIIDGICRGLMYLHRDSRLKIIHRDLKASNILLDENLNPKI 653
Query: 640 SDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRR 699
SDFGLARI + G +DE +T RVVGTYGYM PEYAM GLFSEKSDV+S GV+LLEIVSGRR
Sbjct: 654 SDFGLARIFQ-GNEDEVSTVRVVGTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIVSGRR 712
Query: 700 NSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRD 759
NSSF + +L +AWKLW I+L+DP +++ CFE+ + RC+H+GLLCVQ+ D
Sbjct: 713 NSSFYNDGQNPNLSAYAWKLWNTGEDIALVDPVIFEECFENEIRRCVHVGLLCVQDHAND 772
Query: 760 RPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
RP+++TV+ ML SE ++LP P + AF+ ++ + S ESS +S S NNV+L+++ G
Sbjct: 773 RPSVATVIWMLSSENSNLPEPKQPAFIPRRGT-SEVESSGQSDPRASINNVSLTKITG 829
>At1g11295 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 678 bits (1750), Expect = 0.0
Identities = 374/833 (44%), Positives = 521/833 (61%), Gaps = 51/833 (6%)
Query: 13 IITFLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY- 71
I+ L + S ++ S +L D+ETI S+ F+ GFFSP+NSTNRY GIWY
Sbjct: 10 IVHVLSLSCFFLSVSLAHERALFSGTLNDSETIVSSFRTFRFGFFSPVNSTNRYAGIWYN 69
Query: 72 -INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISS--STNST-AQ 127
I IW+AN+D P+ DS+G+++I +DGN ++ + V+ WSTN+S+ S NST A+
Sbjct: 70 SIPVQTVIWVANKDTPINDSSGVISISEDGNLVVTDGQRRVL-WSTNVSTRASANSTVAE 128
Query: 128 LADSGNLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGK-KISFVSRKSDNDPSSG 186
L +SGNL+L+D ++ A +W+SF +P D+ +P M + N TG I+ S + +DPS G
Sbjct: 129 LLESGNLVLKDANTDAYLWESFKYPTDSWLPNMLVGTNARTGGGNITITSWTNPSDPSPG 188
Query: 187 HYSASLERLDAPEVFIWKDKNIH---WRTGPWNGRVFLGSPRMLTE-YLAGWRFDQDTDG 242
Y+A+L PE+FI+ + + + WR+GPWNG +F G P + +L ++ + DT+G
Sbjct: 189 SYTAALVLAPYPELFIFNNNDNNATVWRSGPWNGLMFNGLPDVYPGLFLYRFKVNDDTNG 248
Query: 243 TTYITYNFADKTMFGILSLTPHGTLKLIEYMNKKELFRL--EVDQNECDFYGKCGPFGNC 300
+ T ++A+ + L L G ++ + + L +V ECD Y +CG + C
Sbjct: 249 SA--TMSYANDSTLRHLYLDYRGFAIRRDWSEARRNWTLGSQVPATECDIYSRCGQYTTC 306
Query: 301 DNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYH 360
+ P CSC GF P+N +EW+ GNW+ GC+RK + L+CE N S D F
Sbjct: 307 NPRKNPHCSCIKGFRPRNLIEWNNGNWSGGCIRK--LPLQCERQNNKGSA---DRFLKLQ 361
Query: 361 NMKPPDFNVRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDL 420
MK PDF R + A + +C CL +CSC+A+A+ CM W L+D Q G+DL
Sbjct: 362 RMKMPDF-ARRSEASEPECFMTCLQSCSCIAFAHGLGYGCMIWNRSLVDSQVLSASGMDL 420
Query: 421 FVRVPAELVAVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWR---KCSARHKAREHQQ 477
+R+ A + K ++ ++I + G ++ C L R K A+ K + +Q
Sbjct: 421 SIRL-----AHSEFKTQDRRPILIGTSLAGGIFVVATCVLLARRIVMKKRAKKKGTDAEQ 475
Query: 478 M-------------KLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIA 524
+ KL ELPL++F+ L TAT+ F +N LG+GGFGPVYKG++ +GQEIA
Sbjct: 476 IFKRVEALAGGSREKLKELPLFEFQVLATATDNFSLSNKLGQGGFGPVYKGMLLEGQEIA 535
Query: 525 VKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLF 584
VKRLS+ASGQG+EE + EVVVISKLQHRNLV+L GCC+ E++LVYEFMP KSLD ++F
Sbjct: 536 VKRLSQASGQGLEELVTEVVVISKLQHRNLVKLFGCCIAGEERMLVYEFMPKKSLDFYIF 595
Query: 585 DPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGL 644
DP + K LDW R II GI RG++YLHRDSRLRIIHRDLKASNILLD ++IPKISDFGL
Sbjct: 596 DPREAKLLDWNTRFEIINGICRGLLYLHRDSRLRIIHRDLKASNILLDENLIPKISDFGL 655
Query: 645 ARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFS 704
ARI G +DEANT+RVVGTYGYM PEYAM GLFSEKSDV+S GV+LLEI+SGRRNS
Sbjct: 656 ARIFP-GNEDEANTRRVVGTYGYMAPEYAMGGLFSEKSDVFSLGVILLEIISGRRNSH-- 712
Query: 705 HHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNIS 764
+L+ W +W E I ++DPE++D FE + +C+HI LLCVQ+ DRP++S
Sbjct: 713 -----STLLAHVWSIWNEGEINGMVDPEIFDQLFEKEIRKCVHIALLCVQDAANDRPSVS 767
Query: 765 TVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
TV +ML SE+ +P P + AF+ + E S+ S NNVT+++V G
Sbjct: 768 TVCMMLSSEVADIPEPKQPAFMPRNVGLE-AEFSESIALKASINNVTITDVSG 819
>At1g61610
Length = 842
Score = 659 bits (1699), Expect = 0.0
Identities = 366/854 (42%), Positives = 514/854 (59%), Gaps = 60/854 (7%)
Query: 7 NSNYFFIITFLIF---CTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNST 63
N N + T LIF C+ SC + +++ T + ++++ +++ S + +F+LGFF+P NST
Sbjct: 5 NRNLTLVTTLLIFHQLCSNVSC--STSNSFTRNHTIREGDSLISEDESFELGFFTPKNST 62
Query: 64 NRYLGIWY--INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSS 121
RY+GIWY I +W+ANR++PL D G + I DGN +I+N N I WSTN+
Sbjct: 63 LRYVGIWYKNIEPQTVVWVANREKPLLDHKGALKIADDGNLVIVNGQNETI-WSTNVEPE 121
Query: 122 TNST-AQLADSGNLIL-RDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKS 179
+N+T A L +G+L+L D W+SF +P D +P MR+ N G+ +F+ KS
Sbjct: 122 SNNTVAVLFKTGDLVLCSDSDRRKWYWESFNNPTDTFLPGMRVRVNPSLGENRAFIPWKS 181
Query: 180 DNDPSSGHYSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRML--TEYLAGWRFD 237
++DPS G YS ++ + A E+ IW+ + WR+GPWN +F G P ML T Y+ G++
Sbjct: 182 ESDPSPGKYSMGIDPVGALEIVIWEGEKRKWRSGPWNSAIFTGIPDMLRFTNYIYGFKLS 241
Query: 238 Q--DTDGTTYITYNFADKTMFGILSLTPHGTLKLIEYMNKKELFRLEVDQ----NECDFY 291
D DG+ Y TY +D + F + P G + + K++ + Q EC+ Y
Sbjct: 242 SPPDRDGSVYFTYVASDSSDFLRFWIRPDGVEEQFRW--NKDIRNWNLLQWKPSTECEKY 299
Query: 292 GKCGPFGNCDNSTV---PICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGS 348
+CG + CD+S CSC DGFEP + +W+ +++ GC R+ +N +V
Sbjct: 300 NRCGNYSVCDDSKEFDSGKCSCIDGFEPVHQDQWNNRDFSGGCQRRVPLNCNQSLVAG-- 357
Query: 349 SIVKQDGFKVYHNMKPPDFNVRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELI 408
++DGF V +K PDF + + + C C +CSC AYA I CM WT +LI
Sbjct: 358 ---QEDGFTVLKGIKVPDFGSVVLHNNSETCKDVCARDCSCKAYALVVGIGCMIWTRDLI 414
Query: 409 DLQKFPNGGVDLFVRVPAELVAVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWR---- 464
D++ F GG + +R L K G S L I++ VIGA +L +C ++LW+
Sbjct: 415 DMEHFERGGNSINIR----LAGSKLGGGKENSTLWIIVFSVIGAFLLGLCIWILWKFKKS 470
Query: 465 ---------------------KCSARHKAREHQQMKLDELPLYDFEKLETATNCFHFNNM 503
S+ K Q+ +LP++ F+ + +AT F N
Sbjct: 471 LKAFLWKKKDITVSDIIENRDYSSSPIKVLVGDQVDTPDLPIFSFDSVASATGDFAEENK 530
Query: 504 LGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVE 563
LG+GGFG VYKG +G+EIAVKRLS S QG+EEF NE+++I+KLQHRNLVRLLGCC+E
Sbjct: 531 LGQGGFGTVYKGNFSEGREIAVKRLSGKSKQGLEEFKNEILLIAKLQHRNLVRLLGCCIE 590
Query: 564 RGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRD 623
E++L+YE+MPNKSLD FLFD ++ +LDWRKR +I GIARG++YLHRDSRL+IIHRD
Sbjct: 591 DNEKMLLYEYMPNKSLDRFLFDESKQGSLDWRKRWEVIGGIARGLLYLHRDSRLKIIHRD 650
Query: 624 LKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSD 683
LKASNILLD++M PKISDFG+ARI + D ANT RVVGTYGYM PEYAMEG+FSEKSD
Sbjct: 651 LKASNILLDTEMNPKISDFGMARIFNY-RQDHANTIRVVGTYGYMAPEYAMEGIFSEKSD 709
Query: 684 VYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSML 743
VYSFGVL+LEIVSGR+N SF D SL+G+AW LW + +IDP V D + +
Sbjct: 710 VYSFGVLILEIVSGRKNVSF-RGTDHGSLIGYAWHLWSQGKTKEMIDPIVKDTRDVTEAM 768
Query: 744 RCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQ 803
RCIH+G+LC Q+ RPN+ +V+LML S+ + LPPP + F H + E + H
Sbjct: 769 RCIHVGMLCTQDSVIHRPNMGSVLLMLESQTSQLPPPRQPTF-HSFLNSGDIELNFDGHD 827
Query: 804 SNSNNNVTLSEVQG 817
S N+VT + + G
Sbjct: 828 VASVNDVTFTTIVG 841
>At1g61480 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 809
Score = 628 bits (1619), Expect = e-180
Identities = 343/801 (42%), Positives = 477/801 (58%), Gaps = 40/801 (4%)
Query: 17 LIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INK 74
L+ TI+ +S IT L +T++S+N ++LGFFS NS N+Y+GIW+ I
Sbjct: 12 LLLITIFLSFSYAG--ITRESPLSIGKTLSSSNGVYELGFFSFNNSQNQYVGIWFKGIIP 69
Query: 75 TNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTN-STAQLADSGN 133
+W+ANR++P+ DS +TI +G+ ++ N+ N ++WS + ++N S A+L D+GN
Sbjct: 70 RVVVWVANREKPVTDSAANLTISSNGSLLLFNE-NHSVVWSIGETFASNGSRAELTDNGN 128
Query: 134 LILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLE 193
L++ D +SG T+W+SF H D +P + N TG+K S KS DPS G ++ +
Sbjct: 129 LVVIDNNSGRTLWESFEHFGDTMLPFSNLMYNLATGEKRVLTSWKSHTDPSPGDFTVQIT 188
Query: 194 RLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYNFADK 253
+ + +WR+GPW F G P M Y + + QDT+G+ TY F
Sbjct: 189 PQVPSQACTMRGSKTYWRSGPWAKTRFTGIPVMDDTYTSPFSLQQDTNGSGSFTY-FERN 247
Query: 254 TMFGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCFDG 313
+ +T G+LK+ ++ E +N CD YG CGPFG C S P C CF G
Sbjct: 248 FKLSYIMITSEGSLKIFQHNGMDWELNFEAPENSCDIYGFCGPFGICVMSVPPKCKCFKG 307
Query: 314 FEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYHNMKPPDFNVRTNN 373
F PK+ EW GNWT+GCVR L C+ NG ++ +GF N+KPPDF +
Sbjct: 308 FVPKSIEEWKRGNWTDGCVRHT--ELHCQGNTNGKTV---NGFYHVANIKPPDFYEFASF 362
Query: 374 ADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAELVAVKK 433
D + C CL NCSCLA+AY I C+ W +L+D +F GG L +R+ +
Sbjct: 363 VDAEGCYQICLHNCSCLAFAYINGIGCLMWNQDLMDAVQFSAGGEILSIRLASS------ 416
Query: 434 EKGHNKSFLIIVIAGVIGALILVICAYLLWRKCSARHKAREHQQMKLDE----------- 482
E G NK IIV A ++ + VI A+ + C R+K + K+ +
Sbjct: 417 ELGGNKRNKIIV-ASIVSLSLFVILAFAAF--CFLRYKVKHTVSAKISKIASKEAWNNDL 473
Query: 483 -------LPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQG 535
L ++ ++TAT+ F +N LG+GGFG VYKG ++DG+EIAVKRLS +SGQG
Sbjct: 474 EPQDVSGLKFFEMNTIQTATDNFSLSNKLGQGGFGSVYKGKLQDGKEIAVKRLSSSSGQG 533
Query: 536 IEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWR 595
EEFMNE+V+ISKLQH+NLVR+LGCC+E E++LVYEF+ NKSLD FLFD ++ +DW
Sbjct: 534 KEEFMNEIVLISKLQHKNLVRILGCCIEGEERLLVYEFLLNKSLDTFLFDSRKRLEIDWP 593
Query: 596 KRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDE 655
KR NIIEGIARG+ YLHRDS LR+IHRDLK SNILLD M PKISDFGLAR+ + G + +
Sbjct: 594 KRFNIIEGIARGLHYLHRDSCLRVIHRDLKVSNILLDEKMNPKISDFGLARMYQ-GTEYQ 652
Query: 656 ANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGF 715
NT+RV GT GYM PEYA G+FSEKSD+YSFGV+LLEI++G + S FS+ +L+ +
Sbjct: 653 DNTRRVAGTLGYMAPEYAWTGMFSEKSDIYSFGVILLEIITGEKISRFSYGRQGKTLLAY 712
Query: 716 AWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEIT 775
AW+ W E I L+D +V D+C + RC+ IGLLCVQ P DRPN ++ ML +
Sbjct: 713 AWESWCESGGIDLLDKDVADSCHPLEVERCVQIGLLCVQHQPADRPNTMELLSMLTTTSD 772
Query: 776 HLPPPGRVAFVHKQSSKSTTE 796
P VH + +S ++
Sbjct: 773 LTSPKQPTFVVHTRDEESLSQ 793
>At1g61430 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 806
Score = 625 bits (1612), Expect = e-179
Identities = 353/820 (43%), Positives = 491/820 (59%), Gaps = 43/820 (5%)
Query: 16 FLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--IN 73
+L F TI+ +S IT +T++S+N ++LGFFS NS N+YLGIW+ I
Sbjct: 11 YLPFFTIFMSFSFAG--ITKESPFSIGQTLSSSNGVYELGFFSLNNSQNQYLGIWFKSII 68
Query: 74 KTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWST-NISSSTNSTAQLADSG 132
+W+ANR++P+ DS + I +G+ ++ N +GV+ WST +I +S S A+L D G
Sbjct: 69 PQVVVWVANREKPVTDSAANLGISSNGSLLLSNGKHGVV-WSTGDIFASNGSRAELTDHG 127
Query: 133 NLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASL 192
NL+ D SG T+W SF H + +PT + N V G+K + KS DPS G + A +
Sbjct: 128 NLVFIDKVSGRTLWQSFEHLGNTLLPTSIMMYNLVAGEKRGLTAWKSYTDPSPGEFVALI 187
Query: 193 ERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYNFAD 252
+ I + ++RTGPW F GSP+M Y + + QD +G+ Y ++F +
Sbjct: 188 TPQVPSQGIIMRGSTRYYRTGPWAKTRFTGSPQMDESYTSPFILTQDVNGSGY--FSFVE 245
Query: 253 KTMFGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCFD 312
+ + LT GT+K++ + E N CD YG CGPFG C S P C CF
Sbjct: 246 RGKPSRMILTSEGTMKVLVHNGMDWESTYEGPANSCDIYGVCGPFGLCVVSIPPKCKCFK 305
Query: 313 GFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYHNMKPPDFNVRTN 372
GF PK + EW GNWT+GCVR+ L C+ +G + F N+KPPDF N
Sbjct: 306 GFVPKFAKEWKKGNWTSGCVRRT--ELHCQGNSSGKDA---NVFYTVPNIKPPDFYEYAN 360
Query: 373 NADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAELVAVK 432
+ + ++C +CL NCSCLA++Y P I C+ W+ +L+D ++F G L +R+ + V
Sbjct: 361 SQNAEECHQNCLHNCSCLAFSYIPGIGCLMWSKDLMDTRQFSAAGELLSIRLARSELDVN 420
Query: 433 KEKGHNKSFLIIVIAGVIGALILVI---CAYLLWRKCSARHKAREH--------QQMKLD 481
K K + ++A + + VI A+ WR C H A Q +
Sbjct: 421 KRK-------MTIVASTVSLTLFVIFGFAAFGFWR-CRVEHNAHISNDAWRNFLQSQDVP 472
Query: 482 ELPLYDFEKLETATNCFHFNNMLGKGGFGPVYK---GVMEDGQEIAVKRLSKASGQGIEE 538
L ++ ++TATN F +N LG GGFG VYK G ++DG+EIAVKRLS +SGQG +E
Sbjct: 473 GLEFFEMNAIQTATNNFSLSNKLGPGGFGSVYKARNGKLQDGREIAVKRLSSSSGQGKQE 532
Query: 539 FMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRS 598
FMNE+V+ISKLQHRNLVR+LGCCVE E++L+Y F+ NKSLD F+FD +K LDW KR
Sbjct: 533 FMNEIVLISKLQHRNLVRVLGCCVEGTEKLLIYGFLKNKSLDTFVFDARKKLELDWPKRF 592
Query: 599 NIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANT 658
IIEGIARG++YLHRDSRLR+IHRDLK SNILLD M PKISDFGLAR+ + G + T
Sbjct: 593 EIIEGIARGLLYLHRDSRLRVIHRDLKVSNILLDEKMNPKISDFGLARMFQ-GTQYQEKT 651
Query: 659 KRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWK 718
+RVVGT GYM PEYA G+FSEKSD+YSFGVLLLEI+SG++ SSFS+ E+ +L+ +AW+
Sbjct: 652 RRVVGTLGYMSPEYAWTGVFSEKSDIYSFGVLLLEIISGKKISSFSYGEEGKALLAYAWE 711
Query: 719 LWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLP 778
W E ++ +D + D+ S + RC+ IGLLCVQ P DRPN ++ ML + + LP
Sbjct: 712 CWCETREVNFLDQALADSSHPSEVGRCVQIGLLCVQHEPADRPNTLELLSMLTT-TSDLP 770
Query: 779 PPGRVAF-VHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
P + F VH + +S + S + N +T S +QG
Sbjct: 771 LPKKPTFVVHTRKDESPSNDSM-----ITVNEMTESVIQG 805
>At1g61370 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 814
Score = 623 bits (1606), Expect = e-178
Identities = 342/824 (41%), Positives = 491/824 (59%), Gaps = 35/824 (4%)
Query: 11 FFIITFLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIW 70
FF + SC A IT + L +T++S N ++LGFFSP NS N+Y+GIW
Sbjct: 8 FFASLLFLLIIFPSCAFA---AITRASPLSIGQTLSSPNGTYELGFFSPNNSRNQYVGIW 64
Query: 71 YINKTNNI--WIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNST-AQ 127
+ N T + W+ANRD+P+ ++ +TI+ +G+ I++ + V+ WS + S+N A+
Sbjct: 65 FKNITPRVVVWVANRDKPVTNNAANLTINSNGSLILVEREQNVV-WSIGETFSSNELRAE 123
Query: 128 LADSGNLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGH 187
L ++GNL+L D S +W+SF H D + + + KK S K+ DPS G
Sbjct: 124 LLENGNLVLIDGVSERNLWESFEHLGDTMLLESSVMYDVPNNKKRVLSSWKNPTDPSPGE 183
Query: 188 YSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTD-GTTYI 246
+ A L P+ FI + +WR GPW F G P M +++ + QD GT +
Sbjct: 184 FVAELTTQVPPQGFIMRGSRPYWRGGPWARVRFTGIPEMDGSHVSKFDISQDVAAGTGSL 243
Query: 247 TYNFADKTM-FGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTV 305
TY+ + +LT G+LK+I + LE + CD Y CGPFG C S
Sbjct: 244 TYSLERRNSNLSYTTLTSAGSLKIIWNNGSGWVTDLEAPVSSCDVYNTCGPFGLCIRSNP 303
Query: 306 PICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQ-DGFKVYHNMKP 364
P C C GF PK+ EW+ NWT GC+R+ NL C++ + ++ D F + N+KP
Sbjct: 304 PKCECLKGFVPKSDEEWNKRNWTGGCMRRT--NLSCDVNSSATAQANNGDIFDIVANVKP 361
Query: 365 PDFNVRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRV 424
PDF + +++ C CL NCSC A++Y I C+ W EL+D+ +F GG L +R+
Sbjct: 362 PDFYEYLSLINEEDCQQRCLGNCSCTAFSYIEQIGCLVWNRELVDVMQFVAGGETLSIRL 421
Query: 425 PAELVAVKKEKGHNKSFLIIV-IAGVIGALILVICAYLLWRKCSARHKARE--------- 474
+ +A G N+ +I+ I + +ILV +Y WR + ++ +
Sbjct: 422 ASSELA-----GSNRVKIIVASIVSISVFMILVFASYWYWRYKAKQNDSNPIPLETSQDA 476
Query: 475 -HQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASG 533
+Q+K ++ +D + + T TN F N LG+GGFGPVYKG ++DG+EIA+KRLS SG
Sbjct: 477 WREQLKPQDVNFFDMQTILTITNNFSMENKLGQGGFGPVYKGNLQDGKEIAIKRLSSTSG 536
Query: 534 QGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLD 593
QG+EEFMNE+++ISKLQHRNLVRLLGCC+E E++L+YEFM NKSL+ F+FD +K LD
Sbjct: 537 QGLEEFMNEIILISKLQHRNLVRLLGCCIEGEEKLLIYEFMANKSLNTFIFDSTKKLELD 596
Query: 594 WRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGED 653
W KR II+GIA G++YLHRDS LR++HRD+K SNILLD +M PKISDFGLAR+ + G
Sbjct: 597 WPKRFEIIQGIACGLLYLHRDSCLRVVHRDMKVSNILLDEEMNPKISDFGLARMFQ-GTQ 655
Query: 654 DEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLV 713
+ANT+RVVGT GYM PEYA G+FSEKSD+Y+FGVLLLEI++G+R SSF+ E+ +L+
Sbjct: 656 HQANTRRVVGTLGYMSPEYAWTGMFSEKSDIYAFGVLLLEIITGKRISSFTIGEEGKTLL 715
Query: 714 GFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSE 773
FAW W E L+D ++ + ES + RC+ IGLLC+Q+ DRPNI+ V+ ML +
Sbjct: 716 EFAWDSWCESGGSDLLDQDISSSGSESEVARCVQIGLLCIQQQAGDRPNIAQVMSMLTTT 775
Query: 774 ITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
+ LP P + F + ES +S S NN+T + + G
Sbjct: 776 M-DLPKPKQPVF-----AMQVQESDSESKTMYSVNNITQTAIVG 813
>At1g61440 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 792
Score = 618 bits (1593), Expect = e-177
Identities = 334/769 (43%), Positives = 468/769 (60%), Gaps = 25/769 (3%)
Query: 33 ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INKTNNIWIANRDQPLKDS 90
IT L +T++S+N ++LGFFS NS N+Y+GIW+ I +W+ANR++P+ DS
Sbjct: 19 ITKESPLSIGQTLSSSNGVYELGFFSFNNSQNQYVGIWFKGIIPRVVVWVANREKPVTDS 78
Query: 91 NGIVTIHKDGNFIILNKPNGVIIWSTN-ISSSTNSTAQLADSGNLILRDISSGATIWDSF 149
+ I G+ +++N + V+ WST IS+S S A+L+D GNL+++D +G T+W+SF
Sbjct: 79 AANLVISSSGSLLLINGKHDVV-WSTGEISASKGSHAELSDYGNLMVKDNVTGRTLWESF 137
Query: 150 THPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLERLDAPEVFIWKDKNIH 209
H + +P + N VTG+K S KS DPS G + + + F+ + +
Sbjct: 138 EHLGNTLLPLSTMMYNLVTGEKRGLSSWKSYTDPSPGDFWVQITPQVPSQGFVMRGSTPY 197
Query: 210 WRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYNFADKTMFGILSLTPHGTLKL 269
+RTGPW + G P+M Y + + QD +G+ Y +Y D + I+ LT G++K+
Sbjct: 198 YRTGPWAKTRYTGIPQMDESYTSPFSLHQDVNGSGYFSYFERDYKLSRIM-LTSEGSMKV 256
Query: 270 IEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTN 329
+ Y E N CD YG CGPFG C S P C CF GF PK+ EW GNWT+
Sbjct: 257 LRYNGLDWKSSYEGPANSCDIYGVCGPFGFCVISDPPKCKCFKGFVPKSIEEWKRGNWTS 316
Query: 330 GCVRKEGMNLKCEMVKNGSSIVKQDGFKVYHNMKPPDFNVRTNNADQDKCGADCLANCSC 389
GC R+ L C+ G + F N+KPPDF N+ D + C CL NCSC
Sbjct: 317 GCARRT--ELHCQGNSTGKDA---NVFHTVPNIKPPDFYEYANSVDAEGCYQSCLHNCSC 371
Query: 390 LAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAELVAVKKEKGHNKSFLIIVIAGV 449
LA+AY P I C+ W+ +L+D +F GG L +R+ + V K K + ++A
Sbjct: 372 LAFAYIPGIGCLMWSKDLMDTMQFSAGGEILSIRLAHSELDVHKRK-------MTIVAST 424
Query: 450 IGALILVICAYL---LWRKCSARHKA--REHQQMKLDELPLYDFEKLETATNCFHFNNML 504
+ + VI + WR H A + Q + L ++ ++TAT+ F +N L
Sbjct: 425 VSLTLFVILGFATFGFWRNRVKHHDAWRNDLQSQDVPGLEFFEMNTIQTATSNFSLSNKL 484
Query: 505 GKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVER 564
G GGFG VYKG ++DG+EIAVKRLS +S QG +EFMNE+V+ISKLQHRNLVR+LGCCVE
Sbjct: 485 GHGGFGSVYKGKLQDGREIAVKRLSSSSEQGKQEFMNEIVLISKLQHRNLVRVLGCCVEG 544
Query: 565 GEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDL 624
E++L+YEFM NKSLD F+F ++ LDW KR +II+GI RG++YLHRDSRLR+IHRDL
Sbjct: 545 KEKLLIYEFMKNKSLDTFVFGSRKRLELDWPKRFDIIQGIVRGLLYLHRDSRLRVIHRDL 604
Query: 625 KASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDV 684
K SNILLD M PKISDFGLAR+ + G + T+RVVGT GYM PEYA G+FSEKSD+
Sbjct: 605 KVSNILLDEKMNPKISDFGLARLFQ-GSQYQDKTRRVVGTLGYMSPEYAWTGVFSEKSDI 663
Query: 685 YSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACFESSMLR 744
YSFGVLLLEI+SG + S FS+ E+ +L+ + W+ W E ++L+D + D+ + + R
Sbjct: 664 YSFGVLLLEIISGEKISRFSYGEEGKALLAYVWECWCETRGVNLLDQALDDSSHPAEVGR 723
Query: 745 CIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAF-VHKQSSK 792
C+ IGLLCVQ P DRPN ++ ML + + LP P + F VH ++ +
Sbjct: 724 CVQIGLLCVQHQPADRPNTLELLSMLTT-TSDLPLPKQPTFAVHTRNDE 771
>At1g61500
Length = 804
Score = 616 bits (1588), Expect = e-176
Identities = 344/817 (42%), Positives = 483/817 (59%), Gaps = 35/817 (4%)
Query: 13 IITFLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYI 72
+ T +F + SA+ IT+ L +T++S N ++LGFFSP N+ ++Y+GIW+
Sbjct: 10 LFTMFLFTLLSGSSSAV---ITTESPLSMGQTLSSANEVYELGFFSPNNTQDQYVGIWFK 66
Query: 73 NKTNNI--WIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNIS-SSTNSTAQLA 129
+ + W+ANR++P+ DS + I G+ ++LN +G + WS+ ++ SS+ A+L+
Sbjct: 67 DTIPRVVVWVANREKPVTDSTAYLAISSSGSLLLLNGKHGTV-WSSGVTFSSSGCRAELS 125
Query: 130 DSGNLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYS 189
DSGNL + D S +W SF H D + T + N T +K S KS DPS G +
Sbjct: 126 DSGNLKVIDNVSERALWQSFDHLGDTLLHTSSLTYNLATAEKRVLTSWKSYTDPSPGDFL 185
Query: 190 ASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYN 249
+ + F+ + +WR+GPW F G P M Y + QD +G+ Y+TY
Sbjct: 186 GQITPQVPSQGFVMRGSTPYWRSGPWAKTRFTGIPFMDESYTGPFTLHQDVNGSGYLTYF 245
Query: 250 FADKTMFGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICS 309
D + I +LT G++K+ E + CDFYG CGPFG C S P+C
Sbjct: 246 QRDYKLSRI-TLTSEGSIKMFRDNGMGWELYYEAPKKLCDFYGACGPFGLCVMSPSPMCK 304
Query: 310 CFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYHNMKPPDFNV 369
CF GF PK+ EW GNWT GCVR L C G D F N+KPPDF
Sbjct: 305 CFRGFVPKSVEEWKRGNWTGGCVRHT--ELDCLGNSTGEDA---DDFHQIANIKPPDFYE 359
Query: 370 RTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVP-AEL 428
++ + ++C C+ NCSCLA+AY I C+ W +L+D +F G L +R+ +EL
Sbjct: 360 FASSVNAEECHQRCVHNCSCLAFAYIKGIGCLVWNQDLMDAVQFSATGELLSIRLARSEL 419
Query: 429 VAVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWRKCSARHKAREHQQ-----MKLDEL 483
K++K ++ I + +IL A+ +WR C H A + +K ++
Sbjct: 420 DGNKRKKT-----IVASIVSLTLFMILGFTAFGVWR-CRVEHIAHISKDAWKNDLKPQDV 473
Query: 484 P---LYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFM 540
P +D ++ ATN F +N LG+GGFG VYKG ++DG+EIAVKRLS +SGQG EEFM
Sbjct: 474 PGLDFFDMHTIQNATNNFSLSNKLGQGGFGSVYKGKLQDGKEIAVKRLSSSSGQGKEEFM 533
Query: 541 NEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNI 600
NE+V+ISKLQHRNLVR+LGCC+E E++L+YEFM NKSLD FLFD ++ +DW KR +I
Sbjct: 534 NEIVLISKLQHRNLVRVLGCCIEEEEKLLIYEFMVNKSLDTFLFDSRKRLEIDWPKRFDI 593
Query: 601 IEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKR 660
I+GIARG++YLH DSRLR+IHRDLK SNILLD M PKISDFGLAR+ + G + + NT+R
Sbjct: 594 IQGIARGLLYLHHDSRLRVIHRDLKVSNILLDEKMNPKISDFGLARMYQ-GTEYQDNTRR 652
Query: 661 VVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLW 720
VVGT GYM PEYA G+FSEKSD+YSFGVL+LEI+SG + S FS+ + +L+ +AW+ W
Sbjct: 653 VVGTLGYMSPEYAWTGMFSEKSDIYSFGVLMLEIISGEKISRFSYGVEGKTLIAYAWESW 712
Query: 721 LEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPP 780
E I L+D ++ D+C + RCI IGLLCVQ P DRPN ++ ML + + LP P
Sbjct: 713 SEYRGIDLLDQDLADSCHPLEVGRCIQIGLLCVQHQPADRPNTLELLAMLTT-TSDLPSP 771
Query: 781 GRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
+ F T + S+ + N +T S + G
Sbjct: 772 KQPTFAF-----HTRDDESLSNDLITVNGMTQSVILG 803
>At1g61420 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 807
Score = 615 bits (1586), Expect = e-176
Identities = 344/807 (42%), Positives = 484/807 (59%), Gaps = 45/807 (5%)
Query: 14 ITFLIFCTIYSCY-SAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY- 71
I F + + S + S+ + IT L +T++S+N ++LGFF+ NS N+Y+GIW+
Sbjct: 6 IVFFAYLLLSSFFISSSSAGITKESPLPIGQTLSSSNGFYELGFFNFNNSQNQYVGIWFK 65
Query: 72 -INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNSTAQLAD 130
I +W+ANR++P+ DS + I +G+ ++ N +GV S S S A+L+D
Sbjct: 66 GIIPRVVVWVANREKPVTDSTANLAISNNGSLLLFNGKHGVAWSSGEALVSNGSRAELSD 125
Query: 131 SGNLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSA 190
+GNLI+ D SG T+W SF H D +P+ + N TG+K S KS DPS G +
Sbjct: 126 TGNLIVIDNFSGRTLWQSFDHLGDTMLPSSTLKYNLATGEKQVLSSWKSYTDPSVGDFVL 185
Query: 191 SLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITY-N 249
+ +V + K ++R+GPW F G P M + QDT+G+ +TY N
Sbjct: 186 QITPQVPTQVLVTKGSTPYYRSGPWAKTRFTGIPLMDDTFTGPVSVQQDTNGSGSLTYLN 245
Query: 250 FADKTMFGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICS 309
D+ +L T GT +L + + ++ CD+YG CGPFG C S P C+
Sbjct: 246 RNDRLQRTML--TSKGTQELSWHNGTDWVLNFVAPEHSCDYYGVCGPFGLCVKSVPPKCT 303
Query: 310 CFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYH---NMKPPD 366
CF GF PK EW GNWT GCVR+ E+ G+S K V+H +KPPD
Sbjct: 304 CFKGFVPKLIEEWKRGNWTGGCVRRT------ELYCQGNSTGKYAN--VFHPVARIKPPD 355
Query: 367 FNVRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPA 426
F + + ++C CL NCSCLA+AY I C+ W +L+D +F GG L +R+
Sbjct: 356 FYEFASFVNVEECQKSCLHNCSCLAFAYIDGIGCLMWNQDLMDAVQFSEGGELLSIRL-- 413
Query: 427 ELVAVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWRKCSARHKAREHQQMKLDE---- 482
+ E G NK I A ++ ++VI A++ + C R++ + + + D
Sbjct: 414 ----ARSELGGNKRKKAIT-ASIVSLSLVVIIAFVAF--CFWRYRVKHNADITTDASQVS 466
Query: 483 ------------LPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSK 530
L +D ++TATN F +N LG+GGFGPVYKG ++DG+EIAVKRLS
Sbjct: 467 WRNDLKPQDVPGLDFFDMHTIQTATNNFSISNKLGQGGFGPVYKGKLQDGKEIAVKRLSS 526
Query: 531 ASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKK 590
+SGQG EEFMNE+V+ISKLQH+NLVR+LGCC+E E++L+YEFM N SLD FLFD ++
Sbjct: 527 SSGQGKEEFMNEIVLISKLQHKNLVRILGCCIEGEEKLLIYEFMLNNSLDTFLFDSRKRL 586
Query: 591 NLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKF 650
+DW KR +II+GIARGI YLHRDS L++IHRDLK SNILLD M PKISDFGLAR+ +
Sbjct: 587 EIDWPKRLDIIQGIARGIHYLHRDSHLKVIHRDLKVSNILLDEKMNPKISDFGLARMYQ- 645
Query: 651 GEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTL 710
G + + NT+RVVGT GYM PEYA G+FSEKSD+YSFGVL+LEI+SG + S FS+ ++
Sbjct: 646 GTEYQDNTRRVVGTLGYMAPEYAWTGMFSEKSDIYSFGVLMLEIISGEKISRFSYGKEEK 705
Query: 711 SLVGFAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLML 770
+L+ +AW+ W + I L+D +V D+C + RC+ IGLLCVQ P DRPN ++ ML
Sbjct: 706 TLIAYAWESWCDTGGIDLLDKDVADSCRPLEVERCVQIGLLCVQHQPADRPNTLELLSML 765
Query: 771 VSEITHLPPPGRVAF-VHKQSSKSTTE 796
+ + LPPP + F VH++ KS++E
Sbjct: 766 TT-TSDLPPPEQPTFVVHRRDDKSSSE 791
>At1g61550
Length = 802
Score = 613 bits (1581), Expect = e-175
Identities = 349/824 (42%), Positives = 484/824 (58%), Gaps = 54/824 (6%)
Query: 18 IFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INKT 75
+F T+ +S T TS S+ +T++S N F+LGFFSP NS N Y+GIW+ I
Sbjct: 8 LFSTLLLSFSYAAITPTSPLSI--GQTLSSPNGIFELGFFSPNNSRNLYVGIWFKGIIPR 65
Query: 76 NNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTN-STAQLADSGNL 134
+W+ANR+ + D+ + I +G+ ++ + + + WST + ++N S+A+L+DSGNL
Sbjct: 66 TVVWVANRENSVTDATADLAISSNGSLLLFDGKHSTV-WSTGETFASNGSSAELSDSGNL 124
Query: 135 ILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLER 194
++ D SG T+W SF H D +P + N TG+K S KS DP G + +
Sbjct: 125 LVIDKVSGITLWQSFEHLGDTMLPYSSLMYNPGTGEKRVLSSWKSYTDPLPGEFVGYITT 184
Query: 195 LDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITY---NFA 251
P+ FI + +WR+GPW F G P Y + QD +G+ Y ++ NF
Sbjct: 185 QVPPQGFIMRGSKPYWRSGPWAKTRFTGVPLTDESYTHPFSVQQDANGSVYFSHLQRNFK 244
Query: 252 DKTMFGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCF 311
+L LT G+LK+ + + ++V N CDFYG CGPFG C S P C CF
Sbjct: 245 RS----LLVLTSEGSLKVTHHNGTDWVLNIDVPANTCDFYGVCGPFGLCVMSIPPKCKCF 300
Query: 312 DGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYH---NMKPPDFN 368
GF P+ S EW GNWT GCVR+ L C+ G + V+H N+KPPDF
Sbjct: 301 KGFVPQFSEEWKRGNWTGGCVRRT--ELLCQGNSTGRHV------NVFHPVANIKPPDFY 352
Query: 369 VRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAEL 428
++ ++C CL NCSCLA+AY I C+ W EL+D+ +F GG L +R+ +
Sbjct: 353 EFVSSGSAEECYQSCLHNCSCLAFAYINGIGCLIWNQELMDVMQFSVGGELLSIRLASS- 411
Query: 429 VAVKKEKGHN--KSFLIIVIAGVIGALILVICAYLLWRKCSARHKA------------RE 474
E G N K +I I + + L A+ WR +H A +
Sbjct: 412 -----EMGGNQRKKTIIASIVSISLFVTLASAAFGFWRY-RLKHNAIVSKVSLQGAWRND 465
Query: 475 HQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQ 534
+ + L ++ + +E ATN F N LG+GGFGPVYKG ++DG+EIAVKRLS +SGQ
Sbjct: 466 LKSEDVSGLYFFEMKTIEIATNNFSLVNKLGQGGFGPVYKGKLQDGKEIAVKRLSSSSGQ 525
Query: 535 GIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDW 594
G EEFMNE+++ISKLQH NLVR+LGCC+E E++LVYEFM NKSLD F+FD ++ +DW
Sbjct: 526 GKEEFMNEILLISKLQHINLVRILGCCIEGEERLLVYEFMVNKSLDTFIFDSRKRVEIDW 585
Query: 595 RKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDD 654
KR +II+GIARG++YLHRDSRLRIIHRD+K SNILLD M PKISDFGLAR+ + G
Sbjct: 586 PKRFSIIQGIARGLLYLHRDSRLRIIHRDVKVSNILLDDKMNPKISDFGLARMYE-GTKY 644
Query: 655 EANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVG 714
+ NT+R+VGT GYM PEYA G+FSEKSD YSFGVLLLE++SG + S FS+ ++ +L+
Sbjct: 645 QDNTRRIVGTLGYMSPEYAWTGVFSEKSDTYSFGVLLLEVISGEKISRFSYDKERKNLLA 704
Query: 715 FAWKLWLEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEI 774
+AW+ W E + +D + D+C S + RC+ IGLLCVQ P DRPN ++ ML +
Sbjct: 705 YAWESWCENGGVGFLDKDATDSCHPSEVGRCVQIGLLCVQHQPADRPNTLELLSMLTT-T 763
Query: 775 THLPPPGRVAF-VHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
+ LP P F VH T++ ++ + N VT S V G
Sbjct: 764 SDLPLPKEPTFAVH------TSDDGSRTSDLITVNEVTQSVVLG 801
>At1g61400 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 821
Score = 608 bits (1567), Expect = e-174
Identities = 341/799 (42%), Positives = 480/799 (59%), Gaps = 31/799 (3%)
Query: 17 LIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INK 74
L++ +I+ +S+ IT L +T++S+N ++LGFFS NS N+Y+GI + I
Sbjct: 22 LLWLSIFISFSSAE--ITEESPLSIGQTLSSSNGVYELGFFSFNNSQNQYVGISFKGIIP 79
Query: 75 TNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNSTAQLADSGNL 134
+W+ANR++P+ DS + I +G+ + N +GV+ S +S S +L DSGNL
Sbjct: 80 RVVVWVANREKPVTDSAANLVISSNGSLQLFNGKHGVVWSSGKALASNGSRVELLDSGNL 139
Query: 135 ILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLER 194
++ + SG T+W+SF H D +P I N TG+K S KS DPS G + +
Sbjct: 140 VVIEKVSGRTLWESFEHLGDTLLPHSTIMYNVHTGEKRGLTSWKSYTDPSPGDFVVLITP 199
Query: 195 LDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYNFADKT 254
+ F+ + ++R+GPW F G P+M Y + + QD +G+ Y +Y D
Sbjct: 200 QVPSQGFLMRGSTPYFRSGPWAKTKFTGLPQMDESYTSPFSLTQDVNGSGYYSYFDRDNK 259
Query: 255 MFGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCFDGF 314
I LTP G++K + Y E N CD YG CGPFG C S P C CF GF
Sbjct: 260 RSRI-RLTPDGSMKALRYNGMDWDTTYEGPANSCDIYGVCGPFGFCVISVPPKCKCFKGF 318
Query: 315 EPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYHNMKPPDFNVRTNNA 374
PK+ EW GNWT+GCVR+ L C+ G + F N+KPPDF ++
Sbjct: 319 IPKSIEEWKTGNWTSGCVRRS--ELHCQGNSTGKDA---NVFHTVPNIKPPDFYEYADSV 373
Query: 375 DQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAELVAVKKE 434
D ++C +CL NCSCLA+AY P I C+ W+ +L+D +F GG L +R+ + V K
Sbjct: 374 DAEECQQNCLNNCSCLAFAYIPGIGCLMWSKDLMDTVQFAAGGELLSIRLARSELDVNKR 433
Query: 435 KGHNKSFLIIVIAGVIGALILVICAYLLWRKCSARHKAREHQQMKLDELP---LYDFEKL 491
K K+ + I ++ + +IL A+ WR+ +++ ++ ++P ++ +
Sbjct: 434 K---KTIIAITVSLTL-FVILGFTAFGFWRRRVEQNEDAWRNDLQTQDVPGLEYFEMNTI 489
Query: 492 ETATNCFHFNNMLGKGGFGPVYK---GVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISK 548
+TATN F +N LG GGFG VYK G ++DG+EIAVKRLS +S QG +EFMNE+V+ISK
Sbjct: 490 QTATNNFSLSNKLGHGGFGSVYKARNGKLQDGREIAVKRLSSSSEQGKQEFMNEIVLISK 549
Query: 549 LQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLF--------DPLQKKNLDWRKRSNI 600
LQHRNLVR+LGCCVE E++L+YEFM NKSLD F+F D ++ +DW KR +I
Sbjct: 550 LQHRNLVRVLGCCVEGTEKLLIYEFMKNKSLDTFVFVFTRCFCLDSKKRLEIDWPKRFDI 609
Query: 601 IEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKR 660
I+GIARG++YLHRDSRLRIIHRDLK SNILLD M PKISDFGLAR+ G + + T+R
Sbjct: 610 IQGIARGLLYLHRDSRLRIIHRDLKVSNILLDEKMNPKISDFGLARMFH-GTEYQDKTRR 668
Query: 661 VVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLW 720
VVGT GYM PEYA G+FSEKSD+YSFGVLLLEI+SG + S FS+ E+ +L+ +AW+ W
Sbjct: 669 VVGTLGYMSPEYAWAGVFSEKSDIYSFGVLLLEIISGEKISRFSYGEEGKTLLAYAWECW 728
Query: 721 LEENIISLIDPEVWDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPP 780
++L+D + D+C + RC+ IGLLCVQ P DRPN ++ ML + + LP P
Sbjct: 729 CGARGVNLLDQALGDSCHPYEVGRCVQIGLLCVQYQPADRPNTLELLSMLTT-TSDLPLP 787
Query: 781 GRVAF-VHKQSSKSTTESS 798
+ F VH + KS + S
Sbjct: 788 KQPTFVVHTRDGKSPSNDS 806
>At1g61490 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 804
Score = 605 bits (1559), Expect = e-173
Identities = 334/779 (42%), Positives = 468/779 (59%), Gaps = 36/779 (4%)
Query: 33 ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INKTNNIWIANRDQPLKDS 90
IT+ L +T++S+N ++LGFFSP NS N Y+GIW+ I +W+ANR+ P D+
Sbjct: 26 ITTESPLSVEQTLSSSNGIYELGFFSPNNSQNLYVGIWFKGIIPRVVVWVANRETPTTDT 85
Query: 91 NGIVTIHKDGNFIILNKPNGVIIWSTNISSSTN-STAQLADSGNLILRDISSGATIWDSF 149
+ + I +G+ ++ N +GV+ WS + ++N S A+L D+GNL++ D +SG T+W+SF
Sbjct: 86 SANLAISSNGSLLLFNGKHGVV-WSIGENFASNGSRAELTDNGNLVVIDNASGRTLWESF 144
Query: 150 THPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLERLDAPEVFIWKDKNIH 209
H D +P + N TG+K S K+D DPS G + + +V I + +
Sbjct: 145 EHFGDTMLPFSSLMYNLATGEKRVLTSWKTDTDPSPGVFVGQITPQVPSQVLIMRGSTRY 204
Query: 210 WRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTDGTTYITYNFADKTMFGILSLTPHGTLKL 269
+RTGPW F G P M Y + + QD +G+ + TY F + ++ G++K
Sbjct: 205 YRTGPWAKTRFTGIPLMDDTYASPFSLQQDANGSGFFTY-FDRSFKLSRIIISSEGSMKR 263
Query: 270 IEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTN 329
+ N CD YG CGPFG C S C C GF P ++ EW GNWT
Sbjct: 264 FRHNGTDWELSYMAPANSCDIYGVCGPFGLCIVSVPLKCKCLKGFVPHSTEEWKRGNWTG 323
Query: 330 GCVRKEGMNLKCEMVKNGSSIVKQDGFKVYH---NMKPPDFNVRTNNADQDKCGADCLAN 386
GC R L C+ G + ++H N+K PDF ++ D ++C CL N
Sbjct: 324 GCARLT--ELHCQGNSTGKDV------NIFHPVTNVKLPDFYEYESSVDAEECHQSCLHN 375
Query: 387 CSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAELVAVKKEKGHNKSFLIIVI 446
CSCLA+AY I C+ W L+D +F GG L +R+ E G NK IIV
Sbjct: 376 CSCLAFAYIHGIGCLIWNQNLMDAVQFSAGGEILSIRL------AHSELGGNKRNKIIVA 429
Query: 447 AGVIGALILVI--CAYLLWRKCSARHKARE-----HQQMKLDELP---LYDFEKLETATN 496
+ V +L +++ A+ WR +HKA +K E+P ++ ++TATN
Sbjct: 430 STVSLSLFVILTSAAFGFWRY-RVKHKAYTLKDAWRNDLKSKEVPGLEFFEMNTIQTATN 488
Query: 497 CFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVR 556
F +N LG+GGFG VYKG ++DG+EIAVK+LS +SGQG EEFMNE+V+ISKLQHRNLVR
Sbjct: 489 NFSLSNKLGQGGFGSVYKGKLQDGKEIAVKQLSSSSGQGKEEFMNEIVLISKLQHRNLVR 548
Query: 557 LLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSR 616
+LGCC+E E++L+YEFM NKSLD F+FD +K +DW KR +I++GIARG++YLHRDSR
Sbjct: 549 VLGCCIEGEEKLLIYEFMLNKSLDTFVFDARKKLEVDWPKRFDIVQGIARGLLYLHRDSR 608
Query: 617 LRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEG 676
L++IHRDLK SNILLD M PKISDFGLAR+ + G + T+RVVGT GYM PEYA G
Sbjct: 609 LKVIHRDLKVSNILLDEKMNPKISDFGLARMYE-GTQCQDKTRRVVGTLGYMSPEYAWTG 667
Query: 677 LFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDA 736
+FSEKSD+YSFGVLLLEI+ G + S FS+ E+ +L+ +AW+ W E I L+D ++ D+
Sbjct: 668 VFSEKSDIYSFGVLLLEIIIGEKISRFSYGEEGKTLLAYAWESWGETKGIDLLDQDLADS 727
Query: 737 CFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAF-VHKQSSKST 794
C + RC+ IGLLCVQ P DRPN ++ ML + + LP P + F VH + +S+
Sbjct: 728 CRPLEVGRCVQIGLLCVQHQPADRPNTLELLAMLTT-TSDLPSPKQPTFVVHSRDDESS 785
>At4g21380 receptor-like serine/threonine protein kinase ARK3
Length = 850
Score = 601 bits (1549), Expect = e-172
Identities = 359/866 (41%), Positives = 501/866 (57%), Gaps = 87/866 (10%)
Query: 6 HNSNYFFIITFLIFCTIYSCYSAINDTITSSKSL--KDNETITSNNTNFKLGFFSPLNST 63
H+ +FF ++F YS +T+++S+SL N TI S F+LGFF P +
Sbjct: 9 HSYTFFFFFLLILF----PAYSISANTLSASESLTISSNNTIVSPGNVFELGFFKPGLDS 64
Query: 64 NRYLGIWY--INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSS 121
YLGIWY I+K +W+ANRD PL S G + I D N ++L++ + +WSTN++
Sbjct: 65 RWYLGIWYKAISKRTYVWVANRDTPLSSSIGTLKI-SDSNLVVLDQSD-TPVWSTNLTGG 122
Query: 122 ---TNSTAQLADSGNLILRDISSGA---TIWDSFTHPADAAVPTMRIAANQVTGKKISFV 175
+ A+L D+GN +LRD + A +W SF P D +P M++ + TG
Sbjct: 123 DVRSPLVAELLDNGNFVLRDSKNSAPDGVLWQSFDFPTDTLLPEMKLGWDAKTGFNRFIR 182
Query: 176 SRKSDNDPSSGHYSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWR 235
S KS +DPSSG +S LE PE+F+W ++ +R+GPWNG F G P M +
Sbjct: 183 SWKSPDDPSSGDFSFKLETEGFPEIFLWNRESRMYRSGPWNGIRFSGVPEMQPFEYMVFN 242
Query: 236 FDQDTDGTTYITYNFADKTMFGILSLTPHGTLKLIEYMNKKELFR--LEVDQNECDFYGK 293
F + TY ++ ++ LS++ G L+ ++ + + +++CD Y +
Sbjct: 243 FTTSKEEVTY-SFRITKSDVYSRLSISSSGLLQRFTWIETAQNWNQFWYAPKDQCDEYKE 301
Query: 294 CGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQ 353
CG +G CD++T P+C+C GF+P+N W L + ++GCVRK ++ G
Sbjct: 302 CGVYGYCDSNTSPVCNCIKGFKPRNPQVWGLRDGSDGCVRK-------TLLSCGGG---- 350
Query: 354 DGFKVYHNMKPPDFNVRTNNADQDK------CGADCLANCSCLAYA----YDPSIFCMYW 403
DGF MK PD T A D+ C CL +C+C A+A C+ W
Sbjct: 351 DGFVRLKKMKLPD----TTTASVDRGIGVKECEQKCLRDCNCTAFANTDIRGSGSGCVTW 406
Query: 404 TGELIDLQKFPNGGVDLFVRVPAELVAVKKEKGHNKSFLIIVIA-GVIGALILVICAYLL 462
TGEL D++ + GG DL+VR L A E N+S II + GV L+L + L
Sbjct: 407 TGELFDIRNYAKGGQDLYVR----LAATDLEDKRNRSAKIIGSSIGVSVLLLLSFIIFFL 462
Query: 463 WRK-----------------------------CSARHKAREHQQMKLDELPLYDFEKLET 493
W++ S RH +RE+ L ELPL +FE++
Sbjct: 463 WKRKQKRSILIETPIVDHQLRSRDLLMNEVVISSRRHISRENNTDDL-ELPLMEFEEVAM 521
Query: 494 ATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRN 553
ATN F N LG+GGFG VYKG + DGQE+AVKRLSK S QG +EF NEV +I++LQH N
Sbjct: 522 ATNNFSNANKLGQGGFGIVYKGKLLDGQEMAVKRLSKTSVQGTDEFKNEVKLIARLQHIN 581
Query: 554 LVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHR 613
LVRLL CCV+ GE++L+YE++ N SLD+ LFD + L+W+ R +II GIARG++YLH+
Sbjct: 582 LVRLLACCVDAGEKMLIYEYLENLSLDSHLFDKSRNSKLNWQMRFDIINGIARGLLYLHQ 641
Query: 614 DSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDD-EANTKRVVGTYGYMPPEY 672
DSR RIIHRDLKASNILLD M PKISDFG+ARI FG D+ EANT++VVGTYGYM PEY
Sbjct: 642 DSRFRIIHRDLKASNILLDKYMTPKISDFGMARI--FGRDETEANTRKVVGTYGYMSPEY 699
Query: 673 AMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPE 732
AM+G+FS KSDV+SFGVLLLEI+S +RN F + + L+L+G W+ W E + +IDP
Sbjct: 700 AMDGIFSMKSDVFSFGVLLLEIISSKRNKGFYNSDRDLNLLGCVWRNWKEGKGLEIIDPI 759
Query: 733 VWDACF---ESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAFVHKQ 789
+ D+ + +LRCI IGLLCVQE DRP +S V+LML SE T +P P + ++
Sbjct: 760 ITDSSSTFRQHEILRCIQIGLLCVQERAEDRPTMSLVILMLGSESTTIPQPKAPGYCLER 819
Query: 790 SSKSTTESS--QKSHQSNSNNNVTLS 813
S T SS Q+ +S + N +T+S
Sbjct: 820 SLLDTDSSSSKQRDDESWTVNQITVS 845
>At1g11340 receptor kinase, putative
Length = 901
Score = 600 bits (1547), Expect = e-171
Identities = 360/859 (41%), Positives = 482/859 (55%), Gaps = 72/859 (8%)
Query: 7 NSNYFFIITFLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRY 66
N+N + F C S DTI +SL+D E I S F GFFS +S RY
Sbjct: 66 NTNMKVVFVIFFFFLFQFCISV--DTIMRRQSLRDGEVILSAGKRFAFGFFSLGDSELRY 123
Query: 67 LGIWY--INKTNNIWIANRDQPLKDSNGIVTIHKDGNFIILNKPNGV-IIWSTNISSST- 122
+GIWY I++ +W+ANRD P+ D++G+V GN + N +IWSTN+S S
Sbjct: 124 VGIWYAQISQQTIVWVANRDHPINDTSGMVKFSNRGNLSVYASDNETELIWSTNVSDSML 183
Query: 123 --NSTAQLADSGNLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSD 180
A L+D GNL+L D +G + W+SF HP D +P MR+ + G S S KS
Sbjct: 184 EPTLVATLSDLGNLVLFDPVTGRSFWESFDHPTDTFLPFMRLGFTRKDGLDRSLTSWKSH 243
Query: 181 NDPSSGHYSASLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDT 240
DP SG +ER P++ ++K WR G W G + G P M Y+ F +
Sbjct: 244 GDPGSGDLILRMERRGFPQLILYKGVTPWWRMGSWTGHRWSGVPEMPIGYIFNNSFVNNE 303
Query: 241 DGTTYITYNFADKTMFGILSLTPHGTLKLIEYMNKKELFR--LEVDQNECDFYGKCGPFG 298
D ++ TY D ++ + GT+ ++ + + + V + +CD Y CGP G
Sbjct: 304 DEVSF-TYGVTDASVITRTMVNETGTMHRFTWIARDKRWNDFWSVPKEQCDNYAHCGPNG 362
Query: 299 NCDNSTVPI--CSCFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGF 356
CD+ + C+C GFEPK W L + + GC +K K S ++DGF
Sbjct: 363 YCDSPSSKTFECTCLPGFEPKFPRHWFLRDSSGGCTKK----------KRASICSEKDGF 412
Query: 357 KVYHNMKPPDFNVRTNNADQD------KCGADCLANCSCLAYA--YDPS----IFCMYWT 404
MK PD T++A D +C CL NCSC+AYA Y S I C+ W
Sbjct: 413 VKLKRMKIPD----TSDASVDMNITLKECKQRCLKNCSCVAYASAYHESKRGAIGCLKWH 468
Query: 405 GELIDLQKFPNGGVDLFVRVPAELVA-----VKKEKGHNKSFLIIVIAGVIGALILVICA 459
G ++D + + N G D ++RV E +++E+ + S+ V V+C
Sbjct: 469 GGMLDARTYLNSGQDFYIRVDKEDAVEQKWLIREEESPSDSYQFDCSRNVTNCHF-VLCC 527
Query: 460 YLLWRKCSARHKAR-----------------EHQQMKLDELPLYDFEKLETATNCFHFNN 502
S RH++ E + + ELPL+D + ATN F N
Sbjct: 528 KGATNAESNRHRSSSANFAPVPFDFDESFRFEQDKARNRELPLFDLNTIVAATNNFSSQN 587
Query: 503 MLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCV 562
LG GGFGPVYKGV+++ EIAVKRLS+ SGQG+EEF NEV +ISKLQHRNLVR+LGCCV
Sbjct: 588 KLGAGGFGPVYKGVLQNRMEIAVKRLSRNSGQGMEEFKNEVKLISKLQHRNLVRILGCCV 647
Query: 563 ERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHR 622
E E++LVYE++PNKSLD F+F Q+ LDW KR I+ GIARGI+YLH+DSRLRIIHR
Sbjct: 648 ELEEKMLVYEYLPNKSLDYFIFHEEQRAELDWPKRMEIVRGIARGILYLHQDSRLRIIHR 707
Query: 623 DLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKS 682
DLKASNILLDS+MIPKISDFG+ARI G E T RVVGT+GYM PEYAMEG FS KS
Sbjct: 708 DLKASNILLDSEMIPKISDFGMARIFG-GNQMEGCTSRVVGTFGYMAPEYAMEGQFSIKS 766
Query: 683 DVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLW----LEENIISLIDPEVWDACF 738
DVYSFGVL+LEI++G++NS+F HE++ +LVG W LW E I +L+D E +D
Sbjct: 767 DVYSFGVLMLEIITGKKNSAF--HEESSNLVGHIWDLWENGEATEIIDNLMDQETYD--- 821
Query: 739 ESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESS 798
E +++CI IGLLCVQE DR ++S+VV+ML T+LP P AF + +
Sbjct: 822 EREVMKCIQIGLLCVQENASDRVDMSSVVIMLGHNATNLPNPKHPAFTSARRRGGENGAC 881
Query: 799 QKSHQSNSNNNVTLSEVQG 817
K S N+VT S++QG
Sbjct: 882 LKGQTGISVNDVTFSDIQG 900
>At1g61380 S-like receptor protein kinase
Length = 805
Score = 600 bits (1546), Expect = e-171
Identities = 338/800 (42%), Positives = 476/800 (59%), Gaps = 23/800 (2%)
Query: 26 YSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INKTNNIWIANR 83
Y+AIN +S L +T++S ++LGFFSP N+ N+Y+GIW+ I +W+ANR
Sbjct: 20 YAAIN----TSSPLSIRQTLSSPGGFYELGFFSPNNTQNQYVGIWFKKIVPRVVVWVANR 75
Query: 84 DQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNST-AQLADSGNLILRDISSG 142
D P+ S +TI +G+ I+L+ VI WST + ++N A+L D+GN ++ D SG
Sbjct: 76 DTPVTSSAANLTISSNGSLILLDGKQDVI-WSTGKAFTSNKCHAELLDTGNFVVIDDVSG 134
Query: 143 ATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLERLDAPEVFI 202
+W SF H + +P + + GKK + KS++DPS G +S + + I
Sbjct: 135 NKLWQSFEHLGNTMLPQSSLMYDTSNGKKRVLTTWKSNSDPSPGEFSLEITPQIPTQGLI 194
Query: 203 WKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDTD-GTTYITYNFADKTMFGILSL 261
+ +WR GPW F G + Y++ + QDT GT +Y+ ++L
Sbjct: 195 RRGSVPYWRCGPWAKTRFSGISGIDASYVSPFSVVQDTAAGTGSFSYSTLRNYNLSYVTL 254
Query: 262 TPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVE 321
TP G +K++ L + +N CD YG+CGP+G C S P C C GF PK+ E
Sbjct: 255 TPEGKMKILWDDGNNWKLHLSLPENPCDLYGRCGPYGLCVRSDPPKCECLKGFVPKSDEE 314
Query: 322 WSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQ-DGFKVYHNMKPPDFNVRTNNADQDKCG 380
W GNWT+GCVR+ L C+ + + K D F ++K PD + + + ++C
Sbjct: 315 WGKGNWTSGCVRRT--KLSCQAKSSMKTQGKDTDIFYRMTDVKTPDLHQFASFLNAEQCY 372
Query: 381 ADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAELVAVKKEKGHNKS 440
CL NCSC A+AY I C+ W GEL D +F + G LF+R+ + +A +
Sbjct: 373 QGCLGNCSCTAFAYISGIGCLVWNGELADTVQFLSSGEFLFIRLASSELAGSSRR----K 428
Query: 441 FLIIVIAGVIGALILVICAYLLWRKCSARHKAREH--QQMKLDELPLYDFEKLETATNCF 498
++ + LILV A +LWR + ++ A ++ ++ + + ++ + TATN F
Sbjct: 429 IIVGTTVSLSIFLILVFAAIMLWRYRAKQNDAWKNGFERQDVSGVNFFEMHTIRTATNNF 488
Query: 499 HFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRNLVRLL 558
+N LG+GGFGPVYKG + DG+EI VKRL+ +SGQG EEFMNE+ +ISKLQHRNLVRLL
Sbjct: 489 SPSNKLGQGGFGPVYKGKLVDGKEIGVKRLASSSGQGTEEFMNEITLISKLQHRNLVRLL 548
Query: 559 GCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLR 618
G C++ E++L+YEFM NKSLD F+FDP K LDW KR NII+GIARG++YLHRDSRLR
Sbjct: 549 GYCIDGEEKLLIYEFMVNKSLDIFIFDPCLKFELDWPKRFNIIQGIARGLLYLHRDSRLR 608
Query: 619 IIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLF 678
+IHRDLK SNILLD M PKISDFGLAR+ + G + NT+RVVGT GYM PEYA GLF
Sbjct: 609 VIHRDLKVSNILLDDRMNPKISDFGLARMFQ-GTQYQDNTRRVVGTLGYMSPEYAWAGLF 667
Query: 679 SEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEVWDACF 738
SEKSD+YSFGVL+LEI+SG+R S F + +++ L+ + W W E +L+D ++ D C
Sbjct: 668 SEKSDIYSFGVLMLEIISGKRISRFIYGDESKGLLAYTWDSWCETGGSNLLDRDLTDTCQ 727
Query: 739 ESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAF-VHKQSSKSTTES 797
+ RC+ IGLLCVQ DRPN V+ ML S T LP P + F VH + ++
Sbjct: 728 AFEVARCVQIGLLCVQHEAVDRPNTLQVLSMLTS-ATDLPVPKQPIFAVHTLNDMPMLQA 786
Query: 798 SQKSHQSNSNNNVTLSEVQG 817
+ S S N +T S +QG
Sbjct: 787 N--SQDFLSVNEMTESMIQG 804
>At1g61360 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 821
Score = 596 bits (1537), Expect = e-170
Identities = 340/826 (41%), Positives = 476/826 (57%), Gaps = 43/826 (5%)
Query: 13 IITFLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYI 72
+ L+ ++S Y IT+S L T++S +++LGFFS NS N+Y+GIW+
Sbjct: 4 VACLLLITALFSSYGYA--AITTSSPLSIGVTLSSPGGSYELGFFSSNNSGNQYVGIWFK 61
Query: 73 NKTNNI--WIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNSTAQLAD 130
T + W+ANR++P+ + +TI +G+ I+L+ ++ S +S A+L D
Sbjct: 62 KVTPRVIVWVANREKPVSSTMANLTISSNGSLILLDSKKDLVWSSGGDPTSNKCRAELLD 121
Query: 131 SGNLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSA 190
+GNL++ D +G +W SF H D +P + + KK S KS+ DPS G + A
Sbjct: 122 TGNLVVVDNVTGNYLWQSFEHLGDTMLPLTSLMYDIPNNKKRVLTSWKSETDPSPGEFVA 181
Query: 191 SLERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQD-TDGTTYITYN 249
+ + I K + +WR+GPW G F G P M Y+ QD +GT +
Sbjct: 182 EITPQVPSQGLIRKGSSPYWRSGPWAGTRFTGIPEMDASYVNPLGMVQDEVNGTGVFAFC 241
Query: 250 FADKTMFGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICS 309
+ LTP G+L++ + E CD YG+CGPFG C S P+C
Sbjct: 242 VLRNFNLSYIKLTPEGSLRITRNNGTDWIKHFEGPLTSCDLYGRCGPFGLCVRSGTPMCQ 301
Query: 310 CFDGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFK---VYH--NMKP 364
C GFEPK+ EW GNW+ GCVR+ NL C+ G+S V+ G YH N+KP
Sbjct: 302 CLKGFEPKSDEEWRSGNWSRGCVRRT--NLSCQ----GNSSVETQGKDRDVFYHVSNIKP 355
Query: 365 PDFNVRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRV 424
PD + +++++C CL NCSC A++Y I C+ W EL+D KF GG L +R+
Sbjct: 356 PDSYELASFSNEEQCHQGCLRNCSCTAFSYVSGIGCLVWNQELLDTVKFIGGGETLSLRL 415
Query: 425 P-AELVAVKKEKGHNKSFLIIVIAGVIGALILVICAYLLWRKCSARHKAR---------- 473
+EL K+ K + + + LILV+ A WR ++ +
Sbjct: 416 AHSELTGRKRIK-----IITVATLSLSVCLILVLVACGCWRYRVKQNGSSLVSKDNVEGA 470
Query: 474 ---EHQQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSK 530
+ Q + L ++ L+TATN F N LG+GGFG VYKG ++DG+EIAVKRL+
Sbjct: 471 WKSDLQSQDVSGLNFFEIHDLQTATNNFSVLNKLGQGGFGTVYKGKLQDGKEIAVKRLTS 530
Query: 531 ASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKK 590
+S QG EEFMNE+ +ISKLQHRNL+RLLGCC++ E++LVYE+M NKSLD F+FD +K
Sbjct: 531 SSVQGTEEFMNEIKLISKLQHRNLLRLLGCCIDGEEKLLVYEYMVNKSLDIFIFDLKKKL 590
Query: 591 NLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKF 650
+DW R NII+GIARG++YLHRDS LR++HRDLK SNILLD M PKISDFGLAR+
Sbjct: 591 EIDWATRFNIIQGIARGLLYLHRDSFLRVVHRDLKVSNILLDEKMNPKISDFGLARLF-H 649
Query: 651 GEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTL 710
G + +T VVGT GYM PEYA G FSEKSD+YSFGVL+LEI++G+ SSFS+ +D
Sbjct: 650 GNQHQDSTGSVVGTLGYMSPEYAWTGTFSEKSDIYSFGVLMLEIITGKEISSFSYGKDNK 709
Query: 711 SLVGFAWKLWLEENIISLIDPEVWDACFESSML--RCIHIGLLCVQELPRDRPNISTVVL 768
+L+ +AW W E ++L+D ++ D+ +S+ RC+HIGLLCVQ DRPNI V+
Sbjct: 710 NLLSYAWDSWSENGGVNLLDQDLDDSDSVNSVEAGRCVHIGLLCVQHQAIDRPNIKQVMS 769
Query: 769 MLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSE 814
ML S T LP P + FV + S E S SH SN+ ++ E
Sbjct: 770 MLTS-TTDLPKPTQPMFVLETSD----EDSSLSHSQRSNDLSSVDE 810
>At1g61390 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 831
Score = 596 bits (1537), Expect = e-170
Identities = 334/805 (41%), Positives = 478/805 (58%), Gaps = 38/805 (4%)
Query: 33 ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYINKTNNI--WIANRDQPLKDS 90
I +S L +T++S + ++LGFFSP NS +Y+GIW+ N + W+ANRD+P+ +
Sbjct: 44 INTSSPLSIGQTLSSPDGVYELGFFSPNNSRKQYVGIWFKNIAPQVVVWVANRDKPVTKT 103
Query: 91 NGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNST-AQLADSGNLILRDISSGATIWDSF 149
+TI +G+ I+L+ VI WST + ++N A+L D+GNL++ D SG T+W SF
Sbjct: 104 AANLTISSNGSLILLDGTQDVI-WSTGEAFTSNKCHAELLDTGNLVVIDDVSGKTLWKSF 162
Query: 150 THPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASLERLDAPEVFIWKDKNIH 209
+ + +P + + GK S +S++DPS G ++ P+ I + + +
Sbjct: 163 ENLGNTMLPQSSVMYDIPRGKNRVLTSWRSNSDPSPGEFTLEFTPQVPPQGLIRRGSSPY 222
Query: 210 WRTGPWNGRVFLGSPRMLTEYLAGWRFDQDT-DGTTYITYNFADKTMFGILSLTPHGTLK 268
WR+GPW F G P + Y++ + QD GT +Y+ ++LT G +K
Sbjct: 223 WRSGPWAKTRFSGIPGIDASYVSPFTVLQDVAKGTASFSYSMLRNYKLSYVTLTSEGKMK 282
Query: 269 LIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCFDGFEPKNSVEWSLGNWT 328
++ K E + CD Y CGPFG C S P C C GF PK+ EW GNWT
Sbjct: 283 ILWNDGKSWKLHFEAPTSSCDLYRACGPFGLCVRSRNPKCICLKGFVPKSDDEWKKGNWT 342
Query: 329 NGCVRKEGMNLKCEMVKNGSSIVKQ-DGFKVYHNMKPPDFNVRTNNADQDKCGADCLANC 387
+GCVR+ L C + + K+ D F +K PD + ++C DCL NC
Sbjct: 343 SGCVRRT--QLSCHTNSSTKTQGKETDSFYHMTRVKTPDLYQLAGFLNAEQCYQDCLGNC 400
Query: 388 SCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPAELVAVKKEKGHNKSFLIIVIA 447
SC A+AY I C+ W EL+D +F + G L +R+ + +A G N++ +I+
Sbjct: 401 SCTAFAYISGIGCLVWNRELVDTVQFLSDGESLSLRLASSELA-----GSNRTKIILGTT 455
Query: 448 GVIGA-LILVICAYLLWRKCSARHK-------------AREHQQMKLDELPLYDFEKLET 493
+ +ILV AY WR + +++ A++ + + + L+D + T
Sbjct: 456 VSLSIFVILVFAAYKSWRYRTKQNEPNPMFIHSSQDAWAKDMEPQDVSGVNLFDMHTIRT 515
Query: 494 ATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAVKRLSKASGQGIEEFMNEVVVISKLQHRN 553
ATN F +N LG+GGFGPVYKG + DG+EIAVKRLS +SGQG +EFMNE+ +ISKLQH+N
Sbjct: 516 ATNNFSSSNKLGQGGFGPVYKGKLVDGKEIAVKRLSSSSGQGTDEFMNEIRLISKLQHKN 575
Query: 554 LVRLLGCCVERGEQILVYEFMPNKSLDAFLFDPLQKKNLDWRKRSNIIEGIARGIMYLHR 613
LVRLLGCC++ E++L+YE++ NKSLD FLFD K +DW+KR NII+G+ARG++YLHR
Sbjct: 576 LVRLLGCCIKGEEKLLIYEYLVNKSLDVFLFDSTLKFEIDWQKRFNIIQGVARGLLYLHR 635
Query: 614 DSRLRIIHRDLKASNILLDSDMIPKISDFGLARIVKFGEDDEANTKRVVGTYGYMPPEYA 673
DSRLR+IHRDLK SNILLD MIPKISDFGLAR+ + G + NT+RVVGT GYM PEYA
Sbjct: 636 DSRLRVIHRDLKVSNILLDEKMIPKISDFGLARMSQ-GTQYQDNTRRVVGTLGYMAPEYA 694
Query: 674 MEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSHHEDTLSLVGFAWKLWLEENIISLIDPEV 733
G+FSEKSD+YSFGVLLLEI+ G + S FS E+ +L+ +AW+ W E + L+D +
Sbjct: 695 WTGVFSEKSDIYSFGVLLLEIIIGEKISRFS--EEGKTLLAYAWESWCETKGVDLLDQAL 752
Query: 734 WDACFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLVSEITHLPPPGRVAF-VHKQSSK 792
D+ + + RC+ IGLLCVQ P DRPN ++ ML + I+ LP P + F VH +
Sbjct: 753 ADSSHPAEVGRCVQIGLLCVQHQPADRPNTLELMSMLTT-ISELPSPKQPTFTVHSRDDD 811
Query: 793 STTESSQKSHQSNSNNNVTLSEVQG 817
ST S+ + N +T S +QG
Sbjct: 812 ST------SNDLITVNEITQSVIQG 830
>At1g11280 serine/threonine kinase like protein
Length = 830
Score = 590 bits (1520), Expect = e-168
Identities = 343/834 (41%), Positives = 479/834 (57%), Gaps = 62/834 (7%)
Query: 16 FLIFCTIYSCYSAINDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYINKT 75
FL SC A IT S L +T++S ++LGFFSP NS N+Y+GIW+ T
Sbjct: 26 FLWLSLFLSCGYA---AITISSPLTLGQTLSSPGGFYELGFFSPNNSQNQYVGIWFKKIT 82
Query: 76 NNI--WIANRDQPLKDSNGIVTIHKDGNFIILNKPNGVIIWSTNISSSTNST-AQLADSG 132
+ W+ANR++P+ +TI ++G+ I+L+ V+ WST S +N A+L D+G
Sbjct: 83 PRVVVWVANREKPITTPVANLTISRNGSLILLDSSKNVV-WSTRRPSISNKCHAKLLDTG 141
Query: 133 NLILRDISSGATIWDSFTHPADAAVPTMRIAANQVTGKKISFVSRKSDNDPSSGHYSASL 192
NL++ D S +W SF +P D +P + N TG+K S KS DPS G + L
Sbjct: 142 NLVIVDDVSENLLWQSFENPGDTMLPYSSLMYNLATGEKRVLSSWKSHTDPSPGDFVVRL 201
Query: 193 ERLDAPEVFIWKDKNIHWRTGPWNGRVFLGSPRMLTEYLAGWRFDQDT-DGTTYITYNFA 251
++ + +++ R+GPW F G P M Y + + QD +GT +Y
Sbjct: 202 TPQVPAQIVTMRGSSVYKRSGPWAKTGFTGVPLMDESYTSPFSLSQDVGNGTGLFSYLQR 261
Query: 252 DKTMFGILSLTPHGTLKLIEYMNKKELFRLEVDQNECDFYGKCGPFGNCDNSTVPICSCF 311
+ ++ +T G LK Y + N CD YG CGPFG C S C C
Sbjct: 262 SSELTRVI-ITSEGYLKTFRYNGTGWVLDFITPANLCDLYGACGPFGLCVTSNPTKCKCM 320
Query: 312 DGFEPKNSVEWSLGNWTNGCVRKEGMNLKCEMVKNGSSIVKQDGFKVYH---NMKPPDFN 368
GF PK EW GN T+GC+R+ L C+ N S+ + G V++ N+KPPD
Sbjct: 321 KGFVPKYKEEWKRGNMTSGCMRRT--ELSCQA--NLSTKTQGKGVDVFYRLANVKPPDLY 376
Query: 369 VRTNNADQDKCGADCLANCSCLAYAYDPSIFCMYWTGELIDLQKFPNGGVDLFVRVPA-E 427
+ D D+C CL+NCSC A+AY I C+ W ELID ++ GG L +R+ + E
Sbjct: 377 EYASFVDADQCHQGCLSNCSCSAFAYITGIGCLLWNHELIDTIRYSVGGEFLSIRLASSE 436
Query: 428 LVAVKKEKGHNKSFLIIVIAGVIGALILVICA---YLLWRKCSARHKAREH--------- 475
L ++ K +I G I I VI A Y WR ++A+++
Sbjct: 437 LAGSRRTK---------IIVGSISLSIFVILAFGSYKYWR-----YRAKQNVGPTWAFFN 482
Query: 476 ----------QQMKLDELPLYDFEKLETATNCFHFNNMLGKGGFGPVYKGVMEDGQEIAV 525
+ ++ L ++ + ATN F+ +N LG+GGFGPVYKG + D ++IAV
Sbjct: 483 NSQDSWKNGLEPQEISGLTFFEMNTIRAATNNFNVSNKLGQGGFGPVYKGTLSDKKDIAV 542
Query: 526 KRLSKASGQGIEEFMNEVVVISKLQHRNLVRLLGCCVERGEQILVYEFMPNKSLDAFLFD 585
KRLS +SGQG EEFMNE+ +ISKLQHRNLVRLLGCC++ E++L+YEF+ NKSLD FLFD
Sbjct: 543 KRLSSSSGQGTEEFMNEIKLISKLQHRNLVRLLGCCIDGEEKLLIYEFLVNKSLDTFLFD 602
Query: 586 PLQKKNLDWRKRSNIIEGIARGIMYLHRDSRLRIIHRDLKASNILLDSDMIPKISDFGLA 645
K +DW KR NII+G++RG++YLHRDS +R+IHRDLK SNILLD M PKISDFGLA
Sbjct: 603 LTLKLQIDWPKRFNIIQGVSRGLLYLHRDSCMRVIHRDLKVSNILLDDKMNPKISDFGLA 662
Query: 646 RIVKFGEDDEANTKRVVGTYGYMPPEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFSH 705
R+ + G + NT++VVGT GYM PEYA G+FSEKSD+Y+FGVLLLEI+SG++ SSF
Sbjct: 663 RMFQ-GTQHQDNTRKVVGTLGYMSPEYAWTGMFSEKSDIYAFGVLLLEIISGKKISSFCC 721
Query: 706 HEDTLSLVGFAWKLWLEENIISLIDPEVWDAC--FESSMLRCIHIGLLCVQELPRDRPNI 763
E+ +L+G AW+ WLE + L+D ++ +C E + RC+ IGLLC+Q+ DRPNI
Sbjct: 722 GEEGKTLLGHAWECWLETGGVDLLDEDISSSCSPVEVEVARCVQIGLLCIQQQAVDRPNI 781
Query: 764 STVVLMLVSEITHLPPPGRVAFVHKQSSKSTTESSQKSHQSNSNNNVTLSEVQG 817
+ VV M+ S T LP P + F + + + S K S N+VT +E+ G
Sbjct: 782 AQVVTMMTS-ATDLPRPKQPLFALQIQDQESVVSVSK-----SVNHVTQTEIYG 829
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.137 0.422
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 19,839,660
Number of Sequences: 26719
Number of extensions: 915384
Number of successful extensions: 5735
Number of sequences better than 10.0: 1009
Number of HSP's better than 10.0 without gapping: 849
Number of HSP's successfully gapped in prelim test: 160
Number of HSP's that attempted gapping in prelim test: 2180
Number of HSP's gapped (non-prelim): 1229
length of query: 817
length of database: 11,318,596
effective HSP length: 107
effective length of query: 710
effective length of database: 8,459,663
effective search space: 6006360730
effective search space used: 6006360730
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144721.7


