

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144720.4 + phase: 0
(83 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
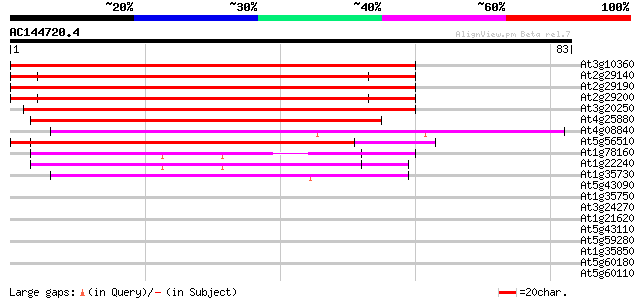
Score E
Sequences producing significant alignments: (bits) Value
At3g10360 putative RNA binding protein 117 1e-27
At2g29140 putative pumilio/Mpt5 family RNA-binding protein 103 1e-23
At2g29190 putative pumilio/Mpt5 family RNA-binding protein 103 2e-23
At2g29200 putative pumilio/Mpt5 family RNA-binding protein 100 1e-22
At3g20250 RNA-binding protein, putative 77 1e-15
At4g25880 pumilio like protein 72 5e-14
At4g08840 putative protein 43 2e-05
At5g56510 putative protein 42 5e-05
At1g78160 putative RNA-binding protein 42 5e-05
At1g22240 hypothetical protein 41 9e-05
At1g35730 hypothetical protein 40 2e-04
At5g43090 putative protein 37 0.002
At1g35750 hypothetical protein 34 0.015
At3g24270 hypothetical protein 33 0.033
At1g21620 hypothetical protein 32 0.043
At5g43110 putative protein 32 0.073
At5g59280 putative protein 31 0.12
At1g35850 hypothetical protein 30 0.21
At5g60180 putative protein 27 2.4
At5g60110 putative protein 27 2.4
>At3g10360 putative RNA binding protein
Length = 1003
Score = 117 bits (293), Expect = 1e-27
Identities = 57/60 (95%), Positives = 59/60 (98%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
+QAMMKDPFGNYVVQKVLETCDDQSL LILSRIKVHLNALKRYTYGKHIV+RVEKLITTG
Sbjct: 928 LQAMMKDPFGNYVVQKVLETCDDQSLALILSRIKVHLNALKRYTYGKHIVARVEKLITTG 987
Score = 34.7 bits (78), Expect = 0.009
Identities = 15/56 (26%), Positives = 30/56 (52%)
Query: 2 QAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLI 57
+ +M D FGNYV+QK E + + + ++ H+ AL YG ++ + +++
Sbjct: 706 RTLMTDVFGNYVIQKFFEHGTTKQRKELAEQVTGHVLALSLQMYGCRVIQKALEVV 761
Score = 32.0 bits (71), Expect = 0.056
Identities = 15/47 (31%), Positives = 25/47 (52%)
Query: 7 DPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
D GN+V+QK +E ++ I+S + AL + YG ++ RV
Sbjct: 783 DQNGNHVIQKCIERLPQDWIQFIISSFYGKVLALSTHPYGCRVIQRV 829
Score = 30.8 bits (68), Expect = 0.12
Identities = 15/52 (28%), Positives = 29/52 (54%), Gaps = 1/52 (1%)
Query: 3 AMMKDPFGNYVVQKVLETCDD-QSLELILSRIKVHLNALKRYTYGKHIVSRV 53
A+ P+G V+Q+VLE DD ++ +I+ I + L + YG +++ +
Sbjct: 815 ALSTHPYGCRVIQRVLEHIDDIETQRIIMEEIMDSVCTLAQDQYGNYVIQHI 866
Score = 30.4 bits (67), Expect = 0.16
Identities = 14/60 (23%), Positives = 32/60 (53%), Gaps = 3/60 (5%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+ +D +GNYV+Q +++ I++++ + + + + ++ VEK +T GG E
Sbjct: 853 LAQDQYGNYVIQHIIQHGKPHERSEIINKLAGQIVKMSQQKFASNV---VEKCLTFGGPE 909
>At2g29140 putative pumilio/Mpt5 family RNA-binding protein
Length = 964
Score = 103 bits (258), Expect = 1e-23
Identities = 49/60 (81%), Positives = 54/60 (89%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
+QAMMKD F NYVVQKVLETCDDQ ELIL+RIKVHLNALK+YTYGKHIV+RVEKL+ G
Sbjct: 890 LQAMMKDQFANYVVQKVLETCDDQQRELILTRIKVHLNALKKYTYGKHIVARVEKLVAAG 949
Score = 39.3 bits (90), Expect = 4e-04
Identities = 17/49 (34%), Positives = 30/49 (60%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+VVQK +E ++++E I+S H+ L + YG ++ RV
Sbjct: 743 VRDQNGNHVVQKCIECVPEENIEFIISTFFGHVVTLSTHPYGCRVIQRV 791
Score = 32.7 bits (73), Expect = 0.033
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+ +D +GNYVVQ VLE +I+ + + + + + ++ VEK +T GG E
Sbjct: 815 LTQDQYGNYVVQHVLEHGKPDERTVIIKELAGKIVQMSQQKFASNV---VEKCLTFGGPE 871
Score = 28.5 bits (62), Expect = 0.62
Identities = 20/51 (39%), Positives = 29/51 (56%), Gaps = 9/51 (17%)
Query: 8 PFGNYVVQKVLETC---DDQS--LELILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q+VLE C D QS +E ILS + + L + YG ++V V
Sbjct: 782 PYGCRVIQRVLEHCHNPDTQSKVMEEILSTVSM----LTQDQYGNYVVQHV 828
Score = 28.1 bits (61), Expect = 0.81
Identities = 15/73 (20%), Positives = 37/73 (50%), Gaps = 1/73 (1%)
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGN 62
A+M D FGNYV+QK E + ++ ++ L YG ++ + +++
Sbjct: 669 ALMTDVFGNYVIQKFFEHGLPPQRRELGEKLIDNVLPLSLQMYGCRVIQKAIEVVDL-DQ 727
Query: 63 ECKLLRYMNSHLL 75
+ ++++ ++ H++
Sbjct: 728 KIQMVKELDGHVM 740
Score = 24.6 bits (52), Expect = 9.0
Identities = 13/45 (28%), Positives = 24/45 (52%), Gaps = 2/45 (4%)
Query: 9 FGNYVVQKVLETCD-DQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+G V+QK +E D DQ ++++ + H+ R G H+V +
Sbjct: 711 YGCRVIQKAIEVVDLDQKIQMV-KELDGHVMRCVRDQNGNHVVQK 754
>At2g29190 putative pumilio/Mpt5 family RNA-binding protein
Length = 972
Score = 103 bits (257), Expect = 2e-23
Identities = 49/60 (81%), Positives = 53/60 (87%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
+QAMMKD F NYVVQKVLETCDDQ ELIL RIKVHLNALK+YTYGKHIV+RVEKL+ G
Sbjct: 898 LQAMMKDQFANYVVQKVLETCDDQQRELILGRIKVHLNALKKYTYGKHIVARVEKLVAAG 957
Score = 36.6 bits (83), Expect = 0.002
Identities = 16/49 (32%), Positives = 30/49 (60%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+VVQK +E ++++E I+S ++ L + YG ++ RV
Sbjct: 751 VRDQNGNHVVQKCIECVPEENIEFIISTFFGNVVTLSTHPYGCRVIQRV 799
Score = 32.3 bits (72), Expect = 0.043
Identities = 17/60 (28%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+ +D +GNYV+Q VLE +I+ + + + + + ++ VEK +T GG E
Sbjct: 823 LAQDQYGNYVIQHVLEHGKPDERTVIIKELAGKIVQMSQQKFASNV---VEKCLTFGGPE 879
Score = 30.8 bits (68), Expect = 0.12
Identities = 16/73 (21%), Positives = 37/73 (49%), Gaps = 1/73 (1%)
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGN 62
A+M D FGNYV+QK E + ++ ++ L YG ++ + +++
Sbjct: 677 ALMTDVFGNYVIQKFFEHGLPPQRRELADKLFDNVLPLSLQMYGCRVIQKAIEVVDL-DQ 735
Query: 63 ECKLLRYMNSHLL 75
+ K+++ ++ H++
Sbjct: 736 KIKMVKELDGHVM 748
Score = 30.4 bits (67), Expect = 0.16
Identities = 14/47 (29%), Positives = 26/47 (54%), Gaps = 1/47 (2%)
Query: 8 PFGNYVVQKVLETCDDQSLE-LILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q+VLE C D + ++ I ++ L + YG +++ V
Sbjct: 790 PYGCRVIQRVLEHCHDPDTQSKVMDEIMSTISMLAQDQYGNYVIQHV 836
>At2g29200 putative pumilio/Mpt5 family RNA-binding protein
Length = 968
Score = 100 bits (250), Expect = 1e-22
Identities = 46/60 (76%), Positives = 53/60 (87%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
+QAMMKD F NYVVQKVLETCDDQ ELIL+RIKVHL ALK+YTYGKH+V+R+EKL+ G
Sbjct: 894 LQAMMKDQFANYVVQKVLETCDDQQRELILTRIKVHLTALKKYTYGKHVVARIEKLVAAG 953
Score = 39.3 bits (90), Expect = 4e-04
Identities = 17/49 (34%), Positives = 30/49 (60%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+VVQK +E ++++E I+S H+ L + YG ++ RV
Sbjct: 747 VRDQNGNHVVQKCIECVPEENIEFIISTFFGHVVTLSTHPYGCRVIQRV 795
Score = 32.7 bits (73), Expect = 0.033
Identities = 18/60 (30%), Positives = 31/60 (51%), Gaps = 3/60 (5%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+ +D +GNYVVQ VLE +I+ + + + + + ++ VEK +T GG E
Sbjct: 819 LAQDQYGNYVVQHVLEHGKPDERTVIIKELAGKIVQMSQQKFASNV---VEKCLTFGGPE 875
Score = 32.0 bits (71), Expect = 0.056
Identities = 16/72 (22%), Positives = 36/72 (49%), Gaps = 1/72 (1%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+M D FGNYV+QK E + ++ H+ L YG ++ + +++ +
Sbjct: 674 LMTDVFGNYVIQKFFEHGLPPQRRELAEKLFDHVLPLSLQMYGCRVIQKAIEVVDL-DQK 732
Query: 64 CKLLRYMNSHLL 75
K+++ ++ H++
Sbjct: 733 IKMVKELDGHVM 744
Score = 30.0 bits (66), Expect = 0.21
Identities = 15/47 (31%), Positives = 26/47 (54%), Gaps = 1/47 (2%)
Query: 8 PFGNYVVQKVLETCDDQSLE-LILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q+VLE C D + ++ I ++ L + YG ++V V
Sbjct: 786 PYGCRVIQRVLEHCHDPDTQSKVMEEILSTVSMLAQDQYGNYVVQHV 832
>At3g20250 RNA-binding protein, putative
Length = 955
Score = 77.4 bits (189), Expect = 1e-15
Identities = 35/58 (60%), Positives = 45/58 (77%)
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTG 60
AMMKD F NYVVQKVLE DQ E+++ R+K+HL +L++YTYGKHIV+R E+L G
Sbjct: 895 AMMKDQFANYVVQKVLEISKDQQREILVQRMKIHLQSLRKYTYGKHIVARFEQLFGEG 952
Score = 34.7 bits (78), Expect = 0.009
Identities = 18/72 (25%), Positives = 39/72 (54%), Gaps = 1/72 (1%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+M D FGNYV+QK +E E ++ ++ + +L YG ++ + ++I +
Sbjct: 673 LMTDVFGNYVIQKFIEHGTPAQREELVKQLAGQMVSLSLQMYGCRVIQKALEVIDV-DQK 731
Query: 64 CKLLRYMNSHLL 75
+L+R ++ ++L
Sbjct: 732 TELIRELDGNVL 743
Score = 31.6 bits (70), Expect = 0.073
Identities = 16/47 (34%), Positives = 24/47 (51%), Gaps = 1/47 (2%)
Query: 8 PFGNYVVQKVLETC-DDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
P+G V+Q++LE C DD+ I+ I AL YG ++ V
Sbjct: 785 PYGCRVIQRILEHCSDDEETHCIIDEILESAFALAHDQYGNYVTQHV 831
Score = 30.4 bits (67), Expect = 0.16
Identities = 13/50 (26%), Positives = 26/50 (52%)
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
A+ D +GNYV Q VLE I+ ++ ++ + ++ Y ++V +
Sbjct: 817 ALAHDQYGNYVTQHVLERGKPDERRQIIEKLTGNVVQMSQHKYASNVVEK 866
Score = 30.4 bits (67), Expect = 0.16
Identities = 11/49 (22%), Positives = 27/49 (54%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+V+QK +E+ + +++ + + L + YG ++ R+
Sbjct: 746 VRDQNGNHVIQKCIESMPAGRIGFVIAAFRGQVATLSTHPYGCRVIQRI 794
Score = 24.6 bits (52), Expect = 9.0
Identities = 20/63 (31%), Positives = 31/63 (48%), Gaps = 6/63 (9%)
Query: 1 MQAMMKDPFGNYVVQKVLETCD-DQSLELILSRIKVHLNALK--RYTYGKHIVSRVEKLI 57
M ++ +G V+QK LE D DQ ELI ++ N LK R G H++ + + +
Sbjct: 706 MVSLSLQMYGCRVIQKALEVIDVDQKTELIR---ELDGNVLKCVRDQNGNHVIQKCIESM 762
Query: 58 TTG 60
G
Sbjct: 763 PAG 765
>At4g25880 pumilio like protein
Length = 861
Score = 72.0 bits (175), Expect = 5e-14
Identities = 31/52 (59%), Positives = 41/52 (78%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEK 55
MMKD +GNYVVQK+ ETC + SR+++H +ALK+YTYGKHIVSR+E+
Sbjct: 799 MMKDQYGNYVVQKIFETCTADQRLTLFSRVRMHASALKKYTYGKHIVSRLEQ 850
Score = 34.3 bits (77), Expect = 0.011
Identities = 18/58 (31%), Positives = 30/58 (51%), Gaps = 3/58 (5%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGG 61
+ KD +GNYV Q VLE + E I ++ H+ L + + ++ +EK + GG
Sbjct: 721 LSKDQYGNYVTQHVLEKGTSEERERIGRKLSGHIVQLSLHKFASNV---IEKCLEYGG 775
Score = 33.1 bits (74), Expect = 0.025
Identities = 20/84 (23%), Positives = 41/84 (48%), Gaps = 2/84 (2%)
Query: 1 MQAMMKDPFGNYVVQKVLETCD-DQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITT 59
+ ++ P+G V+Q++LE C D I I + L + YG ++ V + T+
Sbjct: 681 VSSLSMHPYGCRVIQRLLERCSHDHQCRFITEEILESVCVLSKDQYGNYVTQHVLEKGTS 740
Query: 60 GGNECKLLRYMNSHLLSLNCSDFS 83
E ++ R ++ H++ L+ F+
Sbjct: 741 EERE-RIGRKLSGHIVQLSLHKFA 763
Score = 30.4 bits (67), Expect = 0.16
Identities = 12/49 (24%), Positives = 27/49 (54%)
Query: 5 MKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
++D GN+V+QK +E + +L + +++L + YG ++ R+
Sbjct: 649 VRDQNGNHVIQKCIENIPADKVGFMLYAFRGQVSSLSMHPYGCRVIQRL 697
Score = 29.3 bits (64), Expect = 0.36
Identities = 14/54 (25%), Positives = 26/54 (47%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLI 57
+M D FGNYV+QK E + + + ++ + L YG ++ + +I
Sbjct: 576 LMTDVFGNYVIQKFFEYGNSTQRKELADQLMGQIVPLSLQMYGCRVIQKALDVI 629
>At4g08840 putative protein
Length = 524
Score = 43.1 bits (100), Expect = 2e-05
Identities = 26/90 (28%), Positives = 44/90 (48%), Gaps = 14/90 (15%)
Query: 7 DPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYT---YGKHIVSRVEKLITTG--- 60
DPFGNY++QK++E C+++ IL R+ L + + YG +V ++ + +TT
Sbjct: 288 DPFGNYLIQKLIEVCNEEQRTQILIRLTSKPGLLVKISINNYGTRVVQKLIETVTTKEQI 347
Query: 61 --------GNECKLLRYMNSHLLSLNCSDF 82
L R +N + + LNC F
Sbjct: 348 SLVKSALVPGFLSLFRELNGNHVILNCLKF 377
Score = 27.7 bits (60), Expect = 1.1
Identities = 14/49 (28%), Positives = 26/49 (52%), Gaps = 2/49 (4%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+ +DPFGNY+VQ ++E +L ++ + L +G H+V +
Sbjct: 433 LAQDPFGNYLVQYIIE--KKVGGVNVLFELRGNYVKLATQKFGSHVVEK 479
>At5g56510 putative protein
Length = 604
Score = 42.0 bits (97), Expect = 5e-05
Identities = 21/60 (35%), Positives = 32/60 (53%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNE 63
+M DPFGNY+VQK+LE C++ I+ I L + + H V+K++ T E
Sbjct: 321 LMMDPFGNYLVQKLLEVCNEDQRMQIVHSITRKPGLLIKISCDMHGTRAVQKIVETAKRE 380
Score = 40.8 bits (94), Expect = 1e-04
Identities = 17/51 (33%), Positives = 32/51 (62%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVS 51
+ +M DP+GNYV+Q L+ L++ IK+++++L+ YGK ++S
Sbjct: 548 LDQVMLDPYGNYVIQAALKQSKGNVHALLVDAIKLNISSLRTNPYGKKVLS 598
>At1g78160 putative RNA-binding protein
Length = 626
Score = 42.0 bits (97), Expect = 5e-05
Identities = 23/65 (35%), Positives = 39/65 (59%), Gaps = 13/65 (20%)
Query: 4 MMKDPFGNYVVQKVLETC-DDQSLELIL-------SRIKVHLNALKRYTYGKHIVSRVEK 55
+M DPFGNY++QK+L+ C ++Q +++L I++ LNA YG +V R+ +
Sbjct: 377 LMMDPFGNYLMQKLLDVCTEEQRTQIVLVATEEPGQLIRISLNA-----YGTRVVQRLVE 431
Query: 56 LITTG 60
I +G
Sbjct: 432 TIRSG 436
Score = 38.5 bits (88), Expect = 6e-04
Identities = 17/49 (34%), Positives = 29/49 (58%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+ +DPFGNY VQ V+E ++ ++L+++K H L + H+V R
Sbjct: 525 LAQDPFGNYAVQFVIELRIPSAVAMMLAQLKGHYVQLSMQKFSSHMVER 573
>At1g22240 hypothetical protein
Length = 515
Score = 41.2 bits (95), Expect = 9e-05
Identities = 24/64 (37%), Positives = 38/64 (58%), Gaps = 13/64 (20%)
Query: 4 MMKDPFGNYVVQKVLETC-DDQSLELIL-------SRIKVHLNALKRYTYGKHIVSRVEK 55
+M DPFGNY++QK+L+ C ++Q ++IL I++ LNA YG +V R+ +
Sbjct: 242 LMMDPFGNYLMQKLLDVCNEEQRTQIILMVTSEPGQLIRISLNA-----YGTRVVQRLVE 296
Query: 56 LITT 59
I T
Sbjct: 297 SIKT 300
Score = 40.0 bits (92), Expect = 2e-04
Identities = 18/49 (36%), Positives = 29/49 (58%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+ +DP+GNY VQ VLE D ++ +L+++K H L + H+V R
Sbjct: 390 LAQDPYGNYAVQFVLELRDFSAIAAMLAQLKGHYVELSMQKFSSHMVER 438
Score = 26.6 bits (57), Expect = 2.4
Identities = 14/49 (28%), Positives = 25/49 (50%), Gaps = 1/49 (2%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+++DP+ N+V+Q L ++ I+ H + L+ Y K I SR
Sbjct: 463 LIQDPYANFVIQAALAVTKGSLHATLVEVIRPH-SILRNNPYCKRIFSR 510
>At1g35730 hypothetical protein
Length = 564
Score = 40.0 bits (92), Expect = 2e-04
Identities = 20/56 (35%), Positives = 33/56 (58%), Gaps = 3/56 (5%)
Query: 7 DPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRY---TYGKHIVSRVEKLITT 59
DPFGNY+VQK+ + D++ LI+S + + L R TYG +V ++ + + T
Sbjct: 296 DPFGNYIVQKLFDVSDEEQRTLIVSVLTSNPRELIRICLNTYGTRVVQKMIETVKT 351
Score = 32.0 bits (71), Expect = 0.056
Identities = 14/49 (28%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+ +DPFGNYVVQ +++ S +L + ++H L + H++ +
Sbjct: 441 LSQDPFGNYVVQYLID--QQVSAVKLLVQFRMHYAELATQKFSSHVIEK 487
Score = 25.4 bits (54), Expect = 5.2
Identities = 11/28 (39%), Positives = 17/28 (60%)
Query: 3 AMMKDPFGNYVVQKVLETCDDQSLELIL 30
A++KD GN+V+Q L+T E +L
Sbjct: 368 ALVKDLNGNHVIQSCLQTLGPNDNEFVL 395
Score = 24.6 bits (52), Expect = 9.0
Identities = 11/53 (20%), Positives = 27/53 (50%), Gaps = 4/53 (7%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRI----KVHLNALKRYTYGKHIVSR 52
+++DP+ NYV+Q L +++++ K+H + + + K I+ +
Sbjct: 512 LLQDPYANYVIQTALSVTKGPVRAKLVAKVYRYGKLHSSPYCKKIFSKTILKK 564
>At5g43090 putative protein
Length = 527
Score = 37.0 bits (84), Expect = 0.002
Identities = 20/58 (34%), Positives = 34/58 (58%), Gaps = 3/58 (5%)
Query: 10 GNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITTGGNECKLL 67
GNYVVQ V+E + +L+++++ + L R YG H+ V+KL+ G + KL+
Sbjct: 411 GNYVVQYVVELDNQHETDLLVNKLLRNYAHLARNKYGSHV---VQKLLKLRGIDSKLI 465
Score = 30.8 bits (68), Expect = 0.12
Identities = 14/56 (25%), Positives = 29/56 (51%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKLITT 59
+M DPFGN VV+ ++ C + + LI+ + H++ + ++ L+T+
Sbjct: 252 LMVDPFGNDVVKLLIGKCSSEQIILIVDVVTRHISKFVNICFNPIGTLAIQVLLTS 307
Score = 28.1 bits (61), Expect = 0.81
Identities = 11/53 (20%), Positives = 28/53 (52%)
Query: 1 MQAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+ ++ DPFGNYV+Q + +++ I+ ++ ++ +G I+ ++
Sbjct: 473 IDTLLLDPFGNYVIQTAWFVSKEDVRQMLRYYIERNIRLMRCNKFGNKILEKL 525
>At1g35750 hypothetical protein
Length = 528
Score = 33.9 bits (76), Expect = 0.015
Identities = 20/61 (32%), Positives = 32/61 (51%), Gaps = 13/61 (21%)
Query: 7 DPFGNYVVQKVLETCDDQSLELILS--------RIKVHLNALKRYTYGKHIVSRVEKLIT 58
DP GNY+VQK+L D++ +I+S IK+ LN T G ++ ++ K +
Sbjct: 260 DPLGNYIVQKLLVVSDEEQRTMIVSVLTSKPRELIKICLN-----TNGTRVIQKMIKTVK 314
Query: 59 T 59
T
Sbjct: 315 T 315
Score = 29.6 bits (65), Expect = 0.28
Identities = 15/49 (30%), Positives = 27/49 (54%), Gaps = 2/49 (4%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+ +DPFGNYVVQ +++ S+ L+L + H L + H++ +
Sbjct: 405 LSQDPFGNYVVQCLIDQ-QVSSVNLLLP-FRTHCIELATQKFSSHVIEK 451
Score = 26.6 bits (57), Expect = 2.4
Identities = 13/51 (25%), Positives = 25/51 (48%), Gaps = 1/51 (1%)
Query: 2 QAMMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSR 52
+ +++DP+ NYV+Q L ++ ++K L+ Y K I S+
Sbjct: 474 EQLLQDPYANYVIQTALSVTKGAVRARLVEKVK-RFGKLQSNPYCKKIFSK 523
>At3g24270 hypothetical protein
Length = 137
Score = 32.7 bits (73), Expect = 0.033
Identities = 16/45 (35%), Positives = 24/45 (52%)
Query: 9 FGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
F NY VQK L+ D++ L LI S + L + G ++V R+
Sbjct: 47 FSNYPVQKFLDMLDERCLTLIASEFDSYFENLVKDRVGNYVVQRL 91
>At1g21620 hypothetical protein
Length = 308
Score = 32.3 bits (72), Expect = 0.043
Identities = 15/47 (31%), Positives = 27/47 (56%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIV 50
+ D +GN+V+Q+VL+ D +S I+ ++ H L YG ++V
Sbjct: 177 LSNDAYGNFVIQRVLKLNDLRSKNNIVVSLRGHFVDLSFQKYGSYVV 223
>At5g43110 putative protein
Length = 546
Score = 31.6 bits (70), Expect = 0.073
Identities = 15/48 (31%), Positives = 28/48 (58%)
Query: 9 FGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRVEKL 56
+GNYVVQ ++E + ++ ++ ++ + L R YG H V ++ KL
Sbjct: 401 YGNYVVQYIVELNNRYLIDALVRQLIGNYAHLARNKYGSHAVQKLLKL 448
Score = 25.0 bits (53), Expect = 6.9
Identities = 8/15 (53%), Positives = 12/15 (79%)
Query: 1 MQAMMKDPFGNYVVQ 15
+ ++ DPFGNYV+Q
Sbjct: 464 IDTLLLDPFGNYVIQ 478
>At5g59280 putative protein
Length = 332
Score = 30.8 bits (68), Expect = 0.12
Identities = 16/50 (32%), Positives = 26/50 (52%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+ D +GN+VVQ VL+ D + I ++ + L YG +IV R+
Sbjct: 198 LSNDAYGNFVVQHVLKLRDSRCTRNIADKLCGYCVELSFKKYGSYIVERL 247
>At1g35850 hypothetical protein
Length = 304
Score = 30.0 bits (66), Expect = 0.21
Identities = 16/50 (32%), Positives = 26/50 (52%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+ D +GN+VVQ VL+ D + I ++ + L YG +IV R+
Sbjct: 170 LSNDAYGNFVVQHVLKLHDSRCTGNIADKLCGYCVELSFKKYGSYIVERL 219
>At5g60180 putative protein
Length = 327
Score = 26.6 bits (57), Expect = 2.4
Identities = 15/50 (30%), Positives = 23/50 (46%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+ D GN+VVQ VL D + + I + L YG +IV ++
Sbjct: 195 LSNDASGNFVVQHVLTLYDSRCIHNIAVNLYGQCIELSFKKYGSYIVEKL 244
>At5g60110 putative protein
Length = 327
Score = 26.6 bits (57), Expect = 2.4
Identities = 15/50 (30%), Positives = 23/50 (46%)
Query: 4 MMKDPFGNYVVQKVLETCDDQSLELILSRIKVHLNALKRYTYGKHIVSRV 53
+ D GN+VVQ VL D + + I + L YG +IV ++
Sbjct: 195 LSNDASGNFVVQHVLTLYDSRCIHNIAVNLYGQCIELSFKKYGSYIVEKL 244
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.136 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 1,758,031
Number of Sequences: 26719
Number of extensions: 54505
Number of successful extensions: 205
Number of sequences better than 10.0: 29
Number of HSP's better than 10.0 without gapping: 25
Number of HSP's successfully gapped in prelim test: 4
Number of HSP's that attempted gapping in prelim test: 128
Number of HSP's gapped (non-prelim): 81
length of query: 83
length of database: 11,318,596
effective HSP length: 59
effective length of query: 24
effective length of database: 9,742,175
effective search space: 233812200
effective search space used: 233812200
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 52 (24.6 bits)
Medicago: description of AC144720.4


