

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144721.6 + phase: 0 /pseudo
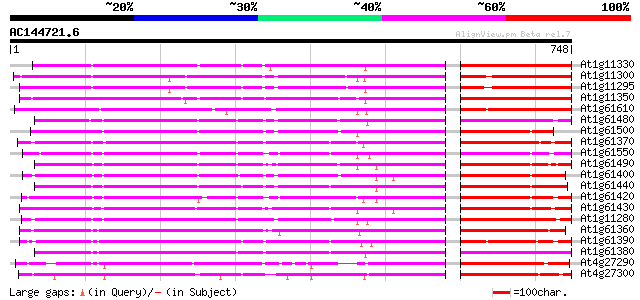
(748 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g11330 receptor-like protein kinase, putative 459 e-129
At1g11300 putative s-locus protein kinase (PPC:1.7.2) 442 e-124
At1g11295 putative s-locus protein kinase (PPC:1.7.2) 434 e-121
At1g11350 putative receptor-like protein kinase gi|4008010; simi... 419 e-117
At1g61610 406 e-113
At1g61480 S-like receptor protein kinase; member of gene cluster... 404 e-113
At1g61500 401 e-112
At1g61370 S-like receptor protein kinase; member of gene cluster... 398 e-111
At1g61550 395 e-110
At1g61490 S-like receptor protein kinase; member of gene cluster... 395 e-110
At1g61400 S-like receptor protein kinase; member of gene cluster... 392 e-109
At1g61440 S-like receptor protein kinase; member of gene cluster... 391 e-109
At1g61420 S-like receptor protein kinase; member of gene cluster... 389 e-108
At1g61430 S-like receptor protein kinase; member of gene cluster... 383 e-106
At1g11280 serine/threonine kinase like protein 373 e-103
At1g61360 S-like receptor protein kinase; member of gene cluster... 371 e-103
At1g61390 S-like receptor protein kinase; member of gene cluster... 370 e-102
At1g61380 S-like receptor protein kinase 369 e-102
At4g27290 putative receptor like kinase 343 2e-94
At4g27300 putative receptor protein kinase 328 5e-90
>At1g11330 receptor-like protein kinase, putative
Length = 840
Score = 459 bits (1180), Expect = e-129
Identities = 255/585 (43%), Positives = 349/585 (59%), Gaps = 42/585 (7%)
Query: 31 DTITSSKSLKDNE--TITSNNTNFKLGFFSPLNSTN--RYLGIWY--INETNNIWIANRD 84
D IT S +KD+E T+ + F+ GFF+P+NST RY+GIWY I +W+AN+D
Sbjct: 31 DRITFSSPIKDSESETLLCKSGIFRFGFFTPVNSTTRLRYVGIWYEKIPIQTVVWVANKD 90
Query: 85 QPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINST-AQLVDVGNLILSD-IN 142
P+ D++G+++I+++GNL + + N ++WST++S P + N+T QL+D GNL+L D N
Sbjct: 91 SPINDTSGVISIYQDGNLAVTDGRN-RLVWSTNVSVPVAPNATWVQLMDSGNLMLQDNRN 149
Query: 143 SRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLERLDAPEV 202
+ +W+SF HP D+ +P M + ++ TG N+ S S +DPS+G+Y + PE+
Sbjct: 150 NGEILWESFKHPYDSFMPRMTLGTDGRTGGNLKLTSWTSHDDPSTGNYTAGIAPFTFPEL 209
Query: 203 FIWYDKRIHWRTGPWNGTVFLGSPRM-LTEYLAGWRFDQDKDGTTYLTYDFAVKAMFGIL 261
IW + WR+GPWNG VF+G P M +L G+ + D GT ++Y A +
Sbjct: 210 LIWKNNVPTWRSGPWNGQVFIGLPNMDSLLFLDGFNLNSDNQGTISMSY--ANDSFMYHF 267
Query: 262 SLTPNGTLKLVEFLNNKEFLSLTVS--QNECDFYGKCGPFGNCDISSVPNICSCFKGFEP 319
+L P G + ++ + + V +CD YG+CG FG+C P C C KGF P
Sbjct: 268 NLDPEGIIYQKDWSTSMRTWRIGVKFPYTDCDAYGRCGRFGSCHAGENPP-CKCVKGFVP 326
Query: 320 KNLVEWSSRNWTNGCVRKEGMNLKCEM---VKNGSSVVKQDKFLVHPNTKPPDFAERSDV 376
KN EW+ NW+NGC+RK L+CE V NG K D FL K P AERS+
Sbjct: 327 KNNTEWNGGNWSNGCMRK--APLQCERQRNVSNGGGGGKADGFLKLQKMKVPISAERSEA 384
Query: 377 SRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVPAELEKEKGN 436
S C CL NCSC AYAYD I CM WS +L+D+Q F SG+DLFIRV K N
Sbjct: 385 SEQVCPKVCLDNCSCTAYAYDRGIGCMLWSGDLVDMQSFLGSGIDLFIRVAHSELKTHSN 444
Query: 437 KSFLIIAIAGGLGAFILVICAYLLWRKWSARHTEQK---------------------EMK 475
+ +I A G+ I +C L RK+ R + + ++K
Sbjct: 445 LAVMIAAPVIGV-MLIAAVCVLLACRKYKKRPAKDRSAELMFKRMEALTSDNESASNQIK 503
Query: 476 LDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEF 535
L ELPL++F L +T+SF N LG+GGFGPVYKG L +GQE+AVKRLS+ SGQG+EE
Sbjct: 504 LKELPLFEFQVLATSTDSFSLRNKLGQGGFGPVYKGKLPEGQEIAVKRLSRKSGQGLEEL 563
Query: 536 MNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
MNEV VISKLQHRNLV+LLGCC+E E+MLVYE+MP KSLDA+LF
Sbjct: 564 MNEVVVISKLQHRNLVKLLGCCIEGEERMLVYEYMPKKSLDAYLF 608
Score = 155 bits (393), Expect = 6e-38
Identities = 76/147 (51%), Positives = 102/147 (68%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAMEG FSEKSDV+S GV+ LEI+SGRRNSS + E++L+L+ +AWKLW + SL
Sbjct: 693 PEYAMEGFFSEKSDVFSLGVIFLEIISGRRNSSSHKEENNLNLLAYAWKLWNDGEAASLA 752
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D V+D FE + +C+HIGLLCVQE+ DRPN+S V+ ML +E L P + AF+ ++
Sbjct: 753 DPAVFDKCFEKEIEKCVHIGLLCVQEVANDRPNVSNVIWMLTTENMSLADPKQPAFIVRR 812
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
+ ESS +S Q S N V+L+ V G
Sbjct: 813 GASEAESSDQSSQKVSINDVSLTAVTG 839
>At1g11300 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 442 bits (1136), Expect = e-124
Identities = 246/603 (40%), Positives = 357/603 (58%), Gaps = 37/603 (6%)
Query: 5 IHNTNHFFIITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTN 64
+H ++ F+ ++ S F+ S + S L D+ETI S+ F+ GFFSP+NST+
Sbjct: 3 LHESSSPFVC-ILVLSCFFLSVSLAQERAFFSGKLNDSETIVSSFRTFRFGFFSPVNSTS 61
Query: 65 RYLGIWY--INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPN 122
RY GIWY ++ IW+AN+D+P+ DS+G++++ ++GNLV+ + + ++WST++S+
Sbjct: 62 RYAGIWYNSVSVQTVIWVANKDKPINDSSGVISVSQDGNLVVTDGQR-RVLWSTNVSTQA 120
Query: 123 SINST-AQLVDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASN-KATGKNISFVSRK 180
S NST A+L+D GNL+L + +S + +W+SF +P D+ +P M + +N + G N++ S K
Sbjct: 121 SANSTVAELLDSGNLVLKEASSDAYLWESFKYPTDSWLPNMLVGTNARIGGGNVTITSWK 180
Query: 181 SENDPSSGHYIGSLERLDAPEVFIWYDKRIH---WRTGPWNGTVFLGSPRMLTEYLAGWR 237
S +DPS G Y +L PE+FI + + WR+GPWNG +F G P + +R
Sbjct: 181 SPSDPSPGSYTAALVLAAYPELFIMNNNNNNSTVWRSGPWNGQMFNGLPDVYAGVFL-YR 239
Query: 238 FDQDKDGTTYLTYDFAVKAMFGILSLTPNGTLKLVEFLNNKE--FLSLTVSQNECDFYGK 295
F + D +T +A + + G++ ++ + + L V ECD Y +
Sbjct: 240 FIVNDDTNGSVTMSYANDSTLRYFYMDYRGSVIRRDWSETRRNWTVGLQVPATECDNYRR 299
Query: 296 CGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVK 355
CG F C+ P +CSC +GF P+NL+EW++ NW+ GC R+ + L+CE N S
Sbjct: 300 CGEFATCNPRKNP-LCSCIRGFRPRNLIEWNNGNWSGGCTRR--VPLQCERQNNNGSA-- 354
Query: 356 QDKFLVHPNTKPPDFAERSDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKF 415
D FL K PDFA RS+ S +C CL CSC+A A+ CM W+ L+D Q+
Sbjct: 355 -DGFLRLRRMKLPDFARRSEASEPECLRTCLQTCSCIAAAHGLGYGCMIWNGSLVDSQEL 413
Query: 416 PTSGVDLFIRVPAELEKEKGNKSFLIIAIAGGLGAFILVICAYLLWR-------KWSARH 468
SG+DL+IR+ K K + LI I G G F++ C L R K R
Sbjct: 414 SASGLDLYIRLAHSEIKTKDKRPILIGTILAG-GIFVVAACVLLARRIVMKKRAKKKGRD 472
Query: 469 TEQ-----------KEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQ 517
EQ + KL ELPL++F L ATN+F N LG+GGFGPVYKG L++GQ
Sbjct: 473 AEQIFERVEALAGGNKGKLKELPLFEFQVLAAATNNFSLRNKLGQGGFGPVYKGKLQEGQ 532
Query: 518 EVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDA 577
E+AVKRLS++SGQG+EE +NEV VISKLQHRNLV+LLGCC+ E+MLVYEFMP KSLD
Sbjct: 533 EIAVKRLSRASGQGLEELVNEVVVISKLQHRNLVKLLGCCIAGEERMLVYEFMPKKSLDY 592
Query: 578 FLF 580
+LF
Sbjct: 593 YLF 595
Score = 151 bits (381), Expect = 1e-36
Identities = 76/147 (51%), Positives = 101/147 (68%), Gaps = 7/147 (4%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM GLFSEKSDV+S GV+LLEI+SGRRNS+ +L+ + W +W E I SL+
Sbjct: 680 PEYAMGGLFSEKSDVFSLGVILLEIISGRRNSNS-------TLLAYVWSIWNEGEINSLV 732
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D E++D FE + +CIHIGLLCVQE DRP++STV ML SEI +P P + AF+ +
Sbjct: 733 DPEIFDLLFEKEIHKCIHIGLLCVQEAANDRPSVSTVCSMLSSEIADIPEPKQPAFISRN 792
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
N ESS+ S +S N+VT+++V G
Sbjct: 793 NVPEAESSENSDLKDSINNVTITDVTG 819
>At1g11295 putative s-locus protein kinase (PPC:1.7.2)
Length = 820
Score = 434 bits (1115), Expect = e-121
Identities = 244/596 (40%), Positives = 353/596 (58%), Gaps = 38/596 (6%)
Query: 13 IITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY- 71
I+ + S F+ S ++ S +L D+ETI S+ F+ GFFSP+NSTNRY GIWY
Sbjct: 10 IVHVLSLSCFFLSVSLAHERALFSGTLNDSETIVSSFRTFRFGFFSPVNSTNRYAGIWYN 69
Query: 72 -INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINST-AQ 129
I IW+AN+D P+ DS+G+++I ++GNLV+ + + ++WST++S+ S NST A+
Sbjct: 70 SIPVQTVIWVANKDTPINDSSGVISISEDGNLVVTDGQR-RVLWSTNVSTRASANSTVAE 128
Query: 130 LVDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGK-NISFVSRKSENDPSSG 188
L++ GNL+L D N+ + +W+SF +P D+ +P M + +N TG NI+ S + +DPS G
Sbjct: 129 LLESGNLVLKDANTDAYLWESFKYPTDSWLPNMLVGTNARTGGGNITITSWTNPSDPSPG 188
Query: 189 HYIGSLERLDAPEVFIWYDKRIH---WRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGT 245
Y +L PE+FI+ + + WR+GPWNG +F G P + +RF + D
Sbjct: 189 SYTAALVLAPYPELFIFNNNDNNATVWRSGPWNGLMFNGLPDVYPGLFL-YRFKVNDDTN 247
Query: 246 TYLTYDFAVKAMFGILSLTPNGTLKLVEFLNNKEFLSL--TVSQNECDFYGKCGPFGNCD 303
T +A + L L G ++ + +L V ECD Y +CG + C+
Sbjct: 248 GSATMSYANDSTLRHLYLDYRGFAIRRDWSEARRNWTLGSQVPATECDIYSRCGQYTTCN 307
Query: 304 ISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHP 363
P+ CSC KGF P+NL+EW++ NW+ GC+RK + L+CE N S D+FL
Sbjct: 308 PRKNPH-CSCIKGFRPRNLIEWNNGNWSGGCIRK--LPLQCERQNNKGSA---DRFLKLQ 361
Query: 364 NTKPPDFAERSDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLF 423
K PDFA RS+ S +C CL +CSC+A+A+ CM W+ L+D Q SG+DL
Sbjct: 362 RMKMPDFARRSEASEPECFMTCLQSCSCIAFAHGLGYGCMIWNRSLVDSQVLSASGMDLS 421
Query: 424 IRVPAELEKEKGNKSFLI-IAIAGGLGAFILVICAYLLWRKWSARHTEQK---------- 472
IR+ K + + LI ++AGG+ F++ C L R + ++K
Sbjct: 422 IRLAHSEFKTQDRRPILIGTSLAGGI--FVVATCVLLARRIVMKKRAKKKGTDAEQIFKR 479
Query: 473 --------EMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRL 524
KL ELPL++F L AT++F SN LG+GGFGPVYKG+L +GQE+AVKRL
Sbjct: 480 VEALAGGSREKLKELPLFEFQVLATATDNFSLSNKLGQGGFGPVYKGMLLEGQEIAVKRL 539
Query: 525 SKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
S++SGQG+EE + EV VISKLQHRNLV+L GCC+ E+MLVYEFMP KSLD ++F
Sbjct: 540 SQASGQGLEELVTEVVVISKLQHRNLVKLFGCCIAGEERMLVYEFMPKKSLDFYIF 595
Score = 134 bits (338), Expect = 1e-31
Identities = 68/147 (46%), Positives = 96/147 (65%), Gaps = 7/147 (4%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM GLFSEKSDV+S GV+LLEI+SGRRNS H+ +L+ W +W E I ++
Sbjct: 680 PEYAMGGLFSEKSDVFSLGVILLEIISGRRNS---HS----TLLAHVWSIWNEGEINGMV 732
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D E++D FE + +C+HI LLCVQ+ DRP++STV +ML SE+ +P P + AF+ +
Sbjct: 733 DPEIFDQLFEKEIRKCVHIALLCVQDAANDRPSVSTVCMMLSSEVADIPEPKQPAFMPRN 792
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
E S+ S N+VT+++V G
Sbjct: 793 VGLEAEFSESIALKASINNVTITDVSG 819
>At1g11350 putative receptor-like protein kinase gi|4008010; similar
to EST gb|AI999419.1
Length = 830
Score = 419 bits (1078), Expect = e-117
Identities = 244/605 (40%), Positives = 354/605 (58%), Gaps = 51/605 (8%)
Query: 13 IITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYI 72
++T I FS C ++ D IT S +D+ET+ SN++ F+ GFFSP+NST RY GIW+
Sbjct: 8 LLTLICFS-LRLCLAT--DVITFSSEFRDSETVVSNHSTFRFGFFSPVNSTGRYAGIWFN 64
Query: 73 N--ETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINST-AQ 129
N +W+AN + P+ DS+G+V+I K GNLV+++ G + WST++ P + N+ A+
Sbjct: 65 NIPVQTVVWVANSNSPINDSSGMVSISKEGNLVVMDGR-GQVHWSTNVLVPVAANTFYAR 123
Query: 130 LVDVGNLILSDINSRST--IWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSS 187
L++ GNL+L + +W+SF HP + +PTM +A++ TG+++ S KS DPS
Sbjct: 124 LLNTGNLVLLGTTNTGDEILWESFEHPQNIYLPTMSLATDTKTGRSLKLRSWKSPFDPSP 183
Query: 188 GHYIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEY---LAGWRFDQDKDG 244
G Y L L PE+ +W D + WR+GPWNG F+G P M +Y L D G
Sbjct: 184 GRYSAGLIPLPFPELVVWKDDLLMWRSGPWNGQYFIGLPNM--DYRINLFELTLSSDNRG 241
Query: 245 TTYLTYDFAVKAMFGILSLTPNGTLKLVEF-LNNKEFLS-LTVSQNECDFYGKCGPFGNC 302
+ ++Y A + L G++ ++ + +E+ + L V +CD Y CG F +C
Sbjct: 242 SVSMSY--AGNTLLYHFLLDSEGSVFQRDWNVAIQEWKTWLKVPSTKCDTYATCGQFASC 299
Query: 303 DIS--SVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFL 360
+ S P C C +GF+P++ EW++ NWT GCVRK L+CE N K D F+
Sbjct: 300 RFNPGSTPP-CMCIRGFKPQSYAEWNNGNWTQGCVRKAP--LQCESRDNNDGSRKSDGFV 356
Query: 361 VHPNTKPPDFAERSDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGV 420
K P +RS + C CL NCSC AY++D I C+ WS L+D+Q+F +GV
Sbjct: 357 RVQKMKVPHNPQRSGANEQDCPESCLKNCSCTAYSFDRGIGCLLWSGNLMDMQEFSGTGV 416
Query: 421 DLFIRVPAELEKEKGNKSFLIIAIAGGLGAFILVICAYL-LWRKWSARHTEQK------- 472
+IR+ K++ N+S ++I + +GAF+ L LW+ A+H E+
Sbjct: 417 VFYIRLADSEFKKRTNRS-IVITVTLLVGAFLFAGTVVLALWK--IAKHREKNRNTRLLN 473
Query: 473 -----------------EMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILED 515
+ KL ELPL++F L ATN+F +N LG+GGFG VYKG L++
Sbjct: 474 ERMEALSSNDVGAILVNQYKLKELPLFEFQVLAVATNNFSITNKLGQGGFGAVYKGRLQE 533
Query: 516 GQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSL 575
G ++AVKRLS++SGQG+EEF+NEV VISKLQHRNLVRLLG C+E E+MLVYEFMP L
Sbjct: 534 GLDIAVKRLSRTSGQGVEEFVNEVVVISKLQHRNLVRLLGFCIEGEERMLVYEFMPENCL 593
Query: 576 DAFLF 580
DA+LF
Sbjct: 594 DAYLF 598
Score = 154 bits (388), Expect = 2e-37
Identities = 74/147 (50%), Positives = 108/147 (73%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAM GLFSEKSDV+S GV+LLEIVSGRRNSSFY++ + +L +AWKLW I+L+
Sbjct: 683 PEYAMGGLFSEKSDVFSLGVILLEIVSGRRNSSFYNDGQNPNLSAYAWKLWNTGEDIALV 742
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D +++ FE+ + RC+H+GLLCVQ+ DRP+++TV+ ML SE ++LP P + AF+ ++
Sbjct: 743 DPVIFEECFENEIRRCVHVGLLCVQDHANDRPSVATVIWMLSSENSNLPEPKQPAFIPRR 802
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
+ ESS +S S N+V+L+++ G
Sbjct: 803 GTSEVESSGQSDPRASINNVSLTKITG 829
>At1g61610
Length = 842
Score = 406 bits (1043), Expect = e-113
Identities = 232/616 (37%), Positives = 348/616 (55%), Gaps = 51/616 (8%)
Query: 7 NTNHFFIITFIIFSTFYSCYS-STNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNR 65
N N + T +IF S S ST+++ T + ++++ +++ S + +F+LGFF+P NST R
Sbjct: 5 NRNLTLVTTLLIFHQLCSNVSCSTSNSFTRNHTIREGDSLISEDESFELGFFTPKNSTLR 64
Query: 66 YLGIWY--INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNS 123
Y+GIWY I +W+ANR++PL D G + I +GNLVI+N +N +I WST++ P S
Sbjct: 65 YVGIWYKNIEPQTVVWVANREKPLLDHKGALKIADDGNLVIVNGQNETI-WSTNVE-PES 122
Query: 124 INSTAQLVDVGNLIL-SDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSE 182
N+ A L G+L+L SD + R W+SF +P D +P MR+ N + G+N +F+ KSE
Sbjct: 123 NNTVAVLFKTGDLVLCSDSDRRKWYWESFNNPTDTFLPGMRVRVNPSLGENRAFIPWKSE 182
Query: 183 NDPSSGHYIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRML--TEYLAGWRFDQ 240
+DPS G Y ++ + A E+ IW ++ WR+GPWN +F G P ML T Y+ G++
Sbjct: 183 SDPSPGKYSMGIDPVGALEIVIWEGEKRKWRSGPWNSAIFTGIPDMLRFTNYIYGFKLSS 242
Query: 241 --DKDGTTYLTYDFAVKAMFGILSLTPNGTLKLVEFLNNKEFLSLTVSQ----NECDFYG 294
D+DG+ Y TY + + F + P+G + +F NK+ + + Q EC+ Y
Sbjct: 243 PPDRDGSVYFTYVASDSSDFLRFWIRPDGVEE--QFRWNKDIRNWNLLQWKPSTECEKYN 300
Query: 295 KCGPFGNCDISSV--PNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSS 352
+CG + CD S CSC GFEP + +W++R+++ GC R+ +N +V
Sbjct: 301 RCGNYSVCDDSKEFDSGKCSCIDGFEPVHQDQWNNRDFSGGCQRRVPLNCNQSLVAG--- 357
Query: 353 VVKQDKFLVHPNTKPPDFAERS-DVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELID 411
++D F V K PDF + + C+ C +CSC AYA I CM W+ +LID
Sbjct: 358 --QEDGFTVLKGIKVPDFGSVVLHNNSETCKDVCARDCSCKAYALVVGIGCMIWTRDLID 415
Query: 412 LQKFPTSGVDLFIRVPAELEKEKGNKSFLIIAIAGGLGAFILVICAYLLWR--------K 463
++ F G + IR+ S L I + +GAF+L +C ++LW+
Sbjct: 416 MEHFERGGNSINIRLAGSKLGGGKENSTLWIIVFSVIGAFLLGLCIWILWKFKKSLKAFL 475
Query: 464 WSARHTEQKEM-------------------KLDELPLYDFVKLENATNSFHNSNMLGKGG 504
W + ++ +LP++ F + +AT F N LG+GG
Sbjct: 476 WKKKDITVSDIIENRDYSSSPIKVLVGDQVDTPDLPIFSFDSVASATGDFAEENKLGQGG 535
Query: 505 FGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQM 564
FG VYKG +G+E+AVKRLS S QG+EEF NE+ +I+KLQHRNLVRLLGCC+E E+M
Sbjct: 536 FGTVYKGNFSEGREIAVKRLSGKSKQGLEEFKNEILLIAKLQHRNLVRLLGCCIEDNEKM 595
Query: 565 LVYEFMPNKSLDAFLF 580
L+YE+MPNKSLD FLF
Sbjct: 596 LLYEYMPNKSLDRFLF 611
Score = 131 bits (329), Expect = 2e-30
Identities = 71/147 (48%), Positives = 92/147 (62%), Gaps = 1/147 (0%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYAMEG+FSEKSDVYSFGVL+LEIVSGR+N SF D SL+G+AW LW + +I
Sbjct: 696 PEYAMEGIFSEKSDVYSFGVLILEIVSGRKNVSF-RGTDHGSLIGYAWHLWSQGKTKEMI 754
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D V D + +RCIH+G+LC Q+ RPN+ +V+LML S+ + LPPP + F
Sbjct: 755 DPIVKDTRDVTEAMRCIHVGMLCTQDSVIHRPNMGSVLLMLESQTSQLPPPRQPTFHSFL 814
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
NS E + S N VT + + G
Sbjct: 815 NSGDIELNFDGHDVASVNDVTFTTIVG 841
>At1g61480 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 809
Score = 404 bits (1039), Expect = e-113
Identities = 232/568 (40%), Positives = 326/568 (56%), Gaps = 31/568 (5%)
Query: 33 ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INETNNIWIANRDQPLKDS 90
IT L +T++S+N ++LGFFS NS N+Y+GIW+ I +W+ANR++P+ DS
Sbjct: 26 ITRESPLSIGKTLSSSNGVYELGFFSFNNSQNQYVGIWFKGIIPRVVVWVANREKPVTDS 85
Query: 91 NGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVGNLILSDINSRSTIWDS 150
+TI NG+L++ N EN S++WS + ++ S A+L D GNL++ D NS T+W+S
Sbjct: 86 AANLTISSNGSLLLFN-ENHSVVWSIGETFASN-GSRAELTDNGNLVVIDNNSGRTLWES 143
Query: 151 FTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLERLDAPEVFIWYDKRI 210
F H D +P + N ATG+ S KS DPS G + + + +
Sbjct: 144 FEHFGDTMLPFSNLMYNLATGEKRVLTSWKSHTDPSPGDFTVQITPQVPSQACTMRGSKT 203
Query: 211 HWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTYDFAVKAMFGILSLTPNGTLK 270
+WR+GPW T F G P M Y + + QD +G+ TY F + +T G+LK
Sbjct: 204 YWRSGPWAKTRFTGIPVMDDTYTSPFSLQQDTNGSGSFTY-FERNFKLSYIMITSEGSLK 262
Query: 271 LVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNW 330
+ + L+ +N CD YG CGPFG C + SVP C CFKGF PK++ EW NW
Sbjct: 263 IFQHNGMDWELNFEAPENSCDIYGFCGPFGIC-VMSVPPKCKCFKGFVPKSIEEWKRGNW 321
Query: 331 TNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHPNTKPPDFAE-RSDVSRDKCRTDCLANC 389
T+GCVR L C+ NG +V + F N KPPDF E S V + C CL NC
Sbjct: 322 TDGCVRHT--ELHCQGNTNGKTV---NGFYHVANIKPPDFYEFASFVDAEGCYQICLHNC 376
Query: 390 SCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVPA-ELEKEKGNKSFLIIAIAGGL 448
SCLA+AY I C+ W+ +L+D +F G L IR+ + EL K NK +I+A L
Sbjct: 377 SCLAFAYINGIGCLMWNQDLMDAVQFSAGGEILSIRLASSELGGNKRNK--IIVASIVSL 434
Query: 449 GAFILVICAYLLWRKWSARHTEQKEMK----------------LDELPLYDFVKLENATN 492
F+++ A + ++ +HT ++ + L ++ ++ AT+
Sbjct: 435 SLFVILAFAAFCFLRYKVKHTVSAKISKIASKEAWNNDLEPQDVSGLKFFEMNTIQTATD 494
Query: 493 SFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVR 552
+F SN LG+GGFG VYKG L+DG+E+AVKRLS SSGQG EEFMNE+ +ISKLQH+NLVR
Sbjct: 495 NFSLSNKLGQGGFGSVYKGKLQDGKEIAVKRLSSSSGQGKEEFMNEIVLISKLQHKNLVR 554
Query: 553 LLGCCVERGEQMLVYEFMPNKSLDAFLF 580
+LGCC+E E++LVYEF+ NKSLD FLF
Sbjct: 555 ILGCCIEGEERLLVYEFLLNKSLDTFLF 582
Score = 111 bits (277), Expect = 2e-24
Identities = 59/147 (40%), Positives = 85/147 (57%), Gaps = 5/147 (3%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+YSFGV+LLEI++G + S F + +L+ +AW+ W E I L+
Sbjct: 667 PEYAWTGMFSEKSDIYSFGVILLEIITGEKISRFSYGRQGKTLLAYAWESWCESGGIDLL 726
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D++V D+ + RC+ IGLLCVQ P DRPN ++ ML + P VH +
Sbjct: 727 DKDVADSCHPLEVERCVQIGLLCVQHQPADRPNTMELLSMLTTTSDLTSPKQPTFVVHTR 786
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
+ +S SQ + N +T S + G
Sbjct: 787 DEES-----LSQGLITVNEMTQSVILG 808
>At1g61500
Length = 804
Score = 401 bits (1030), Expect = e-112
Identities = 232/567 (40%), Positives = 326/567 (56%), Gaps = 25/567 (4%)
Query: 28 STNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYINETNNI--WIANRDQ 85
S++ IT+ L +T++S N ++LGFFSP N+ ++Y+GIW+ + + W+ANR++
Sbjct: 22 SSSAVITTESPLSMGQTLSSANEVYELGFFSPNNTQDQYVGIWFKDTIPRVVVWVANREK 81
Query: 86 PLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVGNLILSDINSRS 145
P+ DS + I +G+L++LN ++G++ WS+ ++ +S A+L D GNL + D S
Sbjct: 82 PVTDSTAYLAISSSGSLLLLNGKHGTV-WSSGVTFSSS-GCRAELSDSGNLKVIDNVSER 139
Query: 146 TIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLERLDAPEVFIW 205
+W SF H D + T + N AT + S KS DPS G ++G + + F+
Sbjct: 140 ALWQSFDHLGDTLLHTSSLTYNLATAEKRVLTSWKSYTDPSPGDFLGQITPQVPSQGFVM 199
Query: 206 YDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTYDFAVKAMFGILSLTP 265
+WR+GPW T F G P M Y + QD +G+ YLTY F ++LT
Sbjct: 200 RGSTPYWRSGPWAKTRFTGIPFMDESYTGPFTLHQDVNGSGYLTY-FQRDYKLSRITLTS 258
Query: 266 NGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEW 325
G++K+ L + CDFYG CGPFG C +S P +C CF+GF PK++ EW
Sbjct: 259 EGSIKMFRDNGMGWELYYEAPKKLCDFYGACGPFGLCVMSPSP-MCKCFRGFVPKSVEEW 317
Query: 326 SSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHPNTKPPDFAE-RSDVSRDKCRTD 384
NWT GCVR L C G D F N KPPDF E S V+ ++C
Sbjct: 318 KRGNWTGGCVRHT--ELDCLGNSTGEDA---DDFHQIANIKPPDFYEFASSVNAEECHQR 372
Query: 385 CLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVP-AELEKEKGNKSFLIIA 443
C+ NCSCLA+AY I C+ W+ +L+D +F +G L IR+ +EL+ K K+ I+A
Sbjct: 373 CVHNCSCLAFAYIKGIGCLVWNQDLMDAVQFSATGELLSIRLARSELDGNKRKKT--IVA 430
Query: 444 IAGGLGAF-ILVICAYLLWR---------KWSARHTEQKEMKLDELPLYDFVKLENATNS 493
L F IL A+ +WR A + K + L +D ++NATN+
Sbjct: 431 SIVSLTLFMILGFTAFGVWRCRVEHIAHISKDAWKNDLKPQDVPGLDFFDMHTIQNATNN 490
Query: 494 FHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRL 553
F SN LG+GGFG VYKG L+DG+E+AVKRLS SSGQG EEFMNE+ +ISKLQHRNLVR+
Sbjct: 491 FSLSNKLGQGGFGSVYKGKLQDGKEIAVKRLSSSSGQGKEEFMNEIVLISKLQHRNLVRV 550
Query: 554 LGCCVERGEQMLVYEFMPNKSLDAFLF 580
LGCC+E E++L+YEFM NKSLD FLF
Sbjct: 551 LGCCIEEEEKLLIYEFMVNKSLDTFLF 577
Score = 110 bits (274), Expect = 4e-24
Identities = 55/125 (44%), Positives = 82/125 (65%), Gaps = 2/125 (1%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+YSFGVL+LEI+SG + S F + + +L+ +AW+ W E I L+
Sbjct: 662 PEYAWTGMFSEKSDIYSFGVLMLEIISGEKISRFSYGVEGKTLIAYAWESWSEYRGIDLL 721
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFV-HK 720
D+++ D+ + RCI IGLLCVQ P DRPN + +L +++ + LP P + F H
Sbjct: 722 DQDLADSCHPLEVGRCIQIGLLCVQHQPADRPN-TLELLAMLTTTSDLPSPKQPTFAFHT 780
Query: 721 KNSKS 725
++ +S
Sbjct: 781 RDDES 785
>At1g61370 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 814
Score = 398 bits (1023), Expect = e-111
Identities = 228/591 (38%), Positives = 337/591 (56%), Gaps = 32/591 (5%)
Query: 11 FFIITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIW 70
FF + F SC + IT + L +T++S N ++LGFFSP NS N+Y+GIW
Sbjct: 8 FFASLLFLLIIFPSCAFAA---ITRASPLSIGQTLSSPNGTYELGFFSPNNSRNQYVGIW 64
Query: 71 YINETNNI--WIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSIS-SPNSINST 127
+ N T + W+ANRD+P+ ++ +TI+ NG+L+++ +E +++WS + S N +
Sbjct: 65 FKNITPRVVVWVANRDKPVTNNAANLTINSNGSLILVEREQ-NVVWSIGETFSSNELR-- 121
Query: 128 AQLVDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSS 187
A+L++ GNL+L D S +W+SF H D + + + K S K+ DPS
Sbjct: 122 AELLENGNLVLIDGVSERNLWESFEHLGDTMLLESSVMYDVPNNKKRVLSSWKNPTDPSP 181
Query: 188 GHYIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQD-KDGTT 246
G ++ L P+ FI R +WR GPW F G P M +++ + QD GT
Sbjct: 182 GEFVAELTTQVPPQGFIMRGSRPYWRGGPWARVRFTGIPEMDGSHVSKFDISQDVAAGTG 241
Query: 247 YLTYDFAVK-AMFGILSLTPNGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDIS 305
LTY + + +LT G+LK++ + L + CD Y CGPFG C I
Sbjct: 242 SLTYSLERRNSNLSYTTLTSAGSLKIIWNNGSGWVTDLEAPVSSCDVYNTCGPFGLC-IR 300
Query: 306 SVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQ-DKFLVHPN 364
S P C C KGF PK+ EW+ RNWT GC+R+ NL C++ + ++ D F + N
Sbjct: 301 SNPPKCECLKGFVPKSDEEWNKRNWTGGCMRRT--NLSCDVNSSATAQANNGDIFDIVAN 358
Query: 365 TKPPDFAER-SDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLF 423
KPPDF E S ++ + C+ CL NCSC A++Y I C+ W+ EL+D+ +F G L
Sbjct: 359 VKPPDFYEYLSLINEEDCQQRCLGNCSCTAFSYIEQIGCLVWNRELVDVMQFVAGGETLS 418
Query: 424 IRVPAELEKEKGNKSFLIIAIAGGLGAF-ILVICAYLLWRKWSARHTE------------ 470
IR+ A E N+ +I+A + F ILV +Y WR + A+ +
Sbjct: 419 IRL-ASSELAGSNRVKIIVASIVSISVFMILVFASYWYWR-YKAKQNDSNPIPLETSQDA 476
Query: 471 -QKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSG 529
++++K ++ +D + TN+F N LG+GGFGPVYKG L+DG+E+A+KRLS +SG
Sbjct: 477 WREQLKPQDVNFFDMQTILTITNNFSMENKLGQGGFGPVYKGNLQDGKEIAIKRLSSTSG 536
Query: 530 QGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
QG+EEFMNE+ +ISKLQHRNLVRLLGCC+E E++L+YEFM NKSL+ F+F
Sbjct: 537 QGLEEFMNEIILISKLQHRNLVRLLGCCIEGEEKLLIYEFMANKSLNTFIF 587
Score = 119 bits (299), Expect = 5e-27
Identities = 63/147 (42%), Positives = 93/147 (62%), Gaps = 5/147 (3%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+Y+FGVLLLEI++G+R SSF E+ +L+ FAW W E L+
Sbjct: 672 PEYAWTGMFSEKSDIYAFGVLLLEIITGKRISSFTIGEEGKTLLEFAWDSWCESGGSDLL 731
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D+++ + ES + RC+ IGLLC+Q+ DRPNI+ V+ ML + + LP P + F +
Sbjct: 732 DQDISSSGSESEVARCVQIGLLCIQQQAGDRPNIAQVMSMLTTTM-DLPKPKQPVFAMQV 790
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
ES +S+ S N++T + + G
Sbjct: 791 Q----ESDSESKTMYSVNNITQTAIVG 813
>At1g61550
Length = 802
Score = 395 bits (1016), Expect = e-110
Identities = 238/584 (40%), Positives = 333/584 (56%), Gaps = 37/584 (6%)
Query: 18 IFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INET 75
+FST +S T TS S+ +T++S N F+LGFFSP NS N Y+GIW+ I
Sbjct: 8 LFSTLLLSFSYAAITPTSPLSI--GQTLSSPNGIFELGFFSPNNSRNLYVGIWFKGIIPR 65
Query: 76 NNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVGN 135
+W+ANR+ + D+ + I NG+L++ + ++ S +WST + ++ S+A+L D GN
Sbjct: 66 TVVWVANRENSVTDATADLAISSNGSLLLFDGKH-STVWSTGETFASN-GSSAELSDSGN 123
Query: 136 LILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLE 195
L++ D S T+W SF H D +P + N TG+ S KS DP G ++G +
Sbjct: 124 LLVIDKVSGITLWQSFEHLGDTMLPYSSLMYNPGTGEKRVLSSWKSYTDPLPGEFVGYIT 183
Query: 196 RLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTYDFAVK 255
P+ FI + +WR+GPW T F G P Y + QD +G+ Y ++ ++
Sbjct: 184 TQVPPQGFIMRGSKPYWRSGPWAKTRFTGVPLTDESYTHPFSVQQDANGSVYFSH---LQ 240
Query: 256 AMF--GILSLTPNGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSC 313
F +L LT G+LK+ L++ V N CDFYG CGPFG C + S+P C C
Sbjct: 241 RNFKRSLLVLTSEGSLKVTHHNGTDWVLNIDVPANTCDFYGVCGPFGLC-VMSIPPKCKC 299
Query: 314 FKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHP--NTKPPDFA 371
FKGF P+ EW NWT GCVR+ E++ G+S + + HP N KPPDF
Sbjct: 300 FKGFVPQFSEEWKRGNWTGGCVRRT------ELLCQGNSTGRHVN-VFHPVANIKPPDFY 352
Query: 372 ER-SDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVPAEL 430
E S S ++C CL NCSCLA+AY I C+ W+ EL+D+ +F G L IR+ A
Sbjct: 353 EFVSSGSAEECYQSCLHNCSCLAFAYINGIGCLIWNQELMDVMQFSVGGELLSIRL-ASS 411
Query: 431 EKEKGNKSFLIIAIAGGLGAFI-LVICAYLLWR----------KWSARHTEQKEMKLDE- 478
E + IIA + F+ L A+ WR K S + + ++K ++
Sbjct: 412 EMGGNQRKKTIIASIVSISLFVTLASAAFGFWRYRLKHNAIVSKVSLQGAWRNDLKSEDV 471
Query: 479 --LPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFM 536
L ++ +E ATN+F N LG+GGFGPVYKG L+DG+E+AVKRLS SSGQG EEFM
Sbjct: 472 SGLYFFEMKTIEIATNNFSLVNKLGQGGFGPVYKGKLQDGKEIAVKRLSSSSGQGKEEFM 531
Query: 537 NEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
NE+ +ISKLQH NLVR+LGCC+E E++LVYEFM NKSLD F+F
Sbjct: 532 NEILLISKLQHINLVRILGCCIEGEERLLVYEFMVNKSLDTFIF 575
Score = 111 bits (277), Expect = 2e-24
Identities = 59/148 (39%), Positives = 90/148 (59%), Gaps = 7/148 (4%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD YSFGVLLLE++SG + S F ++++ +L+ +AW+ W E + +
Sbjct: 660 PEYAWTGVFSEKSDTYSFGVLLLEVISGEKISRFSYDKERKNLLAYAWESWCENGGVGFL 719
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAF-VHK 720
D++ D+ S + RC+ IGLLCVQ P DRPN + +L +++ + LP P + F VH
Sbjct: 720 DKDATDSCHPSEVGRCVQIGLLCVQHQPADRPN-TLELLSMLTTTSDLPLPKEPTFAVH- 777
Query: 721 KNSKSGESSQKSQQSNSNNSVTLSEVQG 748
+ + ++ + N VT S V G
Sbjct: 778 ----TSDDGSRTSDLITVNEVTQSVVLG 801
>At1g61490 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 804
Score = 395 bits (1014), Expect = e-110
Identities = 233/564 (41%), Positives = 325/564 (57%), Gaps = 29/564 (5%)
Query: 33 ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INETNNIWIANRDQPLKDS 90
IT+ L +T++S+N ++LGFFSP NS N Y+GIW+ I +W+ANR+ P D+
Sbjct: 26 ITTESPLSVEQTLSSSNGIYELGFFSPNNSQNLYVGIWFKGIIPRVVVWVANRETPTTDT 85
Query: 91 NGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVGNLILSDINSRSTIWDS 150
+ + I NG+L++ N ++G ++WS + ++ S A+L D GNL++ D S T+W+S
Sbjct: 86 SANLAISSNGSLLLFNGKHG-VVWSIGENFASN-GSRAELTDNGNLVVIDNASGRTLWES 143
Query: 151 FTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLERLDAPEVFIWYDKRI 210
F H D +P + N ATG+ S K++ DPS G ++G + +V I
Sbjct: 144 FEHFGDTMLPFSSLMYNLATGEKRVLTSWKTDTDPSPGVFVGQITPQVPSQVLIMRGSTR 203
Query: 211 HWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTYDFAVKAMFGILSLTPNGTLK 270
++RTGPW T F G P M Y + + QD +G+ + TY F + ++ G++K
Sbjct: 204 YYRTGPWAKTRFTGIPLMDDTYASPFSLQQDANGSGFFTY-FDRSFKLSRIIISSEGSMK 262
Query: 271 LVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNW 330
LS N CD YG CGPFG C I SVP C C KGF P + EW NW
Sbjct: 263 RFRHNGTDWELSYMAPANSCDIYGVCGPFGLC-IVSVPLKCKCLKGFVPHSTEEWKRGNW 321
Query: 331 TNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHP--NTKPPDFAE-RSDVSRDKCRTDCLA 387
T GC R L C+ G+S K D + HP N K PDF E S V ++C CL
Sbjct: 322 TGGCARLT--ELHCQ----GNSTGK-DVNIFHPVTNVKLPDFYEYESSVDAEECHQSCLH 374
Query: 388 NCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVP-AELEKEKGNKSFLIIAIAG 446
NCSCLA+AY I C+ W+ L+D +F G L IR+ +EL K NK +I+A
Sbjct: 375 NCSCLAFAYIHGIGCLIWNQNLMDAVQFSAGGEILSIRLAHSELGGNKRNK--IIVASTV 432
Query: 447 GLGAF-ILVICAYLLWR------KWSARHTEQKEMKLDELPLYDFVKL---ENATNSFHN 496
L F IL A+ WR ++ + + ++K E+P +F ++ + ATN+F
Sbjct: 433 SLSLFVILTSAAFGFWRYRVKHKAYTLKDAWRNDLKSKEVPGLEFFEMNTIQTATNNFSL 492
Query: 497 SNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGC 556
SN LG+GGFG VYKG L+DG+E+AVK+LS SSGQG EEFMNE+ +ISKLQHRNLVR+LGC
Sbjct: 493 SNKLGQGGFGSVYKGKLQDGKEIAVKQLSSSSGQGKEEFMNEIVLISKLQHRNLVRVLGC 552
Query: 557 CVERGEQMLVYEFMPNKSLDAFLF 580
C+E E++L+YEFM NKSLD F+F
Sbjct: 553 CIEGEEKLLIYEFMLNKSLDTFVF 576
Score = 111 bits (278), Expect = 1e-24
Identities = 63/147 (42%), Positives = 93/147 (62%), Gaps = 4/147 (2%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+YSFGVLLLEI+ G + S F + E+ +L+ +AW+ W E I L+
Sbjct: 661 PEYAWTGVFSEKSDIYSFGVLLLEIIIGEKISRFSYGEEGKTLLAYAWESWGETKGIDLL 720
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
D+++ D+ + RC+ IGLLCVQ P DRPN + +L +++ + LP P + FV
Sbjct: 721 DQDLADSCRPLEVGRCVQIGLLCVQHQPADRPN-TLELLAMLTTTSDLPSPKQPTFV--V 777
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
+S+ ESS S+ + N +T S + G
Sbjct: 778 HSRDDESS-LSKDLFTVNEMTQSMILG 803
>At1g61400 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 821
Score = 392 bits (1008), Expect = e-109
Identities = 230/577 (39%), Positives = 343/577 (58%), Gaps = 26/577 (4%)
Query: 17 IIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INE 74
+++ + + +SS IT L +T++S+N ++LGFFS NS N+Y+GI + I
Sbjct: 22 LLWLSIFISFSSAE--ITEESPLSIGQTLSSSNGVYELGFFSFNNSQNQYVGISFKGIIP 79
Query: 75 TNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVG 134
+W+ANR++P+ DS + I NG+L + N ++G ++WS+ + ++ S +L+D G
Sbjct: 80 RVVVWVANREKPVTDSAANLVISSNGSLQLFNGKHG-VVWSSGKALASN-GSRVELLDSG 137
Query: 135 NLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSL 194
NL++ + S T+W+SF H D +P I N TG+ S KS DPS G ++ +
Sbjct: 138 NLVVIEKVSGRTLWESFEHLGDTLLPHSTIMYNVHTGEKRGLTSWKSYTDPSPGDFVVLI 197
Query: 195 ERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTYDFAV 254
+ F+ ++R+GPW T F G P+M Y + + QD +G+ Y +Y F
Sbjct: 198 TPQVPSQGFLMRGSTPYFRSGPWAKTKFTGLPQMDESYTSPFSLTQDVNGSGYYSY-FDR 256
Query: 255 KAMFGILSLTPNGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCF 314
+ LTP+G++K + + + N CD YG CGPFG C IS VP C CF
Sbjct: 257 DNKRSRIRLTPDGSMKALRYNGMDWDTTYEGPANSCDIYGVCGPFGFCVIS-VPPKCKCF 315
Query: 315 KGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHPNTKPPDFAERS 374
KGF PK++ EW + NWT+GCVR+ L C+ G + F PN KPPDF E +
Sbjct: 316 KGFIPKSIEEWKTGNWTSGCVRRS--ELHCQGNSTGKDA---NVFHTVPNIKPPDFYEYA 370
Query: 375 D-VSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVP-AELEK 432
D V ++C+ +CL NCSCLA+AY P I C+ WS +L+D +F G L IR+ +EL+
Sbjct: 371 DSVDAEECQQNCLNNCSCLAFAYIPGIGCLMWSKDLMDTVQFAAGGELLSIRLARSELDV 430
Query: 433 EKGNKSFLIIAIAGGLGAFILV-ICAYLLWRKWSARHTE--QKEMKLDELPLYDFVKL-- 487
K K+ IIAI L F+++ A+ WR+ ++ + + +++ ++P ++ ++
Sbjct: 431 NKRKKT--IIAITVSLTLFVILGFTAFGFWRRRVEQNEDAWRNDLQTQDVPGLEYFEMNT 488
Query: 488 -ENATNSFHNSNMLGKGGFGPVYK---GILEDGQEVAVKRLSKSSGQGIEEFMNEVAVIS 543
+ ATN+F SN LG GGFG VYK G L+DG+E+AVKRLS SS QG +EFMNE+ +IS
Sbjct: 489 IQTATNNFSLSNKLGHGGFGSVYKARNGKLQDGREIAVKRLSSSSEQGKQEFMNEIVLIS 548
Query: 544 KLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
KLQHRNLVR+LGCCVE E++L+YEFM NKSLD F+F
Sbjct: 549 KLQHRNLVRVLGCCVEGTEKLLIYEFMKNKSLDTFVF 585
Score = 111 bits (278), Expect = 1e-24
Identities = 58/141 (41%), Positives = 87/141 (61%), Gaps = 2/141 (1%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+YSFGVLLLEI+SG + S F + E+ +L+ +AW+ W ++L+
Sbjct: 678 PEYAWAGVFSEKSDIYSFGVLLLEIISGEKISRFSYGEEGKTLLAYAWECWCGARGVNLL 737
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAF-VHK 720
D+ + D+ + RC+ IGLLCVQ P DRPN + +L +++ + LP P + F VH
Sbjct: 738 DQALGDSCHPYEVGRCVQIGLLCVQYQPADRPN-TLELLSMLTTTSDLPLPKQPTFVVHT 796
Query: 721 KNSKSGESSQKSQQSNSNNSV 741
++ KS + + SV
Sbjct: 797 RDGKSPSNDSMITVNEMTESV 817
>At1g61440 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 792
Score = 391 bits (1004), Expect = e-109
Identities = 222/557 (39%), Positives = 321/557 (56%), Gaps = 18/557 (3%)
Query: 33 ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INETNNIWIANRDQPLKDS 90
IT L +T++S+N ++LGFFS NS N+Y+GIW+ I +W+ANR++P+ DS
Sbjct: 19 ITKESPLSIGQTLSSSNGVYELGFFSFNNSQNQYVGIWFKGIIPRVVVWVANREKPVTDS 78
Query: 91 NGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVGNLILSDINSRSTIWDS 150
+ I +G+L+++N ++ ++WST S + S A+L D GNL++ D + T+W+S
Sbjct: 79 AANLVISSSGSLLLINGKH-DVVWSTGEISASK-GSHAELSDYGNLMVKDNVTGRTLWES 136
Query: 151 FTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLERLDAPEVFIWYDKRI 210
F H + +P + N TG+ S KS DPS G + + + F+
Sbjct: 137 FEHLGNTLLPLSTMMYNLVTGEKRGLSSWKSYTDPSPGDFWVQITPQVPSQGFVMRGSTP 196
Query: 211 HWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTYDFAVKAMFGILSLTPNGTLK 270
++RTGPW T + G P+M Y + + QD +G+ Y +Y F + LT G++K
Sbjct: 197 YYRTGPWAKTRYTGIPQMDESYTSPFSLHQDVNGSGYFSY-FERDYKLSRIMLTSEGSMK 255
Query: 271 LVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRNW 330
++ + S N CD YG CGPFG C IS P C CFKGF PK++ EW NW
Sbjct: 256 VLRYNGLDWKSSYEGPANSCDIYGVCGPFGFCVISDPPK-CKCFKGFVPKSIEEWKRGNW 314
Query: 331 TNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHPNTKPPDFAERSD-VSRDKCRTDCLANC 389
T+GC R+ L C+ G + F PN KPPDF E ++ V + C CL NC
Sbjct: 315 TSGCARRT--ELHCQGNSTGKDA---NVFHTVPNIKPPDFYEYANSVDAEGCYQSCLHNC 369
Query: 390 SCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVPAELEKEKGNKSFLIIAIAGGLG 449
SCLA+AY P I C+ WS +L+D +F G L IR+ A E + + I+A L
Sbjct: 370 SCLAFAYIPGIGCLMWSKDLMDTMQFSAGGEILSIRL-AHSELDVHKRKMTIVASTVSLT 428
Query: 450 AFILV-ICAYLLWRKWSARHTE-QKEMKLDELPLYDFVKL---ENATNSFHNSNMLGKGG 504
F+++ + WR H + +++ ++P +F ++ + AT++F SN LG GG
Sbjct: 429 LFVILGFATFGFWRNRVKHHDAWRNDLQSQDVPGLEFFEMNTIQTATSNFSLSNKLGHGG 488
Query: 505 FGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQM 564
FG VYKG L+DG+E+AVKRLS SS QG +EFMNE+ +ISKLQHRNLVR+LGCCVE E++
Sbjct: 489 FGSVYKGKLQDGREIAVKRLSSSSEQGKQEFMNEIVLISKLQHRNLVRVLGCCVEGKEKL 548
Query: 565 LVYEFMPNKSLDAFLFG 581
L+YEFM NKSLD F+FG
Sbjct: 549 LIYEFMKNKSLDTFVFG 565
Score = 114 bits (286), Expect = 2e-25
Identities = 59/143 (41%), Positives = 89/143 (61%), Gaps = 2/143 (1%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+YSFGVLLLEI+SG + S F + E+ +L+ + W+ W E ++L+
Sbjct: 649 PEYAWTGVFSEKSDIYSFGVLLLEIISGEKISRFSYGEEGKALLAYVWECWCETRGVNLL 708
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAF-VHK 720
D+ + D+S + + RC+ IGLLCVQ P DRPN + +L +++ + LP P + F VH
Sbjct: 709 DQALDDSSHPAEVGRCVQIGLLCVQHQPADRPN-TLELLSMLTTTSDLPLPKQPTFAVHT 767
Query: 721 KNSKSGESSQKSQQSNSNNSVTL 743
+N + + + SV L
Sbjct: 768 RNDEPPSNDLMITVNEMTESVIL 790
>At1g61420 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 807
Score = 389 bits (999), Expect = e-108
Identities = 234/589 (39%), Positives = 337/589 (56%), Gaps = 43/589 (7%)
Query: 16 FIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--IN 73
+++ S+F+ SS++ IT L +T++S+N ++LGFF+ NS N+Y+GIW+ I
Sbjct: 11 YLLLSSFF--ISSSSAGITKESPLPIGQTLSSSNGFYELGFFNFNNSQNQYVGIWFKGII 68
Query: 74 ETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDV 133
+W+ANR++P+ DS + I NG+L++ N ++G + WS+ + ++ S A+L D
Sbjct: 69 PRVVVWVANREKPVTDSTANLAISNNGSLLLFNGKHG-VAWSSGEALVSN-GSRAELSDT 126
Query: 134 GNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGS 193
GNLI+ D S T+W SF H D +P+ + N ATG+ S KS DPS G ++
Sbjct: 127 GNLIVIDNFSGRTLWQSFDHLGDTMLPSSTLKYNLATGEKQVLSSWKSYTDPSVGDFVLQ 186
Query: 194 LERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTY--- 250
+ +V + ++R+GPW T F G P M + QD +G+ LTY
Sbjct: 187 ITPQVPTQVLVTKGSTPYYRSGPWAKTRFTGIPLMDDTFTGPVSVQQDTNGSGSLTYLNR 246
Query: 251 -DFAVKAMFGILSLTPNGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPN 309
D + M LT GT +L L+ ++ CD+YG CGPFG C + SVP
Sbjct: 247 NDRLQRTM-----LTSKGTQELSWHNGTDWVLNFVAPEHSCDYYGVCGPFGLC-VKSVPP 300
Query: 310 ICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHP--NTKP 367
C+CFKGF PK + EW NWT GCVR+ E+ G+S K + HP KP
Sbjct: 301 KCTCFKGFVPKLIEEWKRGNWTGGCVRRT------ELYCQGNSTGKYAN-VFHPVARIKP 353
Query: 368 PDFAE-RSDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRV 426
PDF E S V+ ++C+ CL NCSCLA+AY I C+ W+ +L+D +F G L IR+
Sbjct: 354 PDFYEFASFVNVEECQKSCLHNCSCLAFAYIDGIGCLMWNQDLMDAVQFSEGGELLSIRL 413
Query: 427 P-AELEKEKGNKSFLIIAIAGGLGAFILVICAYLLWRKWSARHTE-----------QKEM 474
+EL K K+ ++ L I+ A+ WR + +H + ++
Sbjct: 414 ARSELGGNKRKKAITASIVSLSL-VVIIAFVAFCFWR-YRVKHNADITTDASQVSWRNDL 471
Query: 475 KLDELPLYDFVKL---ENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQG 531
K ++P DF + + ATN+F SN LG+GGFGPVYKG L+DG+E+AVKRLS SSGQG
Sbjct: 472 KPQDVPGLDFFDMHTIQTATNNFSISNKLGQGGFGPVYKGKLQDGKEIAVKRLSSSSGQG 531
Query: 532 IEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
EEFMNE+ +ISKLQH+NLVR+LGCC+E E++L+YEFM N SLD FLF
Sbjct: 532 KEEFMNEIVLISKLQHKNLVRILGCCIEGEEKLLIYEFMLNNSLDTFLF 580
Score = 119 bits (299), Expect = 5e-27
Identities = 62/148 (41%), Positives = 95/148 (63%), Gaps = 7/148 (4%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+YSFGVL+LEI+SG + S F + ++ +L+ +AW+ W + I L+
Sbjct: 665 PEYAWTGMFSEKSDIYSFGVLMLEIISGEKISRFSYGKEEKTLIAYAWESWCDTGGIDLL 724
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAF-VHK 720
D++V D+ + RC+ IGLLCVQ P DRPN + +L +++ + LPPP + F VH+
Sbjct: 725 DKDVADSCRPLEVERCVQIGLLCVQHQPADRPN-TLELLSMLTTTSDLPPPEQPTFVVHR 783
Query: 721 KNSKSGESSQKSQQSNSNNSVTLSEVQG 748
++ KS S+ + N +T S + G
Sbjct: 784 RDDKS-----SSEDLITVNEMTKSVILG 806
>At1g61430 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 806
Score = 383 bits (984), Expect = e-106
Identities = 228/585 (38%), Positives = 326/585 (54%), Gaps = 30/585 (5%)
Query: 14 ITFIIFSTFYSCYSSTNDT-ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY- 71
I F + F++ + S + IT +T++S+N ++LGFFS NS N+YLGIW+
Sbjct: 6 IVFFAYLPFFTIFMSFSFAGITKESPFSIGQTLSSSNGVYELGFFSLNNSQNQYLGIWFK 65
Query: 72 -INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWST-SISSPNSINSTAQ 129
I +W+ANR++P+ DS + I NG+L++ N ++G ++WST I + N S A+
Sbjct: 66 SIIPQVVVWVANREKPVTDSAANLGISSNGSLLLSNGKHG-VVWSTGDIFASN--GSRAE 122
Query: 130 LVDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGH 189
L D GNL+ D S T+W SF H + +PT + N G+ + KS DPS G
Sbjct: 123 LTDHGNLVFIDKVSGRTLWQSFEHLGNTLLPTSIMMYNLVAGEKRGLTAWKSYTDPSPGE 182
Query: 190 YIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLT 249
++ + + I ++RTGPW T F GSP+M Y + + QD +G+ Y
Sbjct: 183 FVALITPQVPSQGIIMRGSTRYYRTGPWAKTRFTGSPQMDESYTSPFILTQDVNGSGY-- 240
Query: 250 YDFAVKAMFGILSLTPNGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPN 309
+ F + + LT GT+K++ + N CD YG CGPFG C + S+P
Sbjct: 241 FSFVERGKPSRMILTSEGTMKVLVHNGMDWESTYEGPANSCDIYGVCGPFGLC-VVSIPP 299
Query: 310 ICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDKFLVHPNTKPPD 369
C CFKGF PK EW NWT+GCVR+ L C+ SS + F PN KPPD
Sbjct: 300 KCKCFKGFVPKFAKEWKKGNWTSGCVRR--TELHCQ---GNSSGKDANVFYTVPNIKPPD 354
Query: 370 FAE-RSDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVPA 428
F E + + ++C +CL NCSCLA++Y P I C+ WS +L+D ++F +G L IR+ A
Sbjct: 355 FYEYANSQNAEECHQNCLHNCSCLAFSYIPGIGCLMWSKDLMDTRQFSAAGELLSIRL-A 413
Query: 429 ELEKEKGNKSFLIIAIAGGLGAFILV-ICAYLLWR---------KWSARHTEQKEMKLDE 478
E + + I+A L F++ A+ WR A + +
Sbjct: 414 RSELDVNKRKMTIVASTVSLTLFVIFGFAAFGFWRCRVEHNAHISNDAWRNFLQSQDVPG 473
Query: 479 LPLYDFVKLENATNSFHNSNMLGKGGFGPVYK---GILEDGQEVAVKRLSKSSGQGIEEF 535
L ++ ++ ATN+F SN LG GGFG VYK G L+DG+E+AVKRLS SSGQG +EF
Sbjct: 474 LEFFEMNAIQTATNNFSLSNKLGPGGFGSVYKARNGKLQDGREIAVKRLSSSSGQGKQEF 533
Query: 536 MNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
MNE+ +ISKLQHRNLVR+LGCCVE E++L+Y F+ NKSLD F+F
Sbjct: 534 MNEIVLISKLQHRNLVRVLGCCVEGTEKLLIYGFLKNKSLDTFVF 578
Score = 121 bits (303), Expect = 2e-27
Identities = 64/148 (43%), Positives = 95/148 (63%), Gaps = 6/148 (4%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+YSFGVLLLEI+SG++ SSF + E+ +L+ +AW+ W E ++ +
Sbjct: 663 PEYAWTGVFSEKSDIYSFGVLLLEIISGKKISSFSYGEEGKALLAYAWECWCETREVNFL 722
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAF-VHK 720
D+ + D+S S + RC+ IGLLCVQ P DRPN + +L +++ + LP P K F VH
Sbjct: 723 DQALADSSHPSEVGRCVQIGLLCVQHEPADRPN-TLELLSMLTTTSDLPLPKKPTFVVHT 781
Query: 721 KNSKSGESSQKSQQSNSNNSVTLSEVQG 748
+ + S + + N +T S +QG
Sbjct: 782 RK----DESPSNDSMITVNEMTESVIQG 805
>At1g11280 serine/threonine kinase like protein
Length = 830
Score = 373 bits (958), Expect = e-103
Identities = 225/587 (38%), Positives = 317/587 (53%), Gaps = 33/587 (5%)
Query: 16 FIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYINET 75
F+ S F SC + IT S L +T++S ++LGFFSP NS N+Y+GIW+ T
Sbjct: 26 FLWLSLFLSCGYAA---ITISSPLTLGQTLSSPGGFYELGFFSPNNSQNQYVGIWFKKIT 82
Query: 76 NNI--WIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDV 133
+ W+ANR++P+ +TI +NG+L++L+ +++WST S ++ A+L+D
Sbjct: 83 PRVVVWVANREKPITTPVANLTISRNGSLILLDSSK-NVVWSTRRPSISN-KCHAKLLDT 140
Query: 134 GNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGS 193
GNL++ D S + +W SF +P D +P + N ATG+ S KS DPS G ++
Sbjct: 141 GNLVIVDDVSENLLWQSFENPGDTMLPYSSLMYNLATGEKRVLSSWKSHTDPSPGDFVVR 200
Query: 194 LERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTYDFA 253
L ++ ++ R+GPW T F G P M Y + + QD T L
Sbjct: 201 LTPQVPAQIVTMRGSSVYKRSGPWAKTGFTGVPLMDESYTSPFSLSQDVGNGTGLFSYLQ 260
Query: 254 VKAMFGILSLTPNGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSC 313
+ + +T G LK + L N CD YG CGPFG C ++S P C C
Sbjct: 261 RSSELTRVIITSEGYLKTFRYNGTGWVLDFITPANLCDLYGACGPFGLC-VTSNPTKCKC 319
Query: 314 FKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKN--GSSVVKQDKFLVHPNTKPPDFA 371
KGF PK EW N T+GC+R+ ++ + + G V D F N KPPD
Sbjct: 320 MKGFVPKYKEEWKRGNMTSGCMRRTELSCQANLSTKTQGKGV---DVFYRLANVKPPDLY 376
Query: 372 E-RSDVSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVPAEL 430
E S V D+C CL+NCSC A+AY I C+ W+ ELID ++ G L IR+ +
Sbjct: 377 EYASFVDADQCHQGCLSNCSCSAFAYITGIGCLLWNHELIDTIRYSVGGEFLSIRLASS- 435
Query: 431 EKEKGNKSFLIIAIAGGLGAF-ILVICAYLLWR---------KWSARHTEQKEMK----- 475
+ G++ II + L F IL +Y WR W+ + Q K
Sbjct: 436 -ELAGSRRTKIIVGSISLSIFVILAFGSYKYWRYRAKQNVGPTWAFFNNSQDSWKNGLEP 494
Query: 476 --LDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQGIE 533
+ L ++ + ATN+F+ SN LG+GGFGPVYKG L D +++AVKRLS SSGQG E
Sbjct: 495 QEISGLTFFEMNTIRAATNNFNVSNKLGQGGFGPVYKGTLSDKKDIAVKRLSSSSGQGTE 554
Query: 534 EFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
EFMNE+ +ISKLQHRNLVRLLGCC++ E++L+YEF+ NKSLD FLF
Sbjct: 555 EFMNEIKLISKLQHRNLVRLLGCCIDGEEKLLIYEFLVNKSLDTFLF 601
Score = 117 bits (294), Expect = 2e-26
Identities = 66/149 (44%), Positives = 92/149 (61%), Gaps = 7/149 (4%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+Y+FGVLLLEI+SG++ SSF E+ +L+G AW+ WLE + L+
Sbjct: 686 PEYAWTGMFSEKSDIYAFGVLLLEIISGKKISSFCCGEEGKTLLGHAWECWLETGGVDLL 745
Query: 662 DREVWD--ASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVH 719
D ++ + E + RC+ IGLLC+Q+ DRPNI+ VV M+ S T LP P + F
Sbjct: 746 DEDISSSCSPVEVEVARCVQIGLLCIQQQAVDRPNIAQVVTMMTS-ATDLPRPKQPLFAL 804
Query: 720 KKNSKSGESSQKSQQSNSNNSVTLSEVQG 748
+ + S S S N VT +E+ G
Sbjct: 805 QIQDQESVVS----VSKSVNHVTQTEIYG 829
>At1g61360 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 821
Score = 371 bits (952), Expect = e-103
Identities = 222/593 (37%), Positives = 320/593 (53%), Gaps = 37/593 (6%)
Query: 13 IITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYI 72
+ ++ + +S Y IT+S L T++S +++LGFFS NS N+Y+GIW+
Sbjct: 4 VACLLLITALFSSYGYA--AITTSSPLSIGVTLSSPGGSYELGFFSSNNSGNQYVGIWFK 61
Query: 73 NETNNI--WIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQL 130
T + W+ANR++P+ + +TI NG+L++L+ + ++WS+ P S A+L
Sbjct: 62 KVTPRVIVWVANREKPVSSTMANLTISSNGSLILLDSKK-DLVWSSG-GDPTSNKCRAEL 119
Query: 131 VDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHY 190
+D GNL++ D + + +W SF H D +P + + K S KSE DPS G +
Sbjct: 120 LDTGNLVVVDNVTGNYLWQSFEHLGDTMLPLTSLMYDIPNNKKRVLTSWKSETDPSPGEF 179
Query: 191 IGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKDGTTYLTY 250
+ + + I +WR+GPW GT F G P M Y+ QD+ T + +
Sbjct: 180 VAEITPQVPSQGLIRKGSSPYWRSGPWAGTRFTGIPEMDASYVNPLGMVQDEVNGTGV-F 238
Query: 251 DFAVKAMFGI--LSLTPNGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVP 308
F V F + + LTP G+L++ CD YG+CGPFG C S P
Sbjct: 239 AFCVLRNFNLSYIKLTPEGSLRITRNNGTDWIKHFEGPLTSCDLYGRCGPFGLCVRSGTP 298
Query: 309 NICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQDK----FLVHPN 364
+C C KGFEPK+ EW S NW+ GCVR+ NL C+ SSV Q K F N
Sbjct: 299 -MCQCLKGFEPKSDEEWRSGNWSRGCVRRT--NLSCQ---GNSSVETQGKDRDVFYHVSN 352
Query: 365 TKPPDFAERSDVSRDK-CRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLF 423
KPPD E + S ++ C CL NCSC A++Y I C+ W+ EL+D KF G L
Sbjct: 353 IKPPDSYELASFSNEEQCHQGCLRNCSCTAFSYVSGIGCLVWNQELLDTVKFIGGGETLS 412
Query: 424 IRVP-AELEKEKGNKSFLIIAIAGGLGAFILVICAYLLWRKW---------------SAR 467
+R+ +EL K K + ++ + ILV+ A WR A
Sbjct: 413 LRLAHSELTGRKRIKIITVATLSLSV-CLILVLVACGCWRYRVKQNGSSLVSKDNVEGAW 471
Query: 468 HTEQKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKS 527
++ + + L ++ L+ ATN+F N LG+GGFG VYKG L+DG+E+AVKRL+ S
Sbjct: 472 KSDLQSQDVSGLNFFEIHDLQTATNNFSVLNKLGQGGFGTVYKGKLQDGKEIAVKRLTSS 531
Query: 528 SGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
S QG EEFMNE+ +ISKLQHRNL+RLLGCC++ E++LVYE+M NKSLD F+F
Sbjct: 532 SVQGTEEFMNEIKLISKLQHRNLLRLLGCCIDGEEKLLVYEYMVNKSLDIFIF 584
Score = 120 bits (301), Expect = 3e-27
Identities = 67/142 (47%), Positives = 93/142 (65%), Gaps = 4/142 (2%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G FSEKSD+YSFGVL+LEI++G+ SSF + +D+ +L+ +AW W E ++L+
Sbjct: 669 PEYAWTGTFSEKSDIYSFGVLMLEIITGKEISSFSYGKDNKNLLSYAWDSWSENGGVNLL 728
Query: 662 DREVWDASFESSML--RCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVH 719
D+++ D+ +S+ RC+HIGLLCVQ DRPNI V+ ML S T LP P + FV
Sbjct: 729 DQDLDDSDSVNSVEAGRCVHIGLLCVQHQAIDRPNIKQVMSMLTS-TTDLPKPTQPMFV- 786
Query: 720 KKNSKSGESSQKSQQSNSNNSV 741
+ S S SQ+SN +SV
Sbjct: 787 LETSDEDSSLSHSQRSNDLSSV 808
>At1g61390 S-like receptor protein kinase; member of gene cluster;
similar to EST gb|T41816
Length = 831
Score = 370 bits (951), Expect = e-102
Identities = 224/589 (38%), Positives = 321/589 (54%), Gaps = 33/589 (5%)
Query: 13 IITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWYI 72
++ IIF TF Y+ N +S L +T++S + ++LGFFSP NS +Y+GIW+
Sbjct: 30 LLLLIIFPTFG--YADIN----TSSPLSIGQTLSSPDGVYELGFFSPNNSRKQYVGIWFK 83
Query: 73 NETNNI--WIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQL 130
N + W+ANRD+P+ + +TI NG+L++L+ +IWST + S A+L
Sbjct: 84 NIAPQVVVWVANRDKPVTKTAANLTISSNGSLILLDGTQ-DVIWSTG-EAFTSNKCHAEL 141
Query: 131 VDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHY 190
+D GNL++ D S T+W SF + + +P + + GKN S +S +DPS G +
Sbjct: 142 LDTGNLVVIDDVSGKTLWKSFENLGNTMLPQSSVMYDIPRGKNRVLTSWRSNSDPSPGEF 201
Query: 191 IGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFDQD-KDGTTYLT 249
P+ I +WR+GPW T F G P + Y++ + QD GT +
Sbjct: 202 TLEFTPQVPPQGLIRRGSSPYWRSGPWAKTRFSGIPGIDASYVSPFTVLQDVAKGTASFS 261
Query: 250 YDFAVKAMFGILSLTPNGTLKLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPN 309
Y ++LT G +K++ L + CD Y CGPFG C S P
Sbjct: 262 YSMLRNYKLSYVTLTSEGKMKILWNDGKSWKLHFEAPTSSCDLYRACGPFGLCVRSRNPK 321
Query: 310 ICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQ-DKFLVHPNTKPP 368
C C KGF PK+ EW NWT+GCVR+ L C + + K+ D F K P
Sbjct: 322 -CICLKGFVPKSDDEWKKGNWTSGCVRRT--QLSCHTNSSTKTQGKETDSFYHMTRVKTP 378
Query: 369 DFAERSD-VSRDKCRTDCLANCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVP 427
D + + ++ ++C DCL NCSC A+AY I C+ W+ EL+D +F + G L +R+
Sbjct: 379 DLYQLAGFLNAEQCYQDCLGNCSCTAFAYISGIGCLVWNRELVDTVQFLSDGESLSLRL- 437
Query: 428 AELEKEKGNKSFLIIAIAGGLGAF-ILVICAYLLWRKWSAR--------HTEQKEMKLDE 478
A E N++ +I+ L F ILV AY WR + + H+ Q D
Sbjct: 438 ASSELAGSNRTKIILGTTVSLSIFVILVFAAYKSWRYRTKQNEPNPMFIHSSQDAWAKDM 497
Query: 479 LP-------LYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQG 531
P L+D + ATN+F +SN LG+GGFGPVYKG L DG+E+AVKRLS SSGQG
Sbjct: 498 EPQDVSGVNLFDMHTIRTATNNFSSSNKLGQGGFGPVYKGKLVDGKEIAVKRLSSSSGQG 557
Query: 532 IEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
+EFMNE+ +ISKLQH+NLVRLLGCC++ E++L+YE++ NKSLD FLF
Sbjct: 558 TDEFMNEIRLISKLQHKNLVRLLGCCIKGEEKLLIYEYLVNKSLDVFLF 606
Score = 113 bits (283), Expect = 3e-25
Identities = 64/148 (43%), Positives = 92/148 (61%), Gaps = 9/148 (6%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA G+FSEKSD+YSFGVLLLEI+ G + S F +E+ +L+ +AW+ W E + L+
Sbjct: 691 PEYAWTGVFSEKSDIYSFGVLLLEIIIGEKISRF--SEEGKTLLAYAWESWCETKGVDLL 748
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAF-VHK 720
D+ + D+S + + RC+ IGLLCVQ P DRPN ++ ML + I+ LP P + F VH
Sbjct: 749 DQALADSSHPAEVGRCVQIGLLCVQHQPADRPNTLELMSML-TTISELPSPKQPTFTVHS 807
Query: 721 KNSKSGESSQKSQQSNSNNSVTLSEVQG 748
++ S S + N +T S +QG
Sbjct: 808 RDDDS-----TSNDLITVNEITQSVIQG 830
>At1g61380 S-like receptor protein kinase
Length = 805
Score = 369 bits (947), Expect = e-102
Identities = 216/558 (38%), Positives = 314/558 (55%), Gaps = 16/558 (2%)
Query: 33 ITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYLGIWY--INETNNIWIANRDQPLKDS 90
I +S L +T++S ++LGFFSP N+ N+Y+GIW+ I +W+ANRD P+ S
Sbjct: 23 INTSSPLSIRQTLSSPGGFYELGFFSPNNTQNQYVGIWFKKIVPRVVVWVANRDTPVTSS 82
Query: 91 NGIVTIHKNGNLVILNKENGSIIWSTSISSPNSINSTAQLVDVGNLILSDINSRSTIWDS 150
+TI NG+L++L+ + +IWST + S A+L+D GN ++ D S + +W S
Sbjct: 83 AANLTISSNGSLILLDGKQ-DVIWSTG-KAFTSNKCHAELLDTGNFVVIDDVSGNKLWQS 140
Query: 151 FTHPADAAVPTMRIASNKATGKNISFVSRKSENDPSSGHYIGSLERLDAPEVFIWYDKRI 210
F H + +P + + + GK + KS +DPS G + + + I
Sbjct: 141 FEHLGNTMLPQSSLMYDTSNGKKRVLTTWKSNSDPSPGEFSLEITPQIPTQGLIRRGSVP 200
Query: 211 HWRTGPWNGTVFLGSPRMLTEYLAGWRFDQDKD-GTTYLTYDFAVKAMFGILSLTPNGTL 269
+WR GPW T F G + Y++ + QD GT +Y ++LTP G +
Sbjct: 201 YWRCGPWAKTRFSGISGIDASYVSPFSVVQDTAAGTGSFSYSTLRNYNLSYVTLTPEGKM 260
Query: 270 KLVEFLNNKEFLSLTVSQNECDFYGKCGPFGNCDISSVPNICSCFKGFEPKNLVEWSSRN 329
K++ N L L++ +N CD YG+CGP+G C + S P C C KGF PK+ EW N
Sbjct: 261 KILWDDGNNWKLHLSLPENPCDLYGRCGPYGLC-VRSDPPKCECLKGFVPKSDEEWGKGN 319
Query: 330 WTNGCVRKEGMNLKCEMVKNGSSVVKQ-DKFLVHPNTKPPDFAE-RSDVSRDKCRTDCLA 387
WT+GCVR+ L C+ + + K D F + K PD + S ++ ++C CL
Sbjct: 320 WTSGCVRRT--KLSCQAKSSMKTQGKDTDIFYRMTDVKTPDLHQFASFLNAEQCYQGCLG 377
Query: 388 NCSCLAYAYDPFIRCMYWSSELIDLQKFPTSGVDLFIRVPAELEKEKGNKSFLIIAIAGG 447
NCSC A+AY I C+ W+ EL D +F +SG LFIR+ A E ++ +I+
Sbjct: 378 NCSCTAFAYISGIGCLVWNGELADTVQFLSSGEFLFIRL-ASSELAGSSRRKIIVGTTVS 436
Query: 448 LGAF-ILVICAYLLWRKWSARHTEQKE----MKLDELPLYDFVKLENATNSFHNSNMLGK 502
L F ILV A +LWR + ++ K + + ++ + ATN+F SN LG+
Sbjct: 437 LSIFLILVFAAIMLWRYRAKQNDAWKNGFERQDVSGVNFFEMHTIRTATNNFSPSNKLGQ 496
Query: 503 GGFGPVYKGILEDGQEVAVKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGE 562
GGFGPVYKG L DG+E+ VKRL+ SSGQG EEFMNE+ +ISKLQHRNLVRLLG C++ E
Sbjct: 497 GGFGPVYKGKLVDGKEIGVKRLASSSGQGTEEFMNEITLISKLQHRNLVRLLGYCIDGEE 556
Query: 563 QMLVYEFMPNKSLDAFLF 580
++L+YEFM NKSLD F+F
Sbjct: 557 KLLIYEFMVNKSLDIFIF 574
Score = 115 bits (289), Expect = 7e-26
Identities = 65/147 (44%), Positives = 85/147 (57%), Gaps = 1/147 (0%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA GLFSEKSD+YSFGVL+LEI+SG+R S F + ++S L+ + W W E +L+
Sbjct: 659 PEYAWAGLFSEKSDIYSFGVLMLEIISGKRISRFIYGDESKGLLAYTWDSWCETGGSNLL 718
Query: 662 DREVWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHKK 721
DR++ D + RC+ IGLLCVQ DRPN V+ ML S T LP P + F
Sbjct: 719 DRDLTDTCQAFEVARCVQIGLLCVQHEAVDRPNTLQVLSMLTS-ATDLPVPKQPIFAVHT 777
Query: 722 NSKSGESSQKSQQSNSNNSVTLSEVQG 748
+ SQ S N +T S +QG
Sbjct: 778 LNDMPMLQANSQDFLSVNEMTESMIQG 804
>At4g27290 putative receptor like kinase
Length = 772
Score = 343 bits (881), Expect = 2e-94
Identities = 209/590 (35%), Positives = 328/590 (55%), Gaps = 70/590 (11%)
Query: 8 TNHFFIITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFSPLNSTNRYL 67
TN ++ +FST + D + ++++LKD +TI S S NRYL
Sbjct: 4 TNVLHLLIISLFSTIL--LAQATDILIANQTLKDGDTIVSQG-----------GSRNRYL 50
Query: 68 GIWY--INETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSI- 124
GIWY I+ +W+ANRD PL D +G + + +NG+L + N N IIWS+S SSP+S
Sbjct: 51 GIWYKKISLQTVVWVANRDSPLYDLSGTLKVSENGSLCLFNDRN-HIIWSSS-SSPSSQK 108
Query: 125 ----NSTAQLVDVGNLILSDI-NSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSR 179
N Q++D GNL++ + + + IW S +P D +P M+ N TG N S
Sbjct: 109 ASLRNPIVQILDTGNLVVRNSGDDQDYIWQSLDYPGDMFLPGMKYGLNFVTGLNRFLTSW 168
Query: 180 KSENDPSSGHYIGSLERLDAPEVFIWYDKRIHWRTGPWNGTVFLGSPRMLTEYLAGWRFD 239
++ +DPS+G+Y ++ P+ F+ + + +RTGPWNG F G P + + + +
Sbjct: 169 RAIDDPSTGNYTNKMDPNGVPQFFLKKNSVVVFRTGPWNGLRFTGMPNLKPNPIYRYEYV 228
Query: 240 QDKDGTTYLTYDFAVKAMFGILSLTPNGTLKLVEFLNNKEFLS--LTVSQNECDFYGKCG 297
++ Y TY ++ + L PNG L+ +++N + + L+ + CD Y CG
Sbjct: 229 FTEE-EVYYTYKLENPSVLTRMQLNPNGALQRYTWVDNLQSWNFYLSAMMDSCDQYTLCG 287
Query: 298 PFGNCDISSVPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVKQD 357
+G+C+I+ P C C KGF K W + +W+ GCVR+ + L C ++G + +
Sbjct: 288 SYGSCNINESP-ACRCLKGFVAKTPQAWVAGDWSEGCVRR--VKLDCGKGEDGFLKISKL 344
Query: 358 KFLVHPNTKPPDFAERSDVSRDKCRTDCLANCSCLAYAYDPF-IR-----CMYWSSELID 411
K P+T+ + + D++ +C+ CL NC+C AY+ PF IR C+ W +LID
Sbjct: 345 KL---PDTRTSWYDKNMDLN--ECKKVCLRNCTCSAYS--PFDIRDGGKGCILWFGDLID 397
Query: 412 LQKFPTSGVDLFIRVPA-ELEKEKGNKSFLIIAIAGGLGAFILVICAYLLWRKWSARHTE 470
++++ +G DL++R+ + E+E + S + S+R E
Sbjct: 398 IREYNENGQDLYVRLASSEIETLQRESS------------------------RVSSRKQE 433
Query: 471 QKEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVAVKRLSKSSGQ 530
++++ ELP D + AT+ F N LG+GGFGPVYKG L GQEVAVKRLS++S Q
Sbjct: 434 EEDL---ELPFLDLDTVSEATSGFSAGNKLGQGGFGPVYKGTLACGQEVAVKRLSRTSRQ 490
Query: 531 GIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
G+EEF NE+ +I+KLQHRNLV++LG CV+ E+ML+YE+ PNKSLD+F+F
Sbjct: 491 GVEEFKNEIKLIAKLQHRNLVKILGYCVDEEERMLIYEYQPNKSLDSFIF 540
Score = 127 bits (318), Expect = 3e-29
Identities = 72/148 (48%), Positives = 100/148 (66%), Gaps = 6/148 (4%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEY ++G FS KSDV+SFGVL+LEIVSGRRN F + E L+L+G AW+ +LE+ +I
Sbjct: 625 PEYQIDGYFSLKSDVFSFGVLVLEIVSGRRNRGFRNEEHKLNLLGHAWRQFLEDKAYEII 684
Query: 662 DREVWDASFE-SSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPP--PGKVAFV 718
D V ++ + S +LR IHIGLLCVQ+ P+DRPN+S VVLML SE+ L P PG F
Sbjct: 685 DEAVNESCTDISEVLRVIHIGLLCVQQDPKDRPNMSVVVLMLSSEMLLLDPRQPG---FF 741
Query: 719 HKKNSKSGESSQKSQQSNSNNSVTLSEV 746
+++N ++ + + SNN T+S +
Sbjct: 742 NERNLLFSDTVSINLEIPSNNFQTMSVI 769
>At4g27300 putative receptor protein kinase
Length = 815
Score = 328 bits (842), Expect = 5e-90
Identities = 213/600 (35%), Positives = 327/600 (54%), Gaps = 52/600 (8%)
Query: 12 FIITFIIFSTFYSCYSSTNDTITSSKSLKDNETITSNNTNFKLGFFS---PLNSTNRYLG 68
F ++ + S+ S N IT + LKD +T++S + F+LGFFS +R+LG
Sbjct: 8 FSLSLFLISSSLSVALDYN-VITPKEFLKDGDTLSSPDQVFQLGFFSLDQEEQPQHRFLG 66
Query: 69 IWYINETNNIWIANRDQPLKDSNGIVTIHKNGNLVILNKENGSIIWSTSISSPNSI---- 124
+WY+ +W+ANR+ PL ++G + + G+L + + E+ ++ WS+S SS +
Sbjct: 67 LWYMEPFAVVWVANRNNPLYGTSGFLNLSSLGDLQLFDGEHKAL-WSSSSSSTKASKTAN 125
Query: 125 NSTAQLVDVGNLILSDINSRSTIWDSFTHPADAAVPTMRIASNKATGKNISFVSRKSEND 184
N ++ GNLI SD + +W SF +P + + M++ N T S S K+ D
Sbjct: 126 NPLLKISCSGNLISSD-GEEAVLWQSFDYPMNTILAGMKLGKNFKTQMEWSLSSWKTLKD 184
Query: 185 PSSGHYIGSLERLDAPEVFIWY--DKRIHWRTGPWNGTVFLGSPRMLTEY-LAGWRFDQD 241
PS G + SL+ P++ + D +R G WNG F G+P M E L ++F
Sbjct: 185 PSPGDFTLSLDTRGLPQLILRKNGDSSYSYRLGSWNGLSFTGAPAMGRENSLFDYKFTSS 244
Query: 242 KDGTTYLTYDFAVKAMFGILS-LTPNGTLKLVEFLNNKE---FLSLTVSQNECDFYGKCG 297
Y + I+S L N T KL F+ +K+ L+ T ++ECD+Y CG
Sbjct: 245 AQEVNY-----SWTPRHRIVSRLVLNNTGKLHRFIQSKQNQWILANTAPEDECDYYSICG 299
Query: 298 PFGNCDISS--VPNICSCFKGFEPKNLVEWSSRNWTNGCVRKEGMNLKCEMVKNGSSVVK 355
+ C I+S P+ CSC +GF+PK+ +W+ GCV + N CE K
Sbjct: 300 AYAVCGINSKNTPS-CSCLQGFKPKSGRKWNISRGAYGCVHEIPTN--CE---------K 347
Query: 356 QDKFLVHPNTKPPD-----FAERSDVSRDKCRTDCLANCSCLAYAYDPFIR----CMYWS 406
+D F+ P K PD + +++++ + C+ C +NCSC AYA C+ W
Sbjct: 348 KDAFVKFPGLKLPDTSWSWYDAKNEMTLEDCKIKCSSNCSCTAYANTDIREGGKGCLLWF 407
Query: 407 SELIDLQKFPTSGVDLFIRVPAELEKEKGNKSFLIIAIAGGLGAFILVICAYLLWRKWSA 466
+L+D++++ + G D++IR+ + KG + ++ + A +LV+ +K
Sbjct: 408 GDLVDMREYSSFGQDVYIRMGFAKIEFKGREVVGMVVGSVVAIAVVLVVVFACFRKKIMK 467
Query: 467 RHTEQ------KEMKLDELPLYDFVKLENATNSFHNSNMLGKGGFGPVYKGILEDGQEVA 520
R+ + +E LD LP++D + AT+ F N LG+GGFGPVYKG LEDGQE+A
Sbjct: 468 RYRGENFRKGIEEEDLD-LPIFDRKTISIATDDFSYVNFLGRGGFGPVYKGKLEDGQEIA 526
Query: 521 VKRLSKSSGQGIEEFMNEVAVISKLQHRNLVRLLGCCVERGEQMLVYEFMPNKSLDAFLF 580
VKRLS +SGQG+EEF NEV +I+KLQHRNLVRLLGCC++ E ML+YE+MPNKSLD F+F
Sbjct: 527 VKRLSANSGQGVEEFKNEVKLIAKLQHRNLVRLLGCCIQGEECMLIYEYMPNKSLDFFIF 586
Score = 120 bits (300), Expect = 4e-27
Identities = 64/148 (43%), Positives = 96/148 (64%), Gaps = 5/148 (3%)
Query: 602 PEYAMEGLFSEKSDVYSFGVLLLEIVSGRRNSSFYHNEDSLSLVGFAWKLWLEENIISLI 661
PEYA++G FS KSDV+SFGVL+LEI++G+ N F H + L+L+G WK+W+E+ I +
Sbjct: 671 PEYAIDGHFSVKSDVFSFGVLVLEIITGKTNRGFRHADHDLNLLGHVWKMWVEDREIEVP 730
Query: 662 DRE-VWDASFESSMLRCIHIGLLCVQELPRDRPNISTVVLMLISEITHLPPPGKVAFVHK 720
+ E + + S +LRCIH+ LLCVQ+ P DRP +++VVLM S+ + LP P + F
Sbjct: 731 EEEWLEETSVIPEVLRCIHVALLCVQQKPEDRPTMASVVLMFGSD-SSLPHPTQPGFFTN 789
Query: 721 KNSKSGESSQKSQQSNSNNSVTLSEVQG 748
+N SS + S N V+++ +QG
Sbjct: 790 RNVPDISSSLSLR---SQNEVSITMLQG 814
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.323 0.138 0.430
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 18,053,188
Number of Sequences: 26719
Number of extensions: 818623
Number of successful extensions: 4513
Number of sequences better than 10.0: 776
Number of HSP's better than 10.0 without gapping: 687
Number of HSP's successfully gapped in prelim test: 89
Number of HSP's that attempted gapping in prelim test: 2027
Number of HSP's gapped (non-prelim): 1597
length of query: 748
length of database: 11,318,596
effective HSP length: 107
effective length of query: 641
effective length of database: 8,459,663
effective search space: 5422643983
effective search space used: 5422643983
T: 11
A: 40
X1: 16 ( 7.5 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (22.0 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144721.6


