

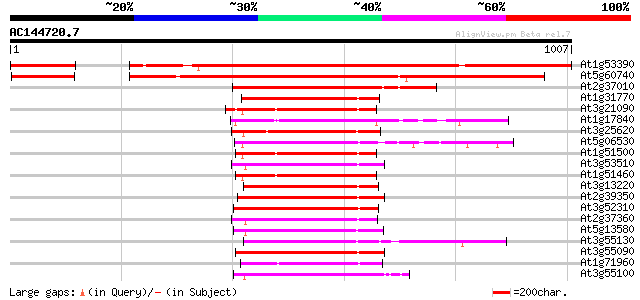
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.7 + phase: 2 /pseudo/partial
(1007 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g53390 putative ABC transporter gb|AAD31586.1 987 0.0
At5g60740 xABC transporter - like protein 981 0.0
At2g37010 putative ABC transporter 507 e-143
At1g31770 unknown protein 207 2e-53
At3g21090 ABC transporter, putative 205 1e-52
At1g17840 putative ABC transporter (At1g17840) 202 8e-52
At3g25620 membrane transporter, putative 201 1e-51
At5g06530 ABC transporter like protein 201 2e-51
At1g51500 ATP-dependent transmembrane transporter, putative 197 2e-50
At3g53510 ABC transporter -like protein 191 2e-48
At1g51460 ATP-dependent transmembrane transporter like protein 191 2e-48
At3g13220 ABC transporter 191 2e-48
At2g39350 ABC transporter like protein 189 9e-48
At3g52310 ABC transporter-like protein 188 1e-47
At2g37360 ABC transporter like protein 188 1e-47
At5g13580 ABC transporter-like protein 187 2e-47
At3g55130 ABC transporter - like protein 187 3e-47
At3g55090 ABC transporter - like protein 187 3e-47
At1g71960 putative ABC transporter 187 3e-47
At3g55100 ABC transporter - like protein 185 1e-46
>At1g53390 putative ABC transporter gb|AAD31586.1
Length = 1096
Score = 987 bits (2551), Expect = 0.0
Identities = 508/798 (63%), Positives = 620/798 (77%), Gaps = 33/798 (4%)
Query: 215 CSDQVLTTRERRVAKSRESAARSARKTANAHQRWKVAKDAAKKGATGLQAQLSRKFSRKK 274
CSDQ+LTTRERR AKSRE+A + AR AH RWK A++AAKK +G++AQ++R FS K+
Sbjct: 300 CSDQILTTRERRQAKSREAAVKKAR----AHHRWKAAREAAKKHVSGIRAQITRTFSGKR 355
Query: 275 DEENLEKVKILNQETSETDVELLPHSQPSNMVASSSAVPTEKGKTPSGLMHMMHEIENDP 334
++ + K+L + S E + S S+ +SS+A EN+
Sbjct: 356 ANQDGDTNKMLGRGDSSEIDEAIDMSTCSSPASSSAA---------------QSSYENED 400
Query: 335 HV----NYNPNTGKE-TRHKSATKEKQPQTNTQIFKYAYAQLEKEKAQQQENKNLTFSGV 389
H N + G E R K T K +T +QIFKYAY ++EKEKA +QENKNLTFSG+
Sbjct: 401 HAAAGSNGRASLGIEGKRVKGQTLAKIKKTQSQIFKYAYDRIEKEKAMEQENKNLTFSGI 460
Query: 390 LKMATNTEKSKRPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTF 449
+KMATN+E KR +E+SF+DLTLTLK+ K +LR VTG +KPGRITA+MGPSGAGKT+
Sbjct: 461 VKMATNSETRKRHLMELSFKDLTLTLKSNGKQVLRCVTGSMKPGRITAVMGPSGAGKTSL 520
Query: 450 LSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRL 509
LSALAGKA+GC ++G ILING+ ESIHS+KKIIGFVPQDDVVHGNLTVEENLWF A+CRL
Sbjct: 521 LSALAGKAVGCKLSGLILINGKQESIHSYKKIIGFVPQDDVVHGNLTVEENLWFHAKCRL 580
Query: 510 SADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLM 569
ADLSK +KVLVVER+I+ LGLQ+VR+S+VGTVEKRG+SGGQRKRVNVGLEMVMEPS+L
Sbjct: 581 PADLSKADKVLVVERIIDSLGLQAVRSSLVGTVEKRGISGGQRKRVNVGLEMVMEPSVLF 640
Query: 570 LDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYH 629
LDEPTSGLDSASSQLLLRALR EALEGVNICMVVHQPSY LF F+DL+LL KGGL VYH
Sbjct: 641 LDEPTSGLDSASSQLLLRALRHEALEGVNICMVVHQPSYTLFKTFNDLVLLAKGGLTVYH 700
Query: 630 GSAKKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPI 689
GS KVEEYFSGLGI+VP+RINPPDYYID+LEG+ G+SG+ Y++LP +WMLH Y +
Sbjct: 701 GSVNKVEEYFSGLGIHVPDRINPPDYYIDVLEGVVISMGNSGIGYKELPQRWMLHKGYSV 760
Query: 690 PLDMRQHAAQFGIPQSVNSANDLGEVGKTFAGELWNDVRSNVELRGEKIRLNFLKSKDLS 749
PLDMR ++A N +TFA ELW DV+SN LR +KIR NFLKS+DLS
Sbjct: 761 PLDMRNNSAAGLETNPDLGTNSPDNAEQTFARELWRDVKSNFRLRRDKIRHNFLKSRDLS 820
Query: 750 NRKTPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSDQTFGASGYT 809
+R+TP + QYKYFL R+ KQR+REA++QA DYLILLLAGACLGS+ K+SD++FGA
Sbjct: 821 HRRTPSTWLQYKYFLGRIAKQRMREAQLQATDYLILLLAGACLGSLIKASDESFGAP--- 877
Query: 810 YTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFNTVIKPVVYLSM 869
+LLCKIAALRSFSLDKLHYWRES SGMSS A FL+KDT+D FN ++KP+VYLSM
Sbjct: 878 ------ALLCKIAALRSFSLDKLHYWRESASGMSSSACFLAKDTIDIFNILVKPLVYLSM 931
Query: 870 FYFLTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWSVLLPVVSTLIATQQK 929
FYF TNPRSTF DNYIVL+CLVYCVTGIAYAL+I +P AQL+SVLLPVV TL+ATQ K
Sbjct: 932 FYFFTNPRSTFFDNYIVLVCLVYCVTGIAYALAIFLQPSTAQLFSVLLPVVLTLVATQPK 991
Query: 930 DSKILKAIANLCYSKWALQALVIANAERYQGVWLITRCGSLLKSGYNLHDWSLCISILIL 989
+S++++ IA+L Y KWAL+A VI NA++Y GVW+ITRCGSL+KSGY+++ WSLCI IL+L
Sbjct: 992 NSELIRIIADLSYPKWALEAFVIGNAQKYYGVWMITRCGSLMKSGYDINKWSLCIMILLL 1051
Query: 990 MGVIGRAIAFFCMVTFKK 1007
+G+ R +AF M+ +K
Sbjct: 1052 VGLTTRGVAFVGMLILQK 1069
Score = 185 bits (469), Expect = 1e-46
Identities = 78/117 (66%), Positives = 94/117 (79%)
Query: 1 DSDWNQAFNFSSDLRFLSSCIKKTKGDISNRLCTAAEVKFYLNSLMEKSSSANYLKPNRN 60
D+DWN+AFNFSS+L FLSSCIKKT+G I R+CTAAE+KFY N K+++ YLKPN N
Sbjct: 80 DADWNRAFNFSSNLNFLSSCIKKTQGSIGKRICTAAEMKFYFNGFFNKTNNPGYLKPNVN 139
Query: 61 CNLTSWVSGCEPGWACSVPSGQKIDLKDSKEMPARTSNCRACCEGFFCPHGITCMIR 117
CNLTSWVSGCEPGW CSV +++DL++SK+ P R NC CCEGFFCP G+TCMIR
Sbjct: 140 CNLTSWVSGCEPGWGCSVDPTEQVDLQNSKDFPERRRNCMPCCEGFFCPRGLTCMIR 196
>At5g60740 xABC transporter - like protein
Length = 1061
Score = 981 bits (2537), Expect = 0.0
Identities = 496/757 (65%), Positives = 598/757 (78%), Gaps = 21/757 (2%)
Query: 215 CSDQVLTTRERRVAKSRESAARSARKTANAHQRWKVAKDAAKKGATGLQAQLSRKFSRKK 274
CSDQVL TRERR AKSRE A +S R + + ++WK AKD AKK AT LQ SR FSR+K
Sbjct: 313 CSDQVLATRERRQAKSREKAVQSVRDS-QSREKWKSAKDIAKKHATELQQSFSRTFSRRK 371
Query: 275 DEENLEKVKILNQETSETDVELLPHSQPSNMVASSSAVPTEKGKTPSGLMHMMHEIENDP 334
+ + ++ L+Q +D L P M+ SSS K K + L M+H+IE +P
Sbjct: 372 SMKQPDLMRGLSQAKPGSDAALPP------MLGSSSDTKKGKKKEKNKLTEMLHDIEQNP 425
Query: 335 H--VNYNPNTGKETRHKSATKEKQPQTNTQIFKYAYAQLEKEKAQQQENKNLTFSGVLKM 392
+N G + K A K K T +Q+F+YAY Q+EKEKA Q++NKNLTFSGV+ M
Sbjct: 426 EDPEGFNLEIGDKNIKKHAPKGKALHTQSQMFRYAYGQIEKEKAMQEQNKNLTFSGVISM 485
Query: 393 ATNTEKSKRPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSA 452
A + + KRP IE++F+DL++TLK +NKH++R VTGK+ PGR++A+MGPSGAGKTTFL+A
Sbjct: 486 ANDIDIRKRPMIEVAFKDLSITLKGKNKHLMRCVTGKLSPGRVSAVMGPSGAGKTTFLTA 545
Query: 453 LAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSAD 512
L GKA GC++TG IL+NG+ ESI S+KKIIGFVPQDD+VHGNLTVEENLWFSA+CRL AD
Sbjct: 546 LTGKAPGCIMTGMILVNGKVESIQSYKKIIGFVPQDDIVHGNLTVEENLWFSARCRLPAD 605
Query: 513 LSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDE 572
L KPEKVLVVERVIE LGLQ VR+S+VGTVEKRG+SGGQRKRVNVGLEMVMEPSLL+LDE
Sbjct: 606 LPKPEKVLVVERVIESLGLQHVRDSLVGTVEKRGISGGQRKRVNVGLEMVMEPSLLILDE 665
Query: 573 PTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSA 632
PTSGLDS+SSQLLLRALRREALEGVNICMVVHQPSY LF MFDDLILL KGGL+ Y G
Sbjct: 666 PTSGLDSSSSQLLLRALRREALEGVNICMVVHQPSYTLFRMFDDLILLAKGGLICYQGPV 725
Query: 633 KKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPIPLD 692
KKVEEYFS LGI VPER+NPPDYYIDILEGI P SSG++Y+ LPV+WMLHN YP+P+D
Sbjct: 726 KKVEEYFSSLGIVVPERVNPPDYYIDILEGILKPSTSSGVTYKQLPVRWMLHNGYPVPMD 785
Query: 693 MRQHAAQFGIPQSVNSAND----------LGEVGKTFAGELWNDVRSNVELRGEKIRLNF 742
M + G+ S + N +G+ G +FAGE W DV++NVE++ + ++ NF
Sbjct: 786 MLKSIE--GMASSASGENSAHGGSAHGSVVGDDGTSFAGEFWQDVKANVEIKKDNLQNNF 843
Query: 743 LKSKDLSNRKTPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSDQT 802
S DLS R+ PGV++QY+YFL R+GKQRLREAR AVDYLILLLAG CLG++ K SD+T
Sbjct: 844 SSSGDLSEREVPGVYQQYRYFLGRLGKQRLREARTLAVDYLILLLAGICLGTLAKVSDET 903
Query: 803 FGASGYTYTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDHFNTVIK 862
FGA GYTYTVIAVSLLCKI ALRSFSLDKLHYWRES +GMSSLAYFL+KDT+DHFNT++K
Sbjct: 904 FGAMGYTYTVIAVSLLCKITALRSFSLDKLHYWRESRAGMSSLAYFLAKDTVDHFNTIVK 963
Query: 863 PVVYLSMFYFLTNPRSTFTDNYIVLLCLVYCVTGIAYALSIVFEPGAAQLWSVLLPVVST 922
P+VYLSMFYF NPRST TDNY+VL+CLVYCVTGIAY L+I+FEPG AQLWSVLLPVV T
Sbjct: 964 PLVYLSMFYFFNNPRSTVTDNYVVLICLVYCVTGIAYTLAILFEPGPAQLWSVLLPVVLT 1023
Query: 923 LIATQQKDSKILKAIANLCYSKWALQALVIANAERYQ 959
LIAT D+KI+ +I+ LCY++WAL+A V++NA+R Q
Sbjct: 1024 LIATSTNDNKIVDSISELCYTRWALEAFVVSNAQRLQ 1060
Score = 160 bits (405), Expect = 3e-39
Identities = 70/115 (60%), Positives = 85/115 (73%), Gaps = 1/115 (0%)
Query: 3 DWNQAFNFSSDLRFLSSCIKKTKGDISNRLCTAAEVKFYLNSLMEKSSSA-NYLKPNRNC 61
D+N+AFNFS+ FL++C K TKGD+ R+CTAAEV+ Y N L+ + A NYLKPN+NC
Sbjct: 79 DYNEAFNFSTKPDFLNACGKTTKGDMMQRICTAAEVRIYFNGLLGGAKRATNYLKPNKNC 138
Query: 62 NLTSWVSGCEPGWACSVPSGQKIDLKDSKEMPARTSNCRACCEGFFCPHGITCMI 116
NL+SW+SGCEPGWAC K+DLKD K +P RT C CC GFFCP GITCMI
Sbjct: 139 NLSSWMSGCEPGWACRTAKDVKVDLKDDKNVPVRTQQCAPCCAGFFCPRGITCMI 193
>At2g37010 putative ABC transporter
Length = 386
Score = 507 bits (1305), Expect = e-143
Identities = 249/366 (68%), Positives = 305/366 (83%), Gaps = 4/366 (1%)
Query: 401 RPFIEISFRDLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGC 460
RP IE++F+DLTLTLK ++KHILR+VTGKI PGR++A+MGPSGAGKTTFLSALAGKA GC
Sbjct: 4 RPVIEVAFKDLTLTLKGKHKHILRSVTGKIMPGRVSAVMGPSGAGKTTFLSALAGKATGC 63
Query: 461 LVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVL 520
TG ILINGRN+SI+S+KKI GFVPQDDVVHGNLTVEENL FSA+CRLSA +SK +KVL
Sbjct: 64 TRTGLILINGRNDSINSYKKITGFVPQDDVVHGNLTVEENLRFSARCRLSAYMSKADKVL 123
Query: 521 VVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSA 580
++ERVIE LGLQ VR+S+VGT+EKRG+SGGQRKRVNVG+EMVMEPSLL+LDEPT+GLDSA
Sbjct: 124 IIERVIESLGLQHVRDSLVGTIEKRGISGGQRKRVNVGVEMVMEPSLLILDEPTTGLDSA 183
Query: 581 SSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFS 640
SSQLLLRALRREALEGVNICMVVHQPSY ++ MFDD+I+L KGGL VYHGS KK+EEYF+
Sbjct: 184 SSQLLLRALRREALEGVNICMVVHQPSYTMYKMFDDMIILAKGGLTVYHGSVKKIEEYFA 243
Query: 641 GLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPIPLDMRQHAAQF 700
+GI VP+R+NPPD+YIDILEGI P G ++ + LPV+WMLHN YP+P DM +
Sbjct: 244 DIGITVPDRVNPPDHYIDILEGIVKPDGD--ITIEQLPVRWMLHNGYPVPHDMLKFCD-- 299
Query: 701 GIPQSVNSANDLGEVGKTFAGELWNDVRSNVELRGEKIRLNFLKSKDLSNRKTPGVFKQY 760
G+P S + +F+ +LW DV++NVE+ ++++ N+ S D SNR TP V +QY
Sbjct: 300 GLPSSSTGSAQEDSTHNSFSNDLWQDVKTNVEITKDQLQHNYSNSHDNSNRVTPTVGRQY 359
Query: 761 KYFLIR 766
+YF+ R
Sbjct: 360 RYFVGR 365
>At1g31770 unknown protein
Length = 648
Score = 207 bits (528), Expect = 2e-53
Identities = 110/248 (44%), Positives = 163/248 (65%), Gaps = 3/248 (1%)
Query: 416 KAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESI 475
K++ K IL +TG + PG A++GPSG+GKTT LSAL G+ L +G ++ NG+ S
Sbjct: 75 KSKEKTILNGITGMVCPGEFLAMLGPSGSGKTTLLSALGGR-LSKTFSGKVMYNGQPFS- 132
Query: 476 HSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVR 535
K+ GFV QDDV++ +LTV E L+F+A RL + L++ EK V+RVI LGL
Sbjct: 133 GCIKRRTGFVAQDDVLYPHLTVWETLFFTALLRLPSSLTRDEKAEHVDRVIAELGLNRCT 192
Query: 536 NSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALE 595
NS++G RG+SGG++KRV++G EM++ PSLL+LDEPTSGLDS ++ ++ ++R A
Sbjct: 193 NSMIGGPLFRGISGGEKKRVSIGQEMLINPSLLLLDEPTSGLDSTTAHRIVTTIKRLASG 252
Query: 596 GVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDY 655
G + +HQPS +++MFD ++LL +G +Y+G+A EYFS LG + +NP D
Sbjct: 253 GRTVVTTIHQPSSRIYHMFDKVVLLSEGS-PIYYGAASSAVEYFSSLGFSTSLTVNPADL 311
Query: 656 YIDILEGI 663
+D+ GI
Sbjct: 312 LLDLANGI 319
>At3g21090 ABC transporter, putative
Length = 680
Score = 205 bits (521), Expect = 1e-52
Identities = 115/276 (41%), Positives = 174/276 (62%), Gaps = 10/276 (3%)
Query: 387 SGVLKMATNTEKSKRPFIEISFRDLTLTLK----AQNKHILRNVTGKIKPGRITAIMGPS 442
SG ++ + E S+ ++ ++ DLT+ + + +L+ + G +PGRI AIMGPS
Sbjct: 8 SGRRQLPSKLEMSRGAYL--AWEDLTVVIPNFSDGPTRRLLQRLNGYAEPGRIMAIMGPS 65
Query: 443 GAGKTTFLSALAGK-ALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENL 501
G+GK+T L +LAG+ A ++TG++L+NG+ + ++ +V Q+DV+ G LTV E +
Sbjct: 66 GSGKSTLLDSLAGRLARNVVMTGNLLLNGKKARLDY--GLVAYVTQEDVLLGTLTVRETI 123
Query: 502 WFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEM 561
+SA RL +D+SK E +VE I LGLQ + V+G RGVSGG+RKRV++ LE+
Sbjct: 124 TYSAHLRLPSDMSKEEVSDIVEGTIMELGLQDCSDRVIGNWHARGVSGGERKRVSIALEI 183
Query: 562 VMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLG 621
+ P +L LDEPTSGLDSAS+ +++ALR A +G + VHQPS +F +FDDL LL
Sbjct: 184 LTRPQILFLDEPTSGLDSASAFFVIQALRNIARDGRTVISSVHQPSSEVFALFDDLFLLS 243
Query: 622 KGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYI 657
G VY G AK E+F+ G P++ NP D+++
Sbjct: 244 SGE-SVYFGEAKSAVEFFAESGFPCPKKRNPSDHFL 278
>At1g17840 putative ABC transporter (At1g17840)
Length = 703
Score = 202 bits (514), Expect = 8e-52
Identities = 158/521 (30%), Positives = 256/521 (48%), Gaps = 54/521 (10%)
Query: 397 EKSKRPFI-----EISFRDLTLTLK---AQNKHILRNVTGKIKPGRITAIMGPSGAGKTT 448
EK+ F+ ++++DLT+ + + +++L +TG +PG +TA+MGPSG+GK+T
Sbjct: 36 EKAPTEFVGDVSARLTWQDLTVMVTMGDGETQNVLEGLTGYAEPGSLTALMGPSGSGKST 95
Query: 449 FLSALAGK-ALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQC 507
L ALA + A ++G++L+NGR + SF +V QDD + G LTV E +W+SA+
Sbjct: 96 MLDALASRLAANAFLSGTVLLNGRKTKL-SFGTA-AYVTQDDNLIGTLTVRETIWYSARV 153
Query: 508 RLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSL 567
RL + + EK +VER I +GLQ ++V+G RG+SGG+++RV++ LE++M P L
Sbjct: 154 RLPDKMLRSEKRALVERTIIEMGLQDCADTVIGNWHLRGISGGEKRRVSIALEILMRPRL 213
Query: 568 LMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMV 627
L LDEPTSGLDSAS+ + + LR + +G + +HQPS +F +FD L LL GG V
Sbjct: 214 LFLDEPTSGLDSASAFFVTQTLRALSRDGRTVIASIHQPSSEVFELFDRLYLL-SGGKTV 272
Query: 628 YHGSAKKVEEYFSGLGINVPERINPPDYYI-----DILEGIAAPGGSSGLSYQ--DLPVK 680
Y G A E+F+ G P NP D+++ D + A GS L ++ D P++
Sbjct: 273 YFGQASDAYEFFAQAGFPCPALRNPSDHFLRCINSDFDKVRATLKGSMKLRFEASDDPLE 332
Query: 681 WMLHNEYPIPLDMRQHAAQFGIPQSVNSANDLGEVGKTFAGELWNDVRSNVELRGEKIRL 740
+ E L H + + +A E F G + + S
Sbjct: 333 KITTAEAIRLLVDYYHTSDY-----YYTAKAKVEEISQFKGTILDSGGSQASF------- 380
Query: 741 NFLKSKDLSNRKTPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSD 800
L++ L+ R + + + Y+ +R+ LI +L C+G+I +
Sbjct: 381 -LLQTYTLTKRSFINMSRDFGYYWLRL---------------LIYILVTVCIGTIYLNVG 424
Query: 801 QTFGA----SGYTYTVIAVSLLCKIAALRSFSLDKLHYWRESDSGMSSLAYFLSKDTMDH 856
++ A V I SF D + RE +G +A F+ +T+
Sbjct: 425 TSYSAILARGSCASFVFGFVTFMSIGGFPSFVEDMKVFQRERLNGHYGVAAFVIANTLSA 484
Query: 857 FNTVIKPVVYLS--MFYFLTNPRSTFTDNYIVLLCLVYCVT 895
+I + ++S + YF+ FT +LCL VT
Sbjct: 485 TPFLIM-ITFISGTICYFMVGLHPGFTHYLFFVLCLYASVT 524
>At3g25620 membrane transporter, putative
Length = 646
Score = 201 bits (512), Expect = 1e-51
Identities = 117/284 (41%), Positives = 178/284 (62%), Gaps = 20/284 (7%)
Query: 398 KSKRPFIEISFRDLTLTLKAQ---------------NKHILRNVTGKIKPGRITAIMGPS 442
+S RP I + F +LT ++K+Q N+ +L+ V+G +KPG + A++GPS
Sbjct: 60 QSLRPII-LKFEELTYSIKSQTGKGSYWFGSQEPKPNRLVLKCVSGIVKPGELLAMLGPS 118
Query: 443 GAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLW 502
G+GKTT ++ALAG+ G L +G++ NG + S K+ GFV QDDV++ +LTV E L
Sbjct: 119 GSGKTTLVTALAGRLQGKL-SGTVSYNGEPFT-SSVKRKTGFVTQDDVLYPHLTVMETLT 176
Query: 503 FSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMV 562
++A RL +L++ EK+ VE V+ LGL NSV+G RG+SGG+RKRV++G EM+
Sbjct: 177 YTALLRLPKELTRKEKLEQVEMVVSDLGLTRCCNSVIGGGLIRGISGGERKRVSIGQEML 236
Query: 563 MEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGK 622
+ PSLL+LDEPTSGLDS ++ ++ LR A G + +HQPS L+ MFD +++L +
Sbjct: 237 VNPSLLLLDEPTSGLDSTTAARIVATLRSLARGGRTVVTTIHQPSSRLYRMFDKVLVLSE 296
Query: 623 GGLMVYHGSAKKVEEYFSGLGINVPER-INPPDYYIDILEGIAA 665
G +Y G + +V EYF +G +NP D+ +D+ GI +
Sbjct: 297 -GCPIYSGDSGRVMEYFGSIGYQPGSSFVNPADFVLDLANGITS 339
>At5g06530 ABC transporter like protein
Length = 627
Score = 201 bits (510), Expect = 2e-51
Identities = 162/523 (30%), Positives = 255/523 (47%), Gaps = 46/523 (8%)
Query: 404 IEISFRDLTLTL------KAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKA 457
I + FRD+T + + K IL ++G + PG + A+MGPSG+GKTT LS LAG+
Sbjct: 31 IFLKFRDVTYKVVIKKLTSSVEKEILTGISGSVNPGEVLALMGPSGSGKTTLLSLLAGRI 90
Query: 458 LGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPE 517
GS+ N + S + K IGFV QDDV+ +LTV+E L ++A+ RL L++ +
Sbjct: 91 SQSSTGGSVTYNDKPYSKY-LKSKIGFVTQDDVLFPHLTVKETLTYAARLRLPKTLTREQ 149
Query: 518 KVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGL 577
K VI+ LGL+ +++++G RGVSGG+RKRV++G E+++ PSLL+LDEPTSGL
Sbjct: 150 KKQRALDVIQELGLERCQDTMIGGAFVRGVSGGERKRVSIGNEIIINPSLLLLDEPTSGL 209
Query: 578 DSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEE 637
DS ++ + L A G + +HQPS LF+ FD LILLG+G L+ Y G + + +
Sbjct: 210 DSTTALRTILMLHDIAEAGKTVITTIHQPSSRLFHRFDKLILLGRGSLL-YFGKSSEALD 268
Query: 638 YFSGLGINVPERINPPDYYIDILEGIAAPGGSSGLSYQDLPVKWMLHNEYPIPLDMRQHA 697
YFS +G + +NP ++ +D+ G + D+ V L + + R+
Sbjct: 269 YFSSIGCSPLIAMNPAEFLLDLANG----------NINDISVPSELDDRVQVGNSGRE-- 316
Query: 698 AQFGIPQSVNSANDLGEVGKTFAGE-----LWNDVRSNVELRGEKIRLNFLKSKDLSNRK 752
Q G P L E +T E L + V + E + KS L +
Sbjct: 317 TQTGKPSPAAVHEYLVEAYETRVAEQEKKKLLDPVPLDEEAKA--------KSTRLKRQW 368
Query: 753 TPGVFKQYKYFLIRVGKQRLREARIQAVDYLILLLAGACLGSITKSSD--QTFGASGYTY 810
++QY R K+R R + +L LG + SD G
Sbjct: 369 GTCWWEQYCILFCRGLKER-RHEYFSWLRVTQVLSTAVILGLLWWQSDIRTPMGLQDQAG 427
Query: 811 TVIAVSLLC----KIAALRSFSLDKLHYWRESDSGMSSL-AYFLSKDTMDHFNTVIKPVV 865
+ +++ A+ +F ++ +E + M L AYFL++ T D I P +
Sbjct: 428 LLFFIAVFWGFFPVFTAIFAFPQERAMLNKERAADMYRLSAYFLARTTSDLPLDFILPSL 487
Query: 866 YLSMFYFLT----NPRSTFTDNYIVLLCLVYCVTGIAYALSIV 904
+L + YF+T +P F V LC++ G+ A+ +
Sbjct: 488 FLLVVYFMTGLRISPYPFFLSMLTVFLCII-AAQGLGLAIGAI 529
>At1g51500 ATP-dependent transmembrane transporter, putative
Length = 687
Score = 197 bits (502), Expect = 2e-50
Identities = 110/258 (42%), Positives = 165/258 (63%), Gaps = 9/258 (3%)
Query: 406 ISFRDLTLTLK----AQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGK-ALGC 460
+++ DLT+ + + +L + G +PGRI AIMGPSG+GK+T L +LAG+ A
Sbjct: 24 LAWEDLTVVIPNFSGGPTRRLLDGLNGHAEPGRIMAIMGPSGSGKSTLLDSLAGRLARNV 83
Query: 461 LVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVL 520
++TG++L+NG+ + ++ +V Q+D++ G LTV E + +SA RLS+DL+K E
Sbjct: 84 IMTGNLLLNGKKARLDY--GLVAYVTQEDILMGTLTVRETITYSAHLRLSSDLTKEEVND 141
Query: 521 VVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSA 580
+VE I LGLQ + V+G RGVSGG+RKRV+V LE++ P +L LDEPTSGLDSA
Sbjct: 142 IVEGTIIELGLQDCADRVIGNWHSRGVSGGERKRVSVALEILTRPQILFLDEPTSGLDSA 201
Query: 581 SSQLLLRALRREALE-GVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYF 639
S+ +++ALR A + G + +HQPS +F +FDDL LL G VY G +K E+F
Sbjct: 202 SAFFVIQALRNIARDGGRTVVSSIHQPSSEVFALFDDLFLLSSGE-TVYFGESKFAVEFF 260
Query: 640 SGLGINVPERINPPDYYI 657
+ G P++ NP D+++
Sbjct: 261 AEAGFPCPKKRNPSDHFL 278
>At3g53510 ABC transporter -like protein
Length = 739
Score = 191 bits (485), Expect = 2e-48
Identities = 108/297 (36%), Positives = 173/297 (57%), Gaps = 25/297 (8%)
Query: 399 SKRPFIEISFRDLTLTLKAQNKH----------------------ILRNVTGKIKPGRIT 436
S PF+ +SF+DLT ++K + K +L ++G+ + G +
Sbjct: 82 SSSPFV-LSFKDLTYSVKIKKKFKPFPCCGNSPFDGNDMEMNTKVLLNGISGEAREGEMM 140
Query: 437 AIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLT 496
A++G SG+GK+T + ALA + + G I +NG K I +V QDD++ LT
Sbjct: 141 AVLGASGSGKSTLIDALANRISKESLRGDITLNGEVLESSLHKVISAYVMQDDLLFPMLT 200
Query: 497 VEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVN 556
VEE L FSA+ RL + LSK +K V+ +I+ LGL++ +V+G RGVSGG+R+RV+
Sbjct: 201 VEETLMFSAEFRLPSSLSKKKKKARVQALIDQLGLRNAAKTVIGDEGHRGVSGGERRRVS 260
Query: 557 VGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDD 616
+G +++ +P +L LDEPTSGLDS S+ ++++ L+R A G + M +HQPSY + + D
Sbjct: 261 IGTDIIHDPIILFLDEPTSGLDSTSAYMVVKVLQRIAQSGSIVIMSIHQPSYRILGLLDK 320
Query: 617 LILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDILEGIA-APGGSSGL 672
LI L +G VY GS + ++FS G +PE N P++ +D++ + +P G+ L
Sbjct: 321 LIFLSRGN-TVYSGSPTHLPQFFSEFGHPIPENENKPEFALDLIRELEDSPEGTKSL 376
>At1g51460 ATP-dependent transmembrane transporter like protein
Length = 678
Score = 191 bits (485), Expect = 2e-48
Identities = 105/257 (40%), Positives = 160/257 (61%), Gaps = 8/257 (3%)
Query: 406 ISFRDLTLTL----KAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCL 461
+++ DLT+ + + K +L V G +P RI AIMGPSG+GK+T L ALAG+ G +
Sbjct: 10 VAWEDLTVVIPNFGEGATKRLLNGVNGCGEPNRILAIMGPSGSGKSTLLDALAGRLAGNV 69
Query: 462 V-TGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVL 520
V +G +L+NG+ + +V Q+DV+ G LTV E++ +SA RL + L++ E
Sbjct: 70 VMSGKVLVNGKKRRLDF--GAAAYVTQEDVLLGTLTVRESISYSAHLRLPSKLTREEISD 127
Query: 521 VVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSA 580
+VE I +GL+ + +G RG+SGG++KR+++ LE++ +PSLL LDEPTSGLDSA
Sbjct: 128 IVEATITDMGLEECSDRTIGNWHLRGISGGEKKRLSIALEVLTKPSLLFLDEPTSGLDSA 187
Query: 581 SSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFS 640
S+ +++ LR A G + +HQPS +F +FDDL+LL GG VY G A+ ++F
Sbjct: 188 SAFFVVQILRNIASSGKTVVSSIHQPSGEVFALFDDLLLL-SGGETVYFGEAESATKFFG 246
Query: 641 GLGINVPERINPPDYYI 657
G P R NP D+++
Sbjct: 247 EAGFPCPSRRNPSDHFL 263
>At3g13220 ABC transporter
Length = 647
Score = 191 bits (484), Expect = 2e-48
Identities = 108/243 (44%), Positives = 154/243 (62%), Gaps = 3/243 (1%)
Query: 420 KHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFK 479
KHIL+ +TG PG I A+MGPSG+GKTT L + G+ L V G + N S S K
Sbjct: 66 KHILKGITGSTGPGEILALMGPSGSGKTTLLKIMGGR-LTDNVKGKLTYNDIPYS-PSVK 123
Query: 480 KIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVV 539
+ IGFV QDDV+ LTVEE L F+A RL + +SK +K +E +I+ LGL+ R + V
Sbjct: 124 RRIGFVTQDDVLLPQLTVEETLAFAAFLRLPSSMSKEQKYAKIEMIIKELGLERCRRTRV 183
Query: 540 GTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNI 599
G +G+SGG+RKR ++ E++++PSLL+LDEPTSGLDS S+ LL L+ A G +
Sbjct: 184 GGGFVKGISGGERKRASIAYEILVDPSLLLLDEPTSGLDSTSATKLLHILQGVAKAGRTV 243
Query: 600 CMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDI 659
+HQPS +F+MFD L+L+ +G ++G A++ EYFS L I +NP ++ +D+
Sbjct: 244 ITTIHQPSSRMFHMFDKLLLISEGH-PAFYGKARESMEYFSSLRILPEIAMNPAEFLLDL 302
Query: 660 LEG 662
G
Sbjct: 303 ATG 305
>At2g39350 ABC transporter like protein
Length = 740
Score = 189 bits (479), Expect = 9e-48
Identities = 96/264 (36%), Positives = 164/264 (61%), Gaps = 2/264 (0%)
Query: 410 DLTLTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILIN 469
++ T + + K +L N++G+ + G I A++G SG+GK+T + ALA + + G++ +N
Sbjct: 97 EIAQTARPKTKTLLNNISGETRDGEIMAVLGASGSGKSTLIDALANRIAKGSLKGTVKLN 156
Query: 470 GRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFL 529
G K I +V QDD++ LTVEE L F+A+ RL L K +K L V+ +I+ L
Sbjct: 157 GETLQSRMLKVISAYVMQDDLLFPMLTVEETLMFAAEFRLPRSLPKSKKKLRVQALIDQL 216
Query: 530 GLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRAL 589
G+++ +++G RG+SGG+R+RV++G++++ +P LL LDEPTSGLDS S+ ++++ L
Sbjct: 217 GIRNAAKTIIGDEGHRGISGGERRRVSIGIDIIHDPILLFLDEPTSGLDSTSAFMVVKVL 276
Query: 590 RREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPER 649
+R A G + M +HQPS+ + + D LI L +G VY GS + +F+ G +PE
Sbjct: 277 KRIAQSGSIVIMSIHQPSHRVLGLLDRLIFLSRGH-TVYSGSPASLPRFFTEFGSPIPEN 335
Query: 650 INPPDYYIDILEGI-AAPGGSSGL 672
N ++ +D++ + + GG+ GL
Sbjct: 336 ENRTEFALDLIRELEGSAGGTRGL 359
>At3g52310 ABC transporter-like protein
Length = 737
Score = 188 bits (478), Expect = 1e-47
Identities = 107/261 (40%), Positives = 163/261 (61%), Gaps = 3/261 (1%)
Query: 403 FIEISFRDLTLTLKAQN-KHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCL 461
FI+I+++ T + + + K IL ++G PG + A+MGPSG+GKTT L+AL G+
Sbjct: 147 FIDITYKVTTKGMTSSSEKSILNGISGSAYPGELLALMGPSGSGKTTLLNALGGRFNQQN 206
Query: 462 VTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLV 521
+ GS+ N + S H K IGFV QDDV+ +LTV+E L ++A RL L++ EK
Sbjct: 207 IGGSVSYNDKPYSKH-LKTRIGFVTQDDVLFPHLTVKETLTYTALLRLPKTLTEQEKEQR 265
Query: 522 VERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSAS 581
VI+ LGL+ +++++G RGVSGG+RKRV +G E++ PSLL+LDEPTS LDS +
Sbjct: 266 AASVIQELGLERCQDTMIGGSFVRGVSGGERKRVCIGNEIMTNPSLLLLDEPTSSLDSTT 325
Query: 582 SQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSG 641
+ +++ L A G I +HQPS LF+ FD L++L +G L+ Y G A + YFS
Sbjct: 326 ALKIVQMLHCIAKAGKTIVTTIHQPSSRLFHRFDKLVVLSRGSLL-YFGKASEAMSYFSS 384
Query: 642 LGINVPERINPPDYYIDILEG 662
+G + +NP ++ +D++ G
Sbjct: 385 IGCSPLLAMNPAEFLLDLVNG 405
Score = 35.4 bits (80), Expect = 0.16
Identities = 47/190 (24%), Positives = 87/190 (45%), Gaps = 15/190 (7%)
Query: 823 ALRSFSLDKLHYWRESDSGMSSL-AYFLSKDTMDHFNTVIKPVVYLSMFYFLTNPR---- 877
A+ +F ++ +E +S M L AYF+++ T D +I PV++L + YF+ R
Sbjct: 552 AIFTFPQERAMLSKERESNMYRLSAYFVARTTSDLPLDLILPVLFLVVVYFMAGLRLRAE 611
Query: 878 STFTDNYIVLLCLVYCV-TGIAYALSIVFEPGAAQLWSVLLPVVSTLIATQQKDSKILKA 936
S F V LC+V G+A S++ A L SV V++ ++A K+
Sbjct: 612 SFFLSVLTVFLCIVAAQGLGLAIGASLMDLKKATTLASV--TVMTFMLAGGYFVKKVPFF 669
Query: 937 IANLCYSKWALQALVIANAERYQGVWLITRCGSLLKSGYNLHDWSLCISILILMGVIGRA 996
IA + + + + +Y+ + + + G ++SG +S L+ M + R
Sbjct: 670 IAWIRFMSFNYHTYKLLVKVQYEEI-MESVNGEEIESGLKE------VSALVAMIIGYRL 722
Query: 997 IAFFCMVTFK 1006
+A+F + K
Sbjct: 723 VAYFSLRRMK 732
>At2g37360 ABC transporter like protein
Length = 755
Score = 188 bits (478), Expect = 1e-47
Identities = 107/281 (38%), Positives = 164/281 (58%), Gaps = 21/281 (7%)
Query: 399 SKRPFIEISFRDLTLTLKAQNKH-------------------ILRNVTGKIKPGRITAIM 439
S PF+ +SF DLT ++K Q K +L ++G+ + G + A++
Sbjct: 92 SSSPFV-LSFTDLTYSVKIQKKFNPLACCRRSGNDSSVNTKILLNGISGEAREGEMMAVL 150
Query: 440 GPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEE 499
G SG+GK+T + ALA + + GSI +NG K I +V QDD++ LTVEE
Sbjct: 151 GASGSGKSTLIDALANRIAKDSLRGSITLNGEVLESSMQKVISAYVMQDDLLFPMLTVEE 210
Query: 500 NLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGL 559
L FSA+ RL LSK +K V+ +I+ LGL+S +V+G RGVSGG+R+RV++G
Sbjct: 211 TLMFSAEFRLPRSLSKKKKKARVQALIDQLGLRSAAKTVIGDEGHRGVSGGERRRVSIGN 270
Query: 560 EMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLIL 619
+++ +P +L LDEPTSGLDS S+ ++++ L+R A G + M +HQPSY + + D LI
Sbjct: 271 DIIHDPIILFLDEPTSGLDSTSAYMVIKVLQRIAQSGSIVIMSIHQPSYRIMGLLDQLIF 330
Query: 620 LGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDIL 660
L KG VY GS + ++FS +PE N ++ +D++
Sbjct: 331 LSKGN-TVYSGSPTHLPQFFSEFKHPIPENENKTEFALDLI 370
>At5g13580 ABC transporter-like protein
Length = 727
Score = 187 bits (476), Expect = 2e-47
Identities = 104/293 (35%), Positives = 169/293 (57%), Gaps = 26/293 (8%)
Query: 402 PFIEISFRDLTLTLKAQNKH------------------------ILRNVTGKIKPGRITA 437
PF+ +SF DLT ++K + K +L +TG+ + G I A
Sbjct: 65 PFV-LSFTDLTYSVKVRRKFTWRRSVSSDPGAPSEGIFSSKTKTLLNGITGEARDGEILA 123
Query: 438 IMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTV 497
++G SG+GK+T + ALA + + G++ +NG + K I +V QDD++ LTV
Sbjct: 124 VLGASGSGKSTLIDALANRIAKGSLKGNVTLNGEVLNSKMQKAISAYVMQDDLLFPMLTV 183
Query: 498 EENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNV 557
EE L F+A+ RL LSK +K L V+ +I+ LGL++ N+V+G RG+SGG+R+RV++
Sbjct: 184 EETLMFAAEFRLPRSLSKSKKSLRVQALIDQLGLRNAANTVIGDEGHRGISGGERRRVSI 243
Query: 558 GLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDL 617
G++++ +P LL LDEPTSGLDS S+ +++ L+R A G + M +HQPSY L + D L
Sbjct: 244 GIDIIHDPILLFLDEPTSGLDSTSALSVIKVLKRIAQSGSMVIMTLHQPSYRLLRLLDRL 303
Query: 618 ILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYIDILEGIAAPGGSS 670
+ L + G V+ GS + +F+ G +PE N ++ +D++ + G +
Sbjct: 304 LFLSR-GQTVFSGSPAMLPRFFAEFGHPIPEHENRTEFALDLIRELEGSAGGT 355
>At3g55130 ABC transporter - like protein
Length = 725
Score = 187 bits (475), Expect = 3e-47
Identities = 133/479 (27%), Positives = 236/479 (48%), Gaps = 24/479 (5%)
Query: 420 KHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNESIHSFK 479
K +L +V+G+ G I A++G SGAGK+T + ALAG+ + GS+ +NG
Sbjct: 97 KTLLDDVSGEASDGDILAVLGASGAGKSTLIDALAGRVAEGSLRGSVTLNGEKVLQSRLL 156
Query: 480 KIIG-FVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQSVRNSV 538
K+I +V QDD++ LTV+E L F+++ RL LSK +K+ VE +I+ LGL++ N+V
Sbjct: 157 KVISAYVMQDDLLFPMLTVKETLMFASEFRLPRSLSKSKKMERVEALIDQLGLRNAANTV 216
Query: 539 VGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREALEGVN 598
+G RGVSGG+R+RV++G++++ +P +L LDEPTSGLDS ++ ++++ L+R A G
Sbjct: 217 IGDEGHRGVSGGERRRVSIGIDIIHDPIVLFLDEPTSGLDSTNAFMVVQVLKRIAQSGSI 276
Query: 599 ICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINPPDYYID 658
+ M +HQPS + + D LI+L + G V++GS + +FS G +PE+ N ++ +D
Sbjct: 277 VIMSIHQPSARIVELLDRLIILSR-GKSVFNGSPASLPGFFSDFGRPIPEKENISEFALD 335
Query: 659 ILEGIAAPGGSSGL-SYQDLPVKWMLHNEYPIPLDMRQHAAQFGIPQSVNSANDLGEVGK 717
++ + G + G + D KW + + + QS N L +
Sbjct: 336 LVREL--EGSNEGTKALVDFNEKWQQN--------------KISLIQSAPQTNKLDQDRS 379
Query: 718 TFAGELWNDVRSNVELRGEKIRLNFLKSKDLSNRKTPGVFKQYKYFLIRVGKQRLREARI 777
E N S +L R N + +S+ P +F+ + R K +R +
Sbjct: 380 LSLKEAINASVSRGKLVSGSSRSNPTSMETVSSYANPSLFETF-ILAKRYMKNWIRMPEL 438
Query: 778 QAVDYLILLLAGACLGSITKSSDQTFGASGYTYT----VIAVSLLCKIAALRSFSLDKLH 833
+++ G L ++ D T + T V+ C + + F ++
Sbjct: 439 VGTRIATVMVTGCLLATVYWKLDHTPRGAQERLTLFAFVVPTMFYCCLDNVPVFIQERYI 498
Query: 834 YWRESDSGMSSLAYFLSKDTMDHFNTVIKPVVYLSMFYFLTNPRSTFTDNYIVLLCLVY 892
+ RE+ + ++ ++ ++ P + S F T S + ++ L+Y
Sbjct: 499 FLRETTHNAYRTSSYVISHSLVSLPQLLAPSLVFSAITFWTVGLSGGLEGFVFYCLLIY 557
>At3g55090 ABC transporter - like protein
Length = 720
Score = 187 bits (475), Expect = 3e-47
Identities = 99/272 (36%), Positives = 169/272 (61%), Gaps = 5/272 (1%)
Query: 405 EISFRDLT---LTLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCL 461
++ F DL T ++ K +L N++G+ + G I A++G SG+GK+T + ALA +
Sbjct: 71 KLDFHDLVPWRRTSFSKTKTLLDNISGETRDGEILAVLGASGSGKSTLIDALANRIAKGS 130
Query: 462 VTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLV 521
+ G++ +NG K I +V QDD++ LTVEE L F+A+ RL L K +K L
Sbjct: 131 LKGTVTLNGEALQSRMLKVISAYVMQDDLLFPMLTVEETLMFAAEFRLPRSLPKSKKKLR 190
Query: 522 VERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSAS 581
V+ +I+ LG+++ +++G RG+SGG+R+RV++G++++ +P +L LDEPTSGLDS S
Sbjct: 191 VQALIDQLGIRNAAKTIIGDEGHRGISGGERRRVSIGIDIIHDPIVLFLDEPTSGLDSTS 250
Query: 582 SQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSG 641
+ ++++ L+R A G I M +HQPS+ + ++ D LI L +G V+ GS + +F+G
Sbjct: 251 AFMVVKVLKRIAESGSIIIMSIHQPSHRVLSLLDRLIFLSRGH-TVFSGSPASLPSFFAG 309
Query: 642 LGINVPERINPPDYYIDILEGI-AAPGGSSGL 672
G +PE N ++ +D++ + + GG+ GL
Sbjct: 310 FGNPIPENENQTEFALDLIRELEGSAGGTRGL 341
>At1g71960 putative ABC transporter
Length = 662
Score = 187 bits (474), Expect = 3e-47
Identities = 104/256 (40%), Positives = 155/256 (59%), Gaps = 3/256 (1%)
Query: 414 TLKAQNKHILRNVTGKIKPGRITAIMGPSGAGKTTFLSALAGKALGCLVTGSILINGRNE 473
T + + IL VTG I PG A++GPSG+GK+T L+A+AG+ G +TG ILIN
Sbjct: 75 TRSTEERTILSGVTGMISPGEFMAVLGPSGSGKSTLLNAVAGRLHGSNLTGKILINDGKI 134
Query: 474 SIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCRLSADLSKPEKVLVVERVIEFLGLQS 533
+ + K+ GFV QDD+++ +LTV E L F A RL L++ K+ E VI LGL
Sbjct: 135 TKQTLKRT-GFVAQDDLLYPHLTVRETLVFVALLRLPRSLTRDVKLRAAESVISELGLTK 193
Query: 534 VRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLLMLDEPTSGLDSASSQLLLRALRREA 593
N+VVG RG+SGG+RKRV++ E+++ PSLL+LDEPTSGLD+ ++ L++ L A
Sbjct: 194 CENTVVGNTFIRGISGGERKRVSIAHELLINPSLLVLDEPTSGLDATAALRLVQTLAGLA 253
Query: 594 L-EGVNICMVVHQPSYALFNMFDDLILLGKGGLMVYHGSAKKVEEYFSGLGINVPERINP 652
+G + +HQPS +F MFD ++LL +G + + G + YF +G + +NP
Sbjct: 254 HGKGKTVVTSIHQPSSRVFQMFDTVLLLSEGKCL-FVGKGRDAMAYFESVGFSPAFPMNP 312
Query: 653 PDYYIDILEGIAAPGG 668
D+ +D+ G+ G
Sbjct: 313 ADFLLDLANGVCQTDG 328
>At3g55100 ABC transporter - like protein
Length = 662
Score = 185 bits (470), Expect = 1e-46
Identities = 112/331 (33%), Positives = 191/331 (56%), Gaps = 21/331 (6%)
Query: 402 PFIEISFRDLTLTLKAQN-------------KHILRNVTGKIKPGRITAIMGPSGAGKTT 448
PF+ ++F DLT + Q K +L +TG+ K G I AI+G SGAGK+T
Sbjct: 19 PFV-LAFNDLTYNVTLQQRFGLRFGHSPAKIKTLLNGITGEAKEGEILAILGASGAGKST 77
Query: 449 FLSALAGKALGCLVTGSILINGRNESIHSFKKIIGFVPQDDVVHGNLTVEENLWFSAQCR 508
+ ALAG+ + G++ +NG + I +V Q+D++ LTVEE L F+A+ R
Sbjct: 78 LIDALAGQIAEGSLKGTVTLNGEALQSRLLRVISAYVMQEDLLFPMLTVEETLMFAAEFR 137
Query: 509 LSADLSKPEKVLVVERVIEFLGLQSVRNSVVGTVEKRGVSGGQRKRVNVGLEMVMEPSLL 568
L LSK +K VE +I+ LGL +V+N+V+G RGVSGG+R+RV++G +++ +P +L
Sbjct: 138 LPRSLSKSKKRNRVETLIDQLGLTTVKNTVIGDEGHRGVSGGERRRVSIGTDIIHDPIVL 197
Query: 569 MLDEPTSGLDSASSQLLLRALRREALEGVNICMVVHQPSYALFNMFDDLILLGKGGLMVY 628
LDEPTSGLDS S+ ++++ L++ A G + M +HQPS + D +I+L G + V+
Sbjct: 198 FLDEPTSGLDSTSAFMVVQVLKKIARSGSIVIMSIHQPSGRIMEFLDRVIVLSSGQI-VF 256
Query: 629 HGSAKKVEEYFSGLGINVPERINPPDYYIDILEGI-AAPGGSSGLSYQDLPVKWMLHNEY 687
S + +FS G +PE+ N ++ +D+++ + +P G+ GL + W H +
Sbjct: 257 SDSPATLPLFFSEFGSPIPEKENIAEFTLDLIKDLEGSPEGTRGL--VEFNRNWQ-HRKL 313
Query: 688 PIPLDMRQHAAQFGIPQSVNSANDLGEVGKT 718
+ + +++ G +++N++ G++ T
Sbjct: 314 RVSQEPHHNSSSLG--EAINASISRGKLVST 342
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.326 0.138 0.420
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 21,701,813
Number of Sequences: 26719
Number of extensions: 913829
Number of successful extensions: 3902
Number of sequences better than 10.0: 150
Number of HSP's better than 10.0 without gapping: 118
Number of HSP's successfully gapped in prelim test: 32
Number of HSP's that attempted gapping in prelim test: 3467
Number of HSP's gapped (non-prelim): 252
length of query: 1007
length of database: 11,318,596
effective HSP length: 109
effective length of query: 898
effective length of database: 8,406,225
effective search space: 7548790050
effective search space used: 7548790050
T: 11
A: 40
X1: 15 ( 7.0 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 65 (29.6 bits)
Medicago: description of AC144720.7


