

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.9 + phase: 0
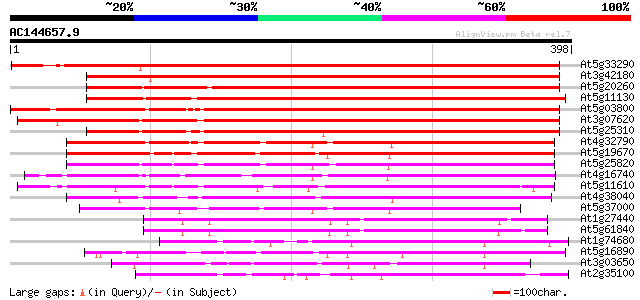
(398 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g33290 unknown protein 494 e-140
At3g42180 unknown protein 457 e-129
At5g20260 putative protein 436 e-122
At5g11130 putative protein 435 e-122
At5g03800 putative protein 425 e-119
At3g07620 hypothetical protein 390 e-109
At5g25310 putative protein 357 8e-99
At4g32790 unknown protein 268 4e-72
At5g19670 putative protein 263 2e-70
At5g25820 putative protein 256 2e-68
At4g16740 limonene cyclase like protein 242 3e-64
At5g11610 unknown protein 240 1e-63
At4g38040 unknown protein 236 2e-62
At5g37000 putative protein 217 1e-56
At1g27440 unknown protein 112 3e-25
At5g61840 unknown protein 107 1e-23
At1g74680 unknown protein 86 5e-17
At5g16890 unknown protein (At5g16890) 85 8e-17
At3g03650 unknown protein 82 5e-16
At2g35100 unknown protein 77 1e-14
>At5g33290 unknown protein
Length = 500
Score = 494 bits (1271), Expect = e-140
Identities = 241/392 (61%), Positives = 295/392 (74%), Gaps = 13/392 (3%)
Query: 2 TSLEKIEEDLAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFKV 61
+ L+KIE DLA+ RA I++A + Q++V S+Y NP AFHQSH EM+ RFKV
Sbjct: 117 SGLDKIESDLAKARAAIKKAAST--------QNYV--SSLYKNPAAFHQSHTEMMNRFKV 166
Query: 62 WVYKEGEQPLVHDGPVNNKYSIEGQFIDEM---DTSNKSPFKATHPELAHVFFLPFSVSK 118
W Y EGE PL HDGPVN+ Y IEGQF+DEM ++S F+A PE AHVFF+PFSV+K
Sbjct: 167 WTYTEGEVPLFHDGPVNDIYGIEGQFMDEMCVDGPKSRSRFRADRPENAHVFFIPFSVAK 226
Query: 119 VIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSY 178
VI +VYKP S ++ RL L+EDY+ +VA K+PYWN SQG DHF++SCHDW P V
Sbjct: 227 VIHFVYKPITSVEGFSRARLHRLIEDYVDVVATKHPYWNRSQGGDHFMVSCHDWAPDVID 286
Query: 179 ANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAG 238
NPKLF+ FIR LCNANTSEGF PN DVSIP++ LP GKLGP + P R+IL FFAG
Sbjct: 287 GNPKLFEKFIRGLCNANTSEGFRPNVDVSIPEIYLPKGKLGPSFLGKSPRVRSILAFFAG 346
Query: 239 GAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAI 298
+HG+IRK L + WK+ D EVQV++ LP G+DYTK MG+SKFCLCPSG EVASPR VEAI
Sbjct: 347 RSHGEIRKILFQHWKEMDNEVQVYDRLPPGKDYTKTMGMSKFCLCPSGWEVASPREVEAI 406
Query: 299 YAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRV 358
YAGCVPVII DNYSLPFSDVLNW FS++I V RI EIKTILQ+++ +Y +Y V V
Sbjct: 407 YAGCVPVIISDNYSLPFSDVLNWDSFSIQIPVSRIKEIKTILQSVSLVRYLKMYKRVLEV 466
Query: 359 RKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
++HF +NRPAKP+D++HM+LHS+WLRRLN RL
Sbjct: 467 KQHFVLNRPAKPYDVMHMMLHSIWLRRLNLRL 498
>At3g42180 unknown protein
Length = 341
Score = 457 bits (1176), Expect = e-129
Identities = 215/339 (63%), Positives = 266/339 (78%), Gaps = 3/339 (0%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSP---FKATHPELAHVFF 111
M+K FKVW YKEGEQPLVHDGPVN+ Y IEGQFIDE+ P F+A+ PE AH FF
Sbjct: 1 MMKTFKVWSYKEGEQPLVHDGPVNDIYGIEGQFIDELSYVMGGPSGRFRASRPEEAHAFF 60
Query: 112 LPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHD 171
LPFSV+ ++ YVY+P S +D+N RL + DY+ +VA+K+P+WN S GADHF++SCHD
Sbjct: 61 LPFSVANIVHYVYQPITSPADFNRARLHRIFNDYVDVVAHKHPFWNQSNGADHFMVSCHD 120
Query: 172 WGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRT 231
W P V + P+ FK+F+R LCNANTSEGF N D SIP++N+P KL PP Q+P NRT
Sbjct: 121 WAPDVPDSKPEFFKNFMRGLCNANTSEGFRRNIDFSIPEINIPKRKLKPPFMGQNPENRT 180
Query: 232 ILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVAS 291
IL FFAG AHG IR+ L WK KD++VQV+++L KGQ+Y +L+G SKFCLCPSG+EVAS
Sbjct: 181 ILAFFAGRAHGYIREVLFSHWKGKDKDVQVYDHLTKGQNYHELIGHSKFCLCPSGYEVAS 240
Query: 292 PRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVL 351
PR VEAIY+GCVPV+I DNYSLPF+DVL+WS+FS+EI VD+IP+IK ILQ I KY +
Sbjct: 241 PREVEAIYSGCVPVVISDNYSLPFNDVLDWSKFSVEIPVDKIPDIKKILQEIPHDKYLRM 300
Query: 352 YSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
Y NV +VR+HF +NRPA+PFD+IHMILHSVWLRRLN RL
Sbjct: 301 YRNVMKVRRHFVVNRPAQPFDVIHMILHSVWLRRLNIRL 339
>At5g20260 putative protein
Length = 334
Score = 436 bits (1120), Expect = e-122
Identities = 207/337 (61%), Positives = 264/337 (77%), Gaps = 5/337 (1%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPF 114
M K+FKVWVY+EGE PLVH GP+NN YSIEGQF+DE++T SPF A +PE AH F LP
Sbjct: 1 MEKKFKVWVYREGETPLVHMGPMNNIYSIEGQFMDEIETG-MSPFAANNPEEAHAFLLPV 59
Query: 115 SVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGP 174
SV+ ++ Y+Y+P + S H++ L DY+ +VA+KYPYWN S GADHF +SCHDW P
Sbjct: 60 SVANIVHYLYRPLVTYSREQLHKVFL---DYVDVVAHKYPYWNRSLGADHFYVSCHDWAP 116
Query: 175 RVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHP-NNRTIL 233
VS +NP+L K+ IR LCNANTSEGF P RDVSIP++N+P G LGPP + ++R IL
Sbjct: 117 DVSGSNPELMKNLIRVLCNANTSEGFMPQRDVSIPEINIPGGHLGPPRLSRSSGHDRPIL 176
Query: 234 TFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPR 293
FFAGG+HG IR+ LL+ WKDKDEEVQVHEYL K +DY KLM ++FCLCPSG+EVASPR
Sbjct: 177 AFFAGGSHGYIRRILLQHWKDKDEEVQVHEYLAKNKDYFKLMATARFCLCPSGYEVASPR 236
Query: 294 VVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYS 353
VV AI GCVPVII D+Y+LPFSDVL+W++F++ + +IPEIKTIL++I+ +YRVL
Sbjct: 237 VVAAINLGCVPVIISDHYALPFSDVLDWTKFTIHVPSKKIPEIKTILKSISWRRYRVLQR 296
Query: 354 NVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
V +V++HF +NRP++PFD++ M+LHSVWLRRLN RL
Sbjct: 297 RVLQVQRHFVINRPSQPFDMLRMLLHSVWLRRLNLRL 333
>At5g11130 putative protein
Length = 336
Score = 435 bits (1119), Expect = e-122
Identities = 200/340 (58%), Positives = 262/340 (76%), Gaps = 4/340 (1%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPF 114
M KRFK+W Y+EGE PL H GP+NN Y+IEGQF+DE++ N S FKA PE A VF++P
Sbjct: 1 MEKRFKIWTYREGEAPLFHKGPLNNIYAIEGQFMDEIENGN-SRFKAASPEEATVFYIPV 59
Query: 115 SVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGP 174
+ +IR+VY+P S Y RLQ +V+DYI +++N+YPYWN S+GADHF LSCHDW P
Sbjct: 60 GIVNIIRFVYRPYTS---YARDRLQNIVKDYISLISNRYPYWNRSRGADHFFLSCHDWAP 116
Query: 175 RVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILT 234
VS +P+L+KHFIRALCNAN+SEGF P RDVS+P++N+P +LG +T + P NR +L
Sbjct: 117 DVSAVDPELYKHFIRALCNANSSEGFTPMRDVSLPEINIPHSQLGFVHTGEPPQNRKLLA 176
Query: 235 FFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRV 294
FFAGG+HG +RK L + WK+KD++V V+E LPK +YTK+M +KFCLCPSG EVASPR+
Sbjct: 177 FFAGGSHGDVRKILFQHWKEKDKDVLVYENLPKTMNYTKMMDKAKFCLCPSGWEVASPRI 236
Query: 295 VEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSN 354
VE++Y+GCVPVII D Y LPFSDVLNW FS+ I + ++P+IK IL+ ITE +Y +
Sbjct: 237 VESLYSGCVPVIIADYYVLPFSDVLNWKTFSVHIPISKMPDIKKILEAITEEEYLNMQRR 296
Query: 355 VRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRLHLKQ 394
V VRKHF +NRP+KP+D++HMI+HS+WLRRLN R+ L Q
Sbjct: 297 VLEVRKHFVINRPSKPYDMLHMIMHSIWLRRLNVRIPLSQ 336
>At5g03800 putative protein
Length = 1388
Score = 425 bits (1092), Expect = e-119
Identities = 207/390 (53%), Positives = 282/390 (72%), Gaps = 9/390 (2%)
Query: 1 MTSLEKIEEDLAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFK 60
+++LEKIE L + RA I+ A +VP G +Y N FH+S+ EM K+FK
Sbjct: 1005 LSNLEKIEFKLQKARASIKAASMDDPVDD---PDYVPLGPMYWNAKVFHRSYLEMEKQFK 1061
Query: 61 VWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVI 120
++VYKEGE PL HDGP + YS+EG FI E++T + F+ +P+ AHVF+LPFSV K++
Sbjct: 1062 IYVYKEGEPPLFHDGPCKSIYSMEGSFIYEIETDTR--FRTNNPDKAHVFYLPFSVVKMV 1119
Query: 121 RYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYAN 180
RYVY+ R SR D++P ++ V+DYI +V +KYPYWN S GADHF+LSCHDWGP S+++
Sbjct: 1120 RYVYE-RNSR-DFSP--IRNTVKDYINLVGDKYPYWNRSIGADHFILSCHDWGPEASFSH 1175
Query: 181 PKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAGGA 240
P L + IRALCNANTSE F P +DVSIP++NL G L P++R IL FFAGG
Sbjct: 1176 PHLGHNSIRALCNANTSERFKPRKDVSIPEINLRTGSLTGLVGGPSPSSRPILAFFAGGV 1235
Query: 241 HGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYA 300
HG +R LL+ W++KD +++VH+YLP+G Y+ +M SKFC+CPSG+EVASPR+VEA+Y+
Sbjct: 1236 HGPVRPVLLQHWENKDNDIRVHKYLPRGTSYSDMMRNSKFCICPSGYEVASPRIVEALYS 1295
Query: 301 GCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRK 360
GCVPV+I Y PFSDVLNW FS+ ++V+ IP +KTIL +I+ +Y +Y V +VR+
Sbjct: 1296 GCVPVLINSGYVPPFSDVLNWRSFSVIVSVEDIPNLKTILTSISPRQYLRMYRRVLKVRR 1355
Query: 361 HFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
HFE+N PAK FD+ HMILHS+W+RRLN ++
Sbjct: 1356 HFEVNSPAKRFDVFHMILHSIWVRRLNVKI 1385
>At3g07620 hypothetical protein
Length = 470
Score = 390 bits (1003), Expect = e-109
Identities = 188/388 (48%), Positives = 266/388 (68%), Gaps = 8/388 (2%)
Query: 6 KIEEDLAQTRALIQRAIRSKKSTTNMK---QSFVPKGSIYLNPHAFHQSHKEMVKRFKVW 62
K+E +LA R LI+ A + STT+ + +VP G IY NP+AFH+S+ M K FK++
Sbjct: 87 KVEAELATARVLIREAQLNYSSTTSSPLGDEDYVPHGDIYRNPYAFHRSYLLMEKMFKIY 146
Query: 63 VYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVIRY 122
VY+EG+ P+ H G + YS+EG F++ M+ ++ ++ P+ AHV+FLPFSV ++ +
Sbjct: 147 VYEEGDPPIFHYGLCKDIYSMEGLFLNFME-NDVLKYRTRDPDKAHVYFLPFSVVMILHH 205
Query: 123 VYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYANPK 182
++ P L+ ++ DY++I++ KYPYWN S G DHF+LSCHDWG R ++ K
Sbjct: 206 LFDPVVRDKAV----LERVIADYVQIISKKYPYWNTSDGFDHFMLSCHDWGHRATWYVKK 261
Query: 183 LFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAGGAHG 242
LF + IR LCNAN SE F P +D P++NL G + P +RT L FFAG +HG
Sbjct: 262 LFFNSIRVLCNANISEYFNPEKDAPFPEINLLTGDINNLTGGLDPISRTTLAFFAGKSHG 321
Query: 243 KIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGC 302
KIR LL WK+KD+++ V+E LP G DYT++M S+FC+CPSGHEVASPRV EAIY+GC
Sbjct: 322 KIRPVLLNHWKEKDKDILVYENLPDGLDYTEMMRKSRFCICPSGHEVASPRVPEAIYSGC 381
Query: 303 VPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRKHF 362
VPV+I +NY LPFSDVLNW +FS+ ++V IPE+K IL +I E +Y LY V++V++H
Sbjct: 382 VPVLISENYVLPFSDVLNWEKFSVSVSVKEIPELKRILMDIPEERYMRLYEGVKKVKRHI 441
Query: 363 EMNRPAKPFDLIHMILHSVWLRRLNFRL 390
+N P K +D+ +MI+HS+WLRRLN +L
Sbjct: 442 LVNDPPKRYDVFNMIIHSIWLRRLNVKL 469
>At5g25310 putative protein
Length = 334
Score = 357 bits (915), Expect = 8e-99
Identities = 173/339 (51%), Positives = 235/339 (69%), Gaps = 9/339 (2%)
Query: 55 MVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPF 114
M KRFKV+VY+EGE PLVHDGP + Y++EG+FI EM+ ++ F+ P A+V+FLPF
Sbjct: 1 MEKRFKVYVYEEGEPPLVHDGPCKSVYAVEGRFITEME-KRRTKFRTYDPNQAYVYFLPF 59
Query: 115 SVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGP 174
SV+ ++RY+Y+ SD P L+ V DYI++V+ +P+WN + GADHF+L+CHDWGP
Sbjct: 60 SVTWLVRYLYE---GNSDAKP--LKTFVSDYIRLVSTNHPFWNRTNGADHFMLTCHDWGP 114
Query: 175 RVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPP---NTDQHPNNRT 231
S AN LF IR +CNAN+SEGF P +DV++P++ L G++ + + R
Sbjct: 115 LTSQANRDLFNTSIRVMCNANSSEGFNPTKDVTLPEIKLYGGEVDHKLRLSKTLSASPRP 174
Query: 232 ILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVAS 291
L FFAGG HG +R LLK WK +D ++ V+EYLPK +Y M SKFC CPSG+EVAS
Sbjct: 175 YLGFFAGGVHGPVRPILLKHWKQRDLDMPVYEYLPKHLNYYDFMRSSKFCFCPSGYEVAS 234
Query: 292 PRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVL 351
PRV+EAIY+ C+PVI+ N+ LPF+DVL W FS+ + V IP +K IL +I+ KY L
Sbjct: 235 PRVIEAIYSECIPVILSVNFVLPFTDVLRWETFSVLVDVSEIPRLKEILMSISNEKYEWL 294
Query: 352 YSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRLNFRL 390
SN+R VR+HFE+N P + FD H+ LHS+WLRRLN +L
Sbjct: 295 KSNLRYVRRHFELNDPPQRFDAFHLTLHSIWLRRLNLKL 333
>At4g32790 unknown protein
Length = 593
Score = 268 bits (685), Expect = 4e-72
Identities = 142/355 (40%), Positives = 219/355 (61%), Gaps = 22/355 (6%)
Query: 41 IYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFK 100
+Y N F +S++ M K+ KV+VY+EG++P++H + Y+ EG F+ ++ +S F
Sbjct: 249 LYWNLSMFKRSYELMEKKLKVYVYREGKRPVLHKPVLKGIYASEGWFMKQLKSSRT--FV 306
Query: 101 ATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQ 160
P AH+F+LPFS + +Y P S SD N L +++Y+ ++++KY +WN +
Sbjct: 307 TKDPRKAHLFYLPFSSKMLEETLYVPG-SHSDKN---LIQFLKNYLDMISSKYSFWNKTG 362
Query: 161 GADHFLLSCHDWGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNL-----PV 215
G+DHFL++CHDW P + + IRALCN++ SEGF +DV++P+ + P+
Sbjct: 363 GSDHFLVACHDWAPSETR---QYMAKCIRALCNSDVSEGFVFGKDVALPETTILVPRRPL 419
Query: 216 GKLGPPNTDQHPNNRTILTFFAGGAHGKIRKKLLKSWK-DKDEEVQVHEYLPKGQ---DY 271
LG Q R IL FFAGG HG +R LL++W ++D ++++ +PK + Y
Sbjct: 420 RALGGKPVSQ----RQILAFFAGGMHGYLRPLLLQNWGGNRDPDMKIFSEIPKSKGKKSY 475
Query: 272 TKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVD 331
+ M SK+C+CP GHEV SPRVVEA++ CVPVII DN+ PF +VLNW F++ +
Sbjct: 476 MEYMKSSKYCICPKGHEVNSPRVVEALFYECVPVIISDNFVPPFFEVLNWESFAVFVLEK 535
Query: 332 RIPEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRL 386
IP++K IL +ITE +YR + V+ V+KHF + + FD+ HMILHS+W R+
Sbjct: 536 DIPDLKNILVSITEERYREMQMRVKMVQKHFLWHSKPERFDIFHMILHSIWYNRV 590
>At5g19670 putative protein
Length = 600
Score = 263 bits (671), Expect = 2e-70
Identities = 138/353 (39%), Positives = 217/353 (61%), Gaps = 18/353 (5%)
Query: 41 IYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFK 100
I+ N F +S++ M + KV+VYKEG +P+ H + Y+ EG F+ M+ + + K
Sbjct: 253 IFRNVSLFKRSYELMERILKVYVYKEGNRPIFHTPILKGLYASEGWFMKLMEGNKQYTVK 312
Query: 101 ATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQ 160
P AH++++PFS ++++ Y R S +N L+ +++Y + +++KYP++N +
Sbjct: 313 --DPRKAHLYYMPFS-ARMLEYTLYVRNS---HNRTNLRQFLKEYTEHISSKYPFFNRTD 366
Query: 161 GADHFLLSCHDWGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGP 220
GADHFL++CHDW P Y +H I+ALCNA+ + GF RD+S+P+ + K
Sbjct: 367 GADHFLVACHDWAP---YETRHHMEHCIKALCNADVTAGFKIGRDISLPETYVRAAK--N 421
Query: 221 PNTD---QHPNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKG----QDYTK 273
P D + P+ R L F+AG HG +R+ LL+ WKDKD ++++ +P G +Y +
Sbjct: 422 PLRDLGGKPPSQRRTLAFYAGSMHGYLRQILLQHWKDKDPDMKIFGRMPFGVASKMNYIE 481
Query: 274 LMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRI 333
M SK+C+CP G+EV SPRVVE+I+ CVPVII DN+ PF +VL+WS FS+ +A I
Sbjct: 482 QMKSSKYCICPKGYEVNSPRVVESIFYECVPVIISDNFVPPFFEVLDWSAFSVIVAEKDI 541
Query: 334 PEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRL 386
P +K IL +I E KY + VR+ ++HF + + +DL HM+LHS+W R+
Sbjct: 542 PRLKDILLSIPEDKYVKMQMAVRKAQRHFLWHAKPEKYDLFHMVLHSIWYNRV 594
>At5g25820 putative protein
Length = 654
Score = 256 bits (654), Expect = 2e-68
Identities = 142/356 (39%), Positives = 212/356 (58%), Gaps = 22/356 (6%)
Query: 41 IYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFK 100
+Y N F +S++ M K KV+ YKEG +P++H + Y+ EG F++ ++ SN + F
Sbjct: 306 LYRNVSMFKRSYELMEKILKVYAYKEGNKPIMHSPILRGIYASEGWFMNIIE-SNNNKFV 364
Query: 101 ATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQ 160
P AH+F+LPFS S+++ + S S N L ++DYI ++ KYP+WN +
Sbjct: 365 TKDPAKAHLFYLPFS-SRMLEVTLYVQDSHSHRN---LIKYLKDYIDFISAKYPFWNRTS 420
Query: 161 GADHFLLSCHDWGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNL-----PV 215
GADHFL +CHDW P + K IRALCN++ EGF +D S+P+ + P+
Sbjct: 421 GADHFLAACHDWAPSETR---KHMAKSIRALCNSDVKEGFVFGKDTSLPETFVRDPKKPL 477
Query: 216 GKLGPPNTDQHPNNRTILTFFAGGA-HGKIRKKLLKSW-KDKDEEVQVHEYLPK---GQD 270
+G + +Q P IL FFAG HG +R LL W +KD ++++ LP+ ++
Sbjct: 478 SNMGGKSANQRP----ILAFFAGKPDHGYLRPILLSYWGNNKDPDLKIFGKLPRTKGNKN 533
Query: 271 YTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAV 330
Y + M SK+C+C G EV SPRVVEAI+ CVPVII DN+ PF +VLNW F++ I
Sbjct: 534 YLQFMKTSKYCICAKGFEVNSPRVVEAIFYDCVPVIISDNFVPPFFEVLNWESFAIFIPE 593
Query: 331 DRIPEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLRRL 386
IP +K IL +I E++YR + V++V+KHF + + +D+ HMILHS+W R+
Sbjct: 594 KDIPNLKKILMSIPESRYRSMQMRVKKVQKHFLWHAKPEKYDMFHMILHSIWYNRV 649
>At4g16740 limonene cyclase like protein
Length = 1024
Score = 242 bits (617), Expect = 3e-64
Identities = 145/387 (37%), Positives = 218/387 (55%), Gaps = 32/387 (8%)
Query: 11 LAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFKVWVYKEGEQP 70
L + IQRA N F P ++ N F +S++ M KV++Y +G++P
Sbjct: 647 LTYAKLEIQRA----PEVINDTDLFAP---LFRNLSVFKRSYELMELILKVYIYPDGDKP 699
Query: 71 LVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVIRYVYKPRKSR 130
+ H+ +N Y+ EG F+ M+ SNK F +PE AH+F++P+SV ++ + K +
Sbjct: 700 IFHEPHLNGIYASEGWFMKLME-SNKQ-FVTKNPERAHLFYMPYSVKQLQK---KTTSTC 754
Query: 131 SDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYANPKLFKHFIRA 190
S N L+ I + Y D WGP +P+L ++ I+A
Sbjct: 755 SPSNTPSGTALMGQIISLSLATIGYRKCFYVKDQ-------WGPYTVNEHPELKRNAIKA 807
Query: 191 LCNANTSEG-FWPNRDVSIPQLNL-----PVGKLGPPNTDQHPNNRTILTFFAGGAHGKI 244
LCNA+ S+G F P +DVS+P+ ++ P+ +G N + R IL FFAG HG++
Sbjct: 808 LCNADLSDGIFVPGKDVSLPETSIRNAGRPLRNIGNGN---RVSQRPILAFFAGNLHGRV 864
Query: 245 RKKLLKSWKDKDEEVQVHEYLP----KGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYA 300
R KLLK W++KDE+++++ LP + Y + M SK+CLCP G+EV SPR+VEAIY
Sbjct: 865 RPKLLKHWRNKDEDMKIYGPLPHNVARKMTYVQHMKSSKYCLCPMGYEVNSPRIVEAIYY 924
Query: 301 GCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRK 360
CVPV+I DN+ LPFSDVL+WS FS+ + IP +K IL I +Y + SNV+ V++
Sbjct: 925 ECVPVVIADNFMLPFSDVLDWSAFSVVVPEKEIPRLKEILLEIPMRRYLKMQSNVKMVQR 984
Query: 361 HFEMNRPAKPFDLIHMILHSVWLRRLN 387
HF + + +D+ HMILHS+W LN
Sbjct: 985 HFLWSPKPRKYDVFHMILHSIWFNLLN 1011
>At5g11610 unknown protein
Length = 546
Score = 240 bits (612), Expect = 1e-63
Identities = 147/393 (37%), Positives = 218/393 (55%), Gaps = 36/393 (9%)
Query: 6 KIEEDLAQTRALIQRAIRSKKSTTNMKQSFVPKGSIYLNPHAFHQSHKEMVKRFKVWVYK 65
K++++L R I++A KK T + P +Y N F +S++ M + KV+VY
Sbjct: 174 KVDQELKTARDKIKKAALVKKDDT----LYAP---LYHNISIFKRSYELMEQTLKVYVYS 226
Query: 66 EGEQPLVH--DGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHVFFLPFSVSKVIRYV 123
EG++P+ H + + Y+ EG F+ M++S++ F P AH+F++PFS S++++
Sbjct: 227 EGDRPIFHQPEAIMEGIYASEGWFMKLMESSHR--FLTKDPTKAHLFYIPFS-SRILQQK 283
Query: 124 YKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGP---RVSYAN 180
S S N L + +YI ++A+ YP WN + G+DHF +CHDW P R Y N
Sbjct: 284 LYVHDSHSRNN---LVKYLGNYIDLIASNYPSWNRTCGSDHFFTACHDWAPTETRGPYIN 340
Query: 181 PKLFKHFIRALCNANTSEGFWPNRDVSIPQ-----LNLPVGKLGPPNTDQHPNNRTILTF 235
IRALCNA+ F +DVS+P+ L P GK+G P+ RTIL F
Sbjct: 341 ------CIRALCNADVGIDFVVGKDVSLPETKVSSLQNPNGKIG----GSRPSKRTILAF 390
Query: 236 FAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVV 295
FAG HG +R LL W + E+ + Y + M S+FC+C G+EV SPRVV
Sbjct: 391 FAGSLHGYVRPILLNQWSSRPEQDMKIFNRIDHKSYIRYMKRSRFCVCAKGYEVNSPRVV 450
Query: 296 EAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNV 355
E+I GCVPVII DN+ PF ++LNW F++ + IP ++ IL +I +Y + V
Sbjct: 451 ESILYGCVPVIISDNFVPPFLEILNWESFAVFVPEKEIPNLRKILISIPVRRYVEMQKRV 510
Query: 356 RRVRKHFEMNRPAKP--FDLIHMILHSVWLRRL 386
+V+KHF M +P +D+ HMILHSVW R+
Sbjct: 511 LKVQKHF-MWHDGEPVRYDIFHMILHSVWYNRV 542
>At4g38040 unknown protein
Length = 425
Score = 236 bits (602), Expect = 2e-62
Identities = 125/349 (35%), Positives = 201/349 (56%), Gaps = 20/349 (5%)
Query: 41 IYLNPHAFHQSHKEMVKRFKVWVYKEGEQPLVHDGP--VNNKYSIEGQFIDEMDTSNKSP 98
+Y +P AF ++ EM KRFKV++Y +G+ + P V KY+ EG F + +S
Sbjct: 86 VYHSPEAFRLNYAEMEKRFKVYIYPDGDPNTFYQTPRKVTGKYASEGYFFQNI---RESR 142
Query: 99 FKATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNI 158
F+ P+ A +FF+P S K+ R + Y + ++V++Y+ + KYPYWN
Sbjct: 143 FRTLDPDEADLFFIPISCHKM-------RGKGTSYE--NMTVIVQNYVDGLIAKYPYWNR 193
Query: 159 SQGADHFLLSCHDWGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGKL 218
+ GADHF ++CHD G R +P L K+ IR +C+ + + GF P++DV++PQ+ P
Sbjct: 194 TLGADHFFVTCHDVGVRAFEGSPLLIKNTIRVVCSPSYNVGFIPHKDVALPQVLQPFAL- 252
Query: 219 GPPNTDQHPNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKGQD---YTKLM 275
P NRT L F+AG + KIR L W++ E + + + Y K
Sbjct: 253 --PAGGNDVENRTTLGFWAGHRNSKIRVILAHVWENDTELDISNNRINRATGHLVYQKRF 310
Query: 276 GLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPE 335
+KFC+CP G +V S R+ ++I+ GC+PVI+ D Y LPF+D+LNW +F++ + +
Sbjct: 311 YRTKFCICPGGSQVNSARITDSIHYGCIPVILSDYYDLPFNDILNWRKFAVVLREQDVYN 370
Query: 336 IKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSVWLR 384
+K IL+NI +++ L++N+ +V+KHF+ N P FD HMI++ +WLR
Sbjct: 371 LKQILKNIPHSEFVSLHNNLVKVQKHFQWNSPPVKFDAFHMIMYELWLR 419
>At5g37000 putative protein
Length = 547
Score = 217 bits (552), Expect = 1e-56
Identities = 114/325 (35%), Positives = 194/325 (59%), Gaps = 29/325 (8%)
Query: 50 QSHKEMVKRFKVWVYKEGEQPLVHDGPVNNKYSIEGQFIDEMDTSNKSPFKATHPELAHV 109
+S+ M ++ K++VYKEG +P+ H Y+ EG F+ M+++ K F P AH+
Sbjct: 231 RSYDLMERKLKIYVYKEGGKPIFHTPMPRGIYASEGWFMKLMESNKK--FVVKDPRKAHL 288
Query: 110 FFLPFSVSKV---IRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFL 166
F++P S+ + + ++ KS +D+ +++Y+ ++A KY +WN + GADHFL
Sbjct: 289 FYIPISIKALRSSLGLDFQTPKSLADH--------LKEYVDLIAGKYKFWNRTGGADHFL 340
Query: 167 LSCHDWGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQLNL-----PVGKLGPP 221
++CHDWG +++ K K+ +R+LCN+N ++GF D ++P + P+ LG
Sbjct: 341 VACHDWGNKLT---TKTMKNSVRSLCNSNVAQGFRIGTDTALPVTYIRSSEAPLEYLGGK 397
Query: 222 NTDQHPNNRTILTFFAGGAHGKIRKKLLKSWKDKDEEVQVHEYLPKG----QDYTKLMGL 277
+ + R IL FFAG HG +R L+K W++K+ ++++ +P+ + Y + M
Sbjct: 398 TSSE----RKILAFFAGSMHGYLRPILVKLWENKEPDMKIFGPMPRDPKSKKQYREYMKS 453
Query: 278 SKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIK 337
S++C+C G+EV +PRVVEAI CVPVII DNY PF +VLNW +F++ + IP ++
Sbjct: 454 SRYCICARGYEVHTPRVVEAIINECVPVIIADNYVPPFFEVLNWEEFAVFVEEKDIPNLR 513
Query: 338 TILQNITETKYRVLYSNVRRVRKHF 362
IL +I E +Y + + V+ V++HF
Sbjct: 514 NILLSIPEDRYIGMQARVKAVQQHF 538
>At1g27440 unknown protein
Length = 412
Score = 112 bits (281), Expect = 3e-25
Identities = 90/321 (28%), Positives = 145/321 (45%), Gaps = 39/321 (12%)
Query: 96 KSPFKATHPELAHVFFLPFSVSKVIR--------YVYKPRKSRSDYNPHRLQL------L 141
K P TH A +F F +S +R + Y P D P L L +
Sbjct: 64 KDPRCLTHMFAAEIFMHRFLLSSPVRTRNPDEADWFYTPIYPTCDLTPTGLPLPFKSPRM 123
Query: 142 VEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYANPKLFKHFIRALCN-ANTSEGF 200
+ I+++++ +PYWN ++GADHF + HD+G Y K + I L A + F
Sbjct: 124 MRSSIQLISSNWPYWNRTEGADHFFVVPHDFGACFHYQEEKAIERGILPLLQRATLVQTF 183
Query: 201 WPNRDVSIPQLNLPVGKLGPPNTDQH----PN-NRTILTFFAG-----------GAHGKI 244
V + + ++ + PP Q P+ R+I +F G G + +
Sbjct: 184 GQRNHVCLDEGSITIPPFAPPQKMQAHFIPPDIPRSIFVYFRGLFYDVNNDPEGGYYARG 243
Query: 245 RKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVP 304
+ + W++ Y + M + FCLCP G SPR+VEA+ GC+P
Sbjct: 244 ARAAV--WENFKNNPLFDISTDHPTTYYEDMQRAIFCLCPLGWAPWSPRLVEAVVFGCIP 301
Query: 305 VIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNI-TET---KYRVLYSNVRRVRK 360
VII D+ LPF+D + W + + +A +PE+ TIL +I TE K R+L + +++
Sbjct: 302 VIIADDIVLPFADAIPWEEIGVFVAEKDVPELDTILTSIPTEVILRKQRLLAN--PSMKR 359
Query: 361 HFEMNRPAKPFDLIHMILHSV 381
+PA+P D H IL+ +
Sbjct: 360 AMLFPQPAQPGDAFHQILNGL 380
>At5g61840 unknown protein
Length = 415
Score = 107 bits (267), Expect = 1e-23
Identities = 84/321 (26%), Positives = 139/321 (43%), Gaps = 39/321 (12%)
Query: 96 KSPFKATHPELAHVFFLPFSVSKVIR--------YVYKPRKSRSDYNPHRLQL------L 141
K P H A ++ F +S +R + Y P + D P+ L L +
Sbjct: 67 KDPRCLNHMFAAEIYMQRFLLSSPVRTLNPEEADWFYVPVYTTCDLTPNGLPLPFKSPRM 126
Query: 142 VEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYANPKLF-KHFIRALCNANTSEGF 200
+ I+++A+ +PYWN ++GADHF + HD+G Y K + + L A + F
Sbjct: 127 MRSAIQLIASNWPYWNRTEGADHFFVVPHDFGACFHYQEEKAIGRGILPLLQRATLVQTF 186
Query: 201 WPNRDVSIPQLNLPVGKLGPPNTDQ-----HPNNRTILTFFAG-----------GAHGKI 244
V + + ++ V PP Q R+I +F G G + +
Sbjct: 187 GQRNHVCLKEGSITVPPYAPPQKMQSHLIPEKTPRSIFVYFRGLFYDVGNDPEGGYYARG 246
Query: 245 RKKLLKSWKDKDEEVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVP 304
+ + W++ + Y + M + FCLCP G SPR+VEA+ GC+P
Sbjct: 247 ARAAV--WENFKDNPLFDISTEHPTTYYEDMQRAIFCLCPLGWAPWSPRLVEAVIFGCIP 304
Query: 305 VIICDNYSLPFSDVLNWSQFSMEIAVDRIPEIKTILQNITE----TKYRVLYSNVRRVRK 360
VII D+ LPF+D + W + + +P + TIL +I K R+L + +
Sbjct: 305 VIIADDIVLPFADAIPWEDIGVFVDEKDVPYLDTILTSIPPEVILRKQRLLANPSMKQAM 364
Query: 361 HFEMNRPAKPFDLIHMILHSV 381
F +PA+P D H +L+ +
Sbjct: 365 LFP--QPAQPGDAFHQVLNGL 383
>At1g74680 unknown protein
Length = 461
Score = 85.5 bits (210), Expect = 5e-17
Identities = 86/310 (27%), Positives = 134/310 (42%), Gaps = 36/310 (11%)
Query: 107 AHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFL 166
A + F+PF S K R + + + LQ + +++K W G DH +
Sbjct: 156 ADIVFVPFFASLSYNRKSKLRGNETSSDDRLLQERLVEFLK----SQDEWKRFDGKDHLI 211
Query: 167 LSCHDWGPRVSYANPKL---------FKHFIRALCNANTSEGFWPNRDVSIPQLNLPVGK 217
++ H + YA L F + A+ N +D+ P ++ V K
Sbjct: 212 VAHHP--NSLLYARNFLGSAMFVLSDFGRYSSAIANLE--------KDIIAPYVH--VVK 259
Query: 218 LGPPNTDQHPNNRTILTFFAGGAH----GKIRKKLLKSWKD-KDEEVQVHEYLPKGQDYT 272
N R +L +F G + G IR++L KD KD G T
Sbjct: 260 TISNNESASFEKRPVLAYFQGAIYRKDGGTIRQELYNLLKDEKDVHFAFGTVRGNGTKQT 319
Query: 273 -KLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEIAVD 331
K M SKFCL +G +S R+ +AI + CVPVII D LPF D L++S FS+ +
Sbjct: 320 GKGMASSKFCLNIAGDTPSSNRLFDAIVSHCVPVIISDQIELPFEDTLDYSGFSVFVHAS 379
Query: 332 RIPE---IKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMILHSV--WLRRL 386
+ + IL+ ITE +++ + ++ V FE P++ D ++MI +V L L
Sbjct: 380 EAVKKEFLVNILRGITEDQWKKKWGRLKEVAGCFEYRFPSQVGDSVNMIWSAVSHKLSSL 439
Query: 387 NFRLHLKQMY 396
F +H K Y
Sbjct: 440 QFDVHRKNRY 449
>At5g16890 unknown protein (At5g16890)
Length = 511
Score = 84.7 bits (208), Expect = 8e-17
Identities = 92/376 (24%), Positives = 162/376 (42%), Gaps = 55/376 (14%)
Query: 54 EMVKRFK---VWV----YKEGEQPLVHDGPVNNKYSIEGQFID----------EMDTSNK 96
EM K+F +W+ YKE + PV+ IE ID E + K
Sbjct: 124 EMPKKFTFDLLWLFHNTYKETSNATSNGSPVHRL--IEQHSIDYWLWADLISPESERRLK 181
Query: 97 SPFKATHPELAHVFFLPFSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKIVANKYPYW 156
S + + A F++PF + + K + + L + +K V ++ P W
Sbjct: 182 SVVRVQKQQDADFFYVPFFTTISFFLLEK----------QQCKALYREALKWVTDQ-PAW 230
Query: 157 NISQGADHFLLSCHDWGPRVSYANPKLFKHFIRALCNANTSEGFWPNRDVSIPQ-LNLPV 215
S+G DH H W + + K K+ I L + +++ ++ VS+ + L LP
Sbjct: 231 KRSEGRDHIFPIHHPWSFK---SVRKFVKNAIWLLPDMDSTGNWYKPGQVSLEKDLILPY 287
Query: 216 GKLGPPNTD--------QHPNNRTILTFFAG----GAHGKIRKKLLKSWKDKDEEVQVHE 263
PN D + RT L FF G A GKIR KL + +
Sbjct: 288 ----VPNVDICDTKCLSESAPMRTTLLFFRGRLKRNAGGKIRAKLGAELSGIKDIIISEG 343
Query: 264 YLPKGQDYTKLMGL--SKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFSDVLNW 321
+G G+ S FCLCP+G +S R+ +AI +GC+PVI+ D PF +L++
Sbjct: 344 TAGEGGKLAAQRGMRRSLFCLCPAGDTPSSARLFDAIVSGCIPVIVSDELEFPFEGILDY 403
Query: 322 SQFSMEIAVDRIPE---IKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHMIL 378
+ ++ ++ + + L+++T + + L +N+ + +HF + PA+P +
Sbjct: 404 KKVAVLVSSSDAIQPGWLVNHLRSLTPFQVKGLQNNLAQYSRHFLYSSPAQPLGPEDLTW 463
Query: 379 HSVWLRRLNFRLHLKQ 394
+ + +N +LH ++
Sbjct: 464 RMIAGKLVNIKLHTRR 479
>At3g03650 unknown protein
Length = 499
Score = 82.0 bits (201), Expect = 5e-16
Identities = 80/319 (25%), Positives = 148/319 (46%), Gaps = 35/319 (10%)
Query: 73 HDGPVNNKYSIEGQF--------IDEMDTSNKSPFKATHPELAHVFFLPFSVSKVIRYVY 124
H G +N ++S+E + E S+++ + + A V F+PF S
Sbjct: 158 HPGGLNLQHSVEYWLTLDLLFSELPEDSRSSRAAIRVKNSSEADVVFVPFFSSLSYNRFS 217
Query: 125 KPRKSRSDYNPHRLQLLVEDYIKIVANKYPYWNISQGADHFLLSCHDWGPRVSYANPKLF 184
K + + LQ E+ +K V ++ W S G DH +++ H +S A KLF
Sbjct: 218 KVNQKQKKSQDKELQ---ENVVKYVTSQKE-WKTSGGKDHVIMAHHP--NSMSTARHKLF 271
Query: 185 KH-FIRALCNANTSEGFWPNRDVSIPQLNLPVGKLGPPNTDQHPNNRTILTFFAGG---- 239
F+ A + ++D+ P +L + N + R IL +F G
Sbjct: 272 PAMFVVADFGRYSPHVANVDKDIVAPYKHLVPSYV---NDTSGFDGRPILLYFQGAIYRK 328
Query: 240 AHGKIRKKLLKSWKDKDE------EVQVHEYLPKGQDYTKLMGLSKFCLCPSGHEVASPR 293
A G +R++L K++ + V+ H G+ M SKFCL +G +S R
Sbjct: 329 AGGFVRQELYNLLKEEKDVHFSFGSVRNHGISKAGEG----MRSSKFCLNIAGDTPSSNR 384
Query: 294 VVEAIYAGCVPVIICDNYSLPFSDVLNWSQFSMEI-AVDRIPE--IKTILQNITETKYRV 350
+ +AI + C+PVII D+ LP+ DVLN+++F + + + D + + + ++++I +Y
Sbjct: 385 LFDAIASHCIPVIISDDIELPYEDVLNYNEFCLFVRSSDALKKGFLMGLVRSIGREEYNK 444
Query: 351 LYSNVRRVRKHFEMNRPAK 369
++ ++ V ++F++ P K
Sbjct: 445 MWLRLKEVERYFDLRFPVK 463
>At2g35100 unknown protein
Length = 460
Score = 77.4 bits (189), Expect = 1e-14
Identities = 73/320 (22%), Positives = 149/320 (45%), Gaps = 38/320 (11%)
Query: 90 EMDTSNKSPFKATHPELAHVFFLP-FSVSKVIRYVYKPRKSRSDYNPHRLQLLVEDYIKI 148
E+D S + + P A +F++P FS +I +P ++ S Y+ ++Q + ++++
Sbjct: 117 EVDRSGSPIVRVSDPADADLFYVPVFSSLSLIVNAGRPVEAGSGYSDEKMQEGLVEWLE- 175
Query: 149 VANKYPYWNISQGADHFLLSCHDWGPRVSYANPKLFKHFIRALCN---ANTSEGFWPNRD 205
+W + G DH + + P Y K+ + + + +G + +D
Sbjct: 176 ---GQEWWRRNAGRDHVIPA---GDPNALYRILDRVKNAVLLVSDFGRLRPDQGSFV-KD 228
Query: 206 VSIP---QLNLPVGKLGPPNTDQHPNNRTILTFFAGGAH----GKIRKKLLKSWKDKDEE 258
V IP ++NL G++G +R L FF G + GK+R L + + +D+
Sbjct: 229 VVIPYSHRVNLFNGEIGV-------EDRNTLLFFMGNRYRKDGGKVRDLLFQVLEKEDDV 281
Query: 259 VQVH--EYLPKGQDYTKLMGLSKFCLCPSGHEVASPRVVEAIYAGCVPVIICDNYSLPFS 316
H + + TK M SKFCL P+G ++ R+ ++I + CVP+I+ D+ LPF
Sbjct: 282 TIKHGTQSRENRRAATKGMHTSKFCLNPAGDTPSACRLFDSIVSLCVPLIVSDSIELPFE 341
Query: 317 DVLNWSQFSMEIAVDRIPEIKTILQNITETKYRVLYSNVRRVRKHFEMNRPAKPFDLIHM 376
DV+++ +FS+ + + + ++Q + + K + + R ++ F+ +
Sbjct: 342 DVIDYRKFSIFVEANAALQPGFLVQMLRKIKTKKILEYQREMKSKFDRTK---------- 391
Query: 377 ILHSVWLRRLNFRLHLKQMY 396
+L S+ ++ HL ++
Sbjct: 392 LLMSMHKPNRSYHFHLNNLF 411
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.321 0.137 0.426
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,622,741
Number of Sequences: 26719
Number of extensions: 431089
Number of successful extensions: 1149
Number of sequences better than 10.0: 48
Number of HSP's better than 10.0 without gapping: 39
Number of HSP's successfully gapped in prelim test: 9
Number of HSP's that attempted gapping in prelim test: 1043
Number of HSP's gapped (non-prelim): 60
length of query: 398
length of database: 11,318,596
effective HSP length: 101
effective length of query: 297
effective length of database: 8,619,977
effective search space: 2560133169
effective search space used: 2560133169
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144657.9


