

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.5 - phase: 0
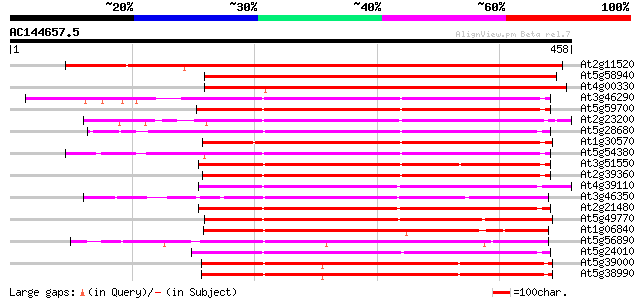
(458 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g11520 putative protein kinase 523 e-149
At5g58940 receptor-like protein kinase precursor - like 337 1e-92
At4g00330 unknown protein 330 8e-91
At3g46290 receptor protein kinase -like 240 1e-63
At5g59700 receptor-like protein kinase precursor - like 239 2e-63
At2g23200 putative protein kinase 235 4e-62
At5g28680 receptor-like protein kinase precursor - like 233 1e-61
At1g30570 putative serine/threonine protein kinase 231 5e-61
At5g54380 receptor-protein kinase-like protein 230 1e-60
At3g51550 receptor protein kinase like protein 229 2e-60
At2g39360 putative protein kinase 229 2e-60
At4g39110 putative receptor-like protein kinase 229 2e-60
At3g46350 receptor-like protein kinase homolog 227 1e-59
At2g21480 putative protein kinase 226 2e-59
At5g49770 receptor protein kinase-like 224 1e-58
At1g06840 receptor protein kinase, putative 223 1e-58
At5g56890 unknown protein 223 2e-58
At5g24010 receptor-protein kinase-like protein 223 2e-58
At5g39000 protein kinase - like protein 221 6e-58
At5g38990 protein kinase - like protein 221 6e-58
>At2g11520 putative protein kinase
Length = 510
Score = 523 bits (1348), Expect = e-149
Identities = 265/414 (64%), Positives = 323/414 (78%), Gaps = 9/414 (2%)
Query: 46 STAGRKLLQKDLNNNSTSQGDPKQVSSSQVGIFAGGALLVCCAVVCPCFYGKRRKATSHA 105
S GR+ L++ +S + + S +V I G LL+CCA+ CPCF+ K RKA SH
Sbjct: 96 SLLGRRFLEEKTVKDSKNSKPKTEYSHVKVSIAGSGFLLLCCALCCPCFH-KERKANSHE 154
Query: 106 VLEKDPNSMELGSSFEPSVS-DKIPASPLRVPPSPSR-------FSMSPKLSRLQSLHLN 157
VL K+ NS+ SSFE S S +KIP SP R PPSPSR ++MSP+ SRL L+L
Sbjct: 155 VLPKESNSVHQVSSFEMSPSSEKIPQSPFRAPPSPSRVPQSPSRYAMSPRPSRLGPLNLT 214
Query: 158 LSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELL 217
+SQ++ AT NF+++ QIGEGGFG V+K LDDG VVA+KRAK+EHFE+LRTEF SEV+LL
Sbjct: 215 MSQINTATGNFADSHQIGEGGFGVVFKGVLDDGQVVAIKRAKKEHFENLRTEFKSEVDLL 274
Query: 218 AKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHG 277
+KI HRNLVKLLGY+DKG+ER++ITE+V NGTLR+HLDG RG L+FNQRLEI IDV HG
Sbjct: 275 SKIGHRNLVKLLGYVDKGDERLIITEYVRNGTLRDHLDGARGTKLNFNQRLEIVIDVCHG 334
Query: 278 LTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGY 337
LTYLH YAE+QIIHRD+KSSNILLT+SMRAKVADFGFA+ GP +++ THI T+VKGTVGY
Sbjct: 335 LTYLHSYAERQIIHRDIKSSNILLTDSMRAKVADFGFARGGPTDSNQTHILTQVKGTVGY 394
Query: 338 LDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVA 397
LDPEYMKTYHLT KSDVYSFGILL+EILTGRRPVE K+ +ER+T+RWAF KYNEG V
Sbjct: 395 LDPEYMKTYHLTAKSDVYSFGILLVEILTGRRPVEAKRLPDERITVRWAFDKYNEGRVFE 454
Query: 398 LLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKSS 451
L+DP +E V + KMF LAF CAAP + +RPDM+ VG+QLWAIR+ YL+ S
Sbjct: 455 LVDPNARERVDEKILRKMFSLAFQCAAPTKKERPDMEAVGKQLWAIRSSYLRRS 508
>At5g58940 receptor-like protein kinase precursor - like
Length = 466
Score = 337 bits (863), Expect = 1e-92
Identities = 161/289 (55%), Positives = 219/289 (75%), Gaps = 2/289 (0%)
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHF-ESLRTEFSSEVELLA 218
Q+ +AT NFS QIGEGGFGTV+K LDDG +VA+KRA++ ++ +S EF +E+ L+
Sbjct: 135 QLQRATANFSSVHQIGEGGFGTVFKGKLDDGTIVAIKRARKNNYGKSWLLEFKNEIYTLS 194
Query: 219 KIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGL 278
KI+H NLVKL G+++ G+E++++ E+VANG LREHLDGLRG L+ +RLEIAIDVAH L
Sbjct: 195 KIEHMNLVKLYGFLEHGDEKVIVVEYVANGNLREHLDGLRGNRLEMAERLEIAIDVAHAL 254
Query: 279 TYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYL 338
TYLH Y + IIHRD+K+SNIL+T +RAKVADFGFA+L + THIST+VKG+ GY+
Sbjct: 255 TYLHTYTDSPIIHRDIKASNILITNKLRAKVADFGFARLVSEDLGATHISTQVKGSAGYV 314
Query: 339 DPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVAL 398
DP+Y++T+ LT KSDVYSFG+LL+EILTGRRP+ELK+ ++R+T++WA R+ + V +
Sbjct: 315 DPDYLRTFQLTDKSDVYSFGVLLVEILTGRRPIELKRPRKDRLTVKWALRRLKDDEAVLI 374
Query: 399 LDP-LMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRAD 446
+DP L + +VA KM LA C P R+ RP MK + E+LWAIR +
Sbjct: 375 MDPFLKRNRAAIEVAEKMLRLASECVTPTRATRPAMKGIAEKLWAIRRE 423
>At4g00330 unknown protein
Length = 411
Score = 330 bits (847), Expect = 8e-91
Identities = 161/300 (53%), Positives = 219/300 (72%), Gaps = 5/300 (1%)
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLR---TEFSSEVEL 216
++ AT+NFS + +IG+GGFGTVYK L DG AVKRAK+ + + EF SE++
Sbjct: 111 EIYDATKNFSPSFRIGQGGFGTVYKVKLRDGKTFAVKRAKKSMHDDRQGADAEFMSEIQT 170
Query: 217 LAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAH 276
LA++ H +LVK G++ +E+IL+ E+VANGTLR+HLD GK LD RL+IA DVAH
Sbjct: 171 LAQVTHLSLVKYYGFVVHNDEKILVVEYVANGTLRDHLDCKEGKTLDMATRLDIATDVAH 230
Query: 277 GLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGP-VNNDHTHISTKVKGTV 335
+TYLH+Y + IIHRD+KSSNILLTE+ RAKVADFGFA+L P ++ TH+ST+VKGT
Sbjct: 231 AITYLHMYTQPPIIHRDIKSSNILLTENYRAKVADFGFARLAPDTDSGATHVSTQVKGTA 290
Query: 336 GYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSV 395
GYLDPEY+ TY LT KSDVYSFG+LL+E+LTGRRP+EL + +ER+T+RWA +K+ G
Sbjct: 291 GYLDPEYLTTYQLTEKSDVYSFGVLLVELLTGRRPIELSRGQKERITIRWAIKKFTSGDT 350
Query: 396 VALLDPLMQEAVKTDVAV-KMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKSSSTT 454
+++LDP +++ ++A+ K+ ++AF C AP R RP MK E LW IR DY + +T+
Sbjct: 351 ISVLDPKLEQNSANNLALEKVLEMAFQCLAPHRRSRPSMKKCSEILWGIRKDYRELLNTS 410
>At3g46290 receptor protein kinase -like
Length = 830
Score = 240 bits (613), Expect = 1e-63
Identities = 164/443 (37%), Positives = 228/443 (51%), Gaps = 39/443 (8%)
Query: 14 LSTIFGSEVVLQT-KGCGDNLVANSYSS-HVNLPSTAGRKLLQKDLNNN----STSQGDP 67
L+ + + V QT KG V+ S+ H + P+ L +NN+ ST P
Sbjct: 335 LAGAYSMDFVTQTPKGSNKVRVSIGPSTVHTDYPNAIVNGLEIMKMNNSKGQLSTGTFVP 394
Query: 68 KQVSSSQ--VGIFAGGALLVCCAVV----CPCFYGKRRKAT---SHAVLEKDPNSMELGS 118
SSS+ +G+ G A+ AVV C Y KR++ S + N +GS
Sbjct: 395 GSSSSSKSNLGLIVGSAIGSLLAVVFLGSCFVLYKKRKRGQDGHSKTWMPFSINGTSMGS 454
Query: 119 SFEPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGG 178
+ S + ++ + + + V AT NF E+ IG GG
Sbjct: 455 KY-------------------SNGTTLTSITTNANYRIPFAAVKDATNNFDESRNIGVGG 495
Query: 179 FGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNER 238
FG VYK L+DG VAVKR + + L EF +E+E+L++ HR+LV L+GY D+ NE
Sbjct: 496 FGKVYKGELNDGTKVAVKRGNPKSQQGL-AEFRTEIEMLSQFRHRHLVSLIGYCDENNEM 554
Query: 239 ILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSN 298
ILI E++ NGT++ HL G L + QRLEI I A GL YLH K +IHRDVKS+N
Sbjct: 555 ILIYEYMENGTVKSHLYGSGLPSLTWKQRLEICIGAARGLHYLHTGDSKPVIHRDVKSAN 614
Query: 299 ILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFG 358
ILL E+ AKVADFG +K GP D TH+ST VKG+ GYLDPEY + LT KSDVYSFG
Sbjct: 615 ILLDENFMAKVADFGLSKTGP-ELDQTHVSTAVKGSFGYLDPEYFRRQQLTDKSDVYSFG 673
Query: 359 ILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDL 418
++L E+L R ++ E WA + +G + ++D ++ ++ D K +
Sbjct: 674 VVLFEVLCARPVIDPTLPREMVNLAEWAMKWQKKGQLDQIIDQSLRGNIRPDSLRKFAET 733
Query: 419 AFNCAAPVRSDRPDMKTVGEQLW 441
C A DRP M G+ LW
Sbjct: 734 GEKCLADYGVDRPSM---GDVLW 753
>At5g59700 receptor-like protein kinase precursor - like
Length = 829
Score = 239 bits (611), Expect = 2e-63
Identities = 132/289 (45%), Positives = 176/289 (60%), Gaps = 5/289 (1%)
Query: 153 SLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSS 212
S + L V +AT +F E IG GGFG VYK L DG VAVKRA + + L EF +
Sbjct: 467 SYRIPLVAVKEATNSFDENRAIGVGGFGKVYKGELHDGTKVAVKRANPKSQQGL-AEFRT 525
Query: 213 EVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAI 272
E+E+L++ HR+LV L+GY D+ NE IL+ E++ NGTL+ HL G L + QRLEI I
Sbjct: 526 EIEMLSQFRHRHLVSLIGYCDENNEMILVYEYMENGTLKSHLYGSGLLSLSWKQRLEICI 585
Query: 273 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVK 332
A GL YLH K +IHRDVKS+NILL E++ AKVADFG +K GP D TH+ST VK
Sbjct: 586 GSARGLHYLHTGDAKPVIHRDVKSANILLDENLMAKVADFGLSKTGP-EIDQTHVSTAVK 644
Query: 333 GTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNE 392
G+ GYLDPEY + LT KSDVYSFG+++ E+L R ++ + E WA + +
Sbjct: 645 GSFGYLDPEYFRRQQLTEKSDVYSFGVVMFEVLCARPVIDPTLTREMVNLAEWAMKWQKK 704
Query: 393 GSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
G + ++DP ++ ++ D K + C A DRP M G+ LW
Sbjct: 705 GQLEHIIDPSLRGKIRPDSLRKFGETGEKCLADYGVDRPSM---GDVLW 750
>At2g23200 putative protein kinase
Length = 834
Score = 235 bits (599), Expect = 4e-62
Identities = 160/412 (38%), Positives = 222/412 (53%), Gaps = 39/412 (9%)
Query: 61 STSQGDPKQVSSSQVGIFAGGALLVCCA------VVCPCFYGKRRKATSHAVLEK---DP 111
S S D SSS+V I G A+ A ++ F +RR + +E P
Sbjct: 386 SKSGSDYSNRSSSRVHIITGCAVAAAAASALVFSLLFMVFLKRRRSKKTKPEVEGTVWSP 445
Query: 112 NSMELGSSFEPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLS----QVSKATRN 167
+ G S SD P S P L++LHL L+ + AT N
Sbjct: 446 LPLHRGGS-----SDNRPISQYHNSP-------------LRNLHLGLTIPFTDILSATNN 487
Query: 168 FSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVK 227
F E L IG+GGFG VYKA L DG A+KR K + + EF +E+++L++I HR+LV
Sbjct: 488 FDEQLLIGKGGFGYVYKAILPDGTKAAIKRGKTGSGQGI-LEFQTEIQVLSRIRHRHLVS 546
Query: 228 LLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLY-AE 286
L GY ++ +E IL+ EF+ GTL+EHL G L + QRLEI I A GL YLH +E
Sbjct: 547 LTGYCEENSEMILVYEFMEKGTLKEHLYGSNLPSLTWKQRLEICIGAARGLDYLHSSGSE 606
Query: 287 KQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTY 346
IIHRDVKS+NILL E AKVADFG +K+ N D ++IS +KGT GYLDPEY++T+
Sbjct: 607 GAIIHRDVKSTNILLDEHNIAKVADFGLSKIH--NQDESNISINIKGTFGYLDPEYLQTH 664
Query: 347 HLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEA 406
LT KSDVY+FG++LLE+L R ++ EE W ++G++ +LDP +
Sbjct: 665 KLTEKSDVYAFGVVLLEVLFARPAIDPYLPHEEVNLSEWVMFCKSKGTIDEILDPSLIGQ 724
Query: 407 VKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIRADYLKSSSTTARRE 458
++T+ K ++A C +RP M+ V +W + L+ T RRE
Sbjct: 725 IETNSLKKFMEIAEKCLKEYGDERPSMRDV---IWDLEY-VLQLQMMTNRRE 772
>At5g28680 receptor-like protein kinase precursor - like
Length = 858
Score = 233 bits (595), Expect = 1e-61
Identities = 145/379 (38%), Positives = 215/379 (56%), Gaps = 17/379 (4%)
Query: 64 QGDPKQVSSSQVGIFAGGALLVCCAVVCPCFYGKRRKATSHAVLEKDPNSMELGSSFEPS 123
QGD K++++ +G G A ++ CA+ C Y ++RK + S SS+ P
Sbjct: 427 QGD-KRITAFVIGSAGGVAAVLFCAL-CFTMYQRKRKFSG---------SDSHTSSWLPI 475
Query: 124 VSDK-IPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTV 182
+ A+ + + S L+ +LS++ T NF E+ IG GGFG V
Sbjct: 476 YGNSHTSATKSTISGKSNNGSHLSNLAAGLCRRFSLSEIKHGTHNFDESNVIGVGGFGKV 535
Query: 183 YKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERILIT 242
YK +D G VA+K++ + L EF +E+ELL+++ H++LV L+GY D+G E LI
Sbjct: 536 YKGVIDGGTKVAIKKSNPNSEQGLN-EFETEIELLSRLRHKHLVSLIGYCDEGGEMCLIY 594
Query: 243 EFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLT 302
++++ GTLREHL + L + +RLEIAI A GL YLH A+ IIHRDVK++NILL
Sbjct: 595 DYMSLGTLREHLYNTKRPQLTWKRRLEIAIGAARGLHYLHTGAKYTIIHRDVKTTNILLD 654
Query: 303 ESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLL 362
E+ AKV+DFG +K GP N + H++T VKG+ GYLDPEY + LT KSDVYSFG++L
Sbjct: 655 ENWVAKVSDFGLSKTGP-NMNGGHVTTVVKGSFGYLDPEYFRRQQLTEKSDVYSFGVVLF 713
Query: 363 EILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDLAFNC 422
E+L R + S E+ WA +G++ ++DP ++ + + K D A C
Sbjct: 714 EVLCARPALNPSLSKEQVSLGDWAMNCKRKGTLEDIIDPNLKGKINPECLKKFADTAEKC 773
Query: 423 AAPVRSDRPDMKTVGEQLW 441
+ DRP T+G+ LW
Sbjct: 774 LSDSGLDRP---TMGDVLW 789
>At1g30570 putative serine/threonine protein kinase
Length = 849
Score = 231 bits (590), Expect = 5e-61
Identities = 123/286 (43%), Positives = 180/286 (62%), Gaps = 5/286 (1%)
Query: 158 LSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELL 217
L+++ AT+NF + L IG GGFG VY+ L+DG ++A+KRA H + EF +E+ +L
Sbjct: 510 LAEIRAATKNFDDGLAIGVGGFGKVYRGELEDGTLIAIKRAT-PHSQQGLAEFETEIVML 568
Query: 218 AKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHG 277
+++ HR+LV L+G+ D+ NE IL+ E++ANGTLR HL G L + QRLE I A G
Sbjct: 569 SRLRHRHLVSLIGFCDEHNEMILVYEYMANGTLRSHLFGSNLPPLSWKQRLEACIGSARG 628
Query: 278 LTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGY 337
L YLH +E+ IIHRDVK++NILL E+ AK++DFG +K GP + DHTH+ST VKG+ GY
Sbjct: 629 LHYLHTGSERGIIHRDVKTTNILLDENFVAKMSDFGLSKAGP-SMDHTHVSTAVKGSFGY 687
Query: 338 LDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVA 397
LDPEY + LT KSDVYSFG++L E + R + ++ WA + ++ +
Sbjct: 688 LDPEYFRRQQLTEKSDVYSFGVVLFEAVCARAVINPTLPKDQINLAEWALSWQKQRNLES 747
Query: 398 LLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAI 443
++D ++ + K ++A C A +RP M GE LW++
Sbjct: 748 IIDSNLRGNYSPESLEKYGEIAEKCLADEGKNRPMM---GEVLWSL 790
>At5g54380 receptor-protein kinase-like protein
Length = 855
Score = 230 bits (587), Expect = 1e-60
Identities = 155/402 (38%), Positives = 211/402 (51%), Gaps = 22/402 (5%)
Query: 46 STAGRKLLQKDLNNNSTSQGDPKQVSSSQVGIFAGGA-LLVCCAVVCPCFYGKRRKATSH 104
S +G ++ L S S+ K V +G G L++ AV C C RK S
Sbjct: 393 SLSGVSSVKSLLPGGSGSKSKKKAVI---IGSLVGAVTLILLIAVCCYCCLVASRKQRS- 448
Query: 105 AVLEKDPNSMELGSSFEPSVSDKIPA-SPLRVPPSPSRFSMSPKLSRLQSLHLN----LS 159
S + G + P + + S + S S + L S HL
Sbjct: 449 -------TSPQEGGNGHPWLPLPLYGLSQTLTKSTASHKSATASCISLASTHLGRCFMFQ 501
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAK 219
++ AT F E+ +G GGFG VYK L+DG VAVKR + + EF +E+E+L+K
Sbjct: 502 EIMDATNKFDESSLLGVGGFGRVYKGTLEDGTKVAVKRGNPRSEQGM-AEFRTEIEMLSK 560
Query: 220 IDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLT 279
+ HR+LV L+GY D+ +E IL+ E++ANG LR HL G L + QRLEI I A GL
Sbjct: 561 LRHRHLVSLIGYCDERSEMILVYEYMANGPLRSHLYGADLPPLSWKQRLEICIGAARGLH 620
Query: 280 YLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLD 339
YLH A + IIHRDVK++NILL E++ AKVADFG +K GP + D TH+ST VKG+ GYLD
Sbjct: 621 YLHTGASQSIIHRDVKTTNILLDENLVAKVADFGLSKTGP-SLDQTHVSTAVKGSFGYLD 679
Query: 340 PEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALL 399
PEY + LT KSDVYSFG++L+E+L R + E+ WA +G + ++
Sbjct: 680 PEYFRRQQLTEKSDVYSFGVVLMEVLCCRPALNPVLPREQVNIAEWAMAWQKKGLLDQIM 739
Query: 400 DPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
D + V K + A C A DRP M G+ LW
Sbjct: 740 DSNLTGKVNPASLKKFGETAEKCLAEYGVDRPSM---GDVLW 778
>At3g51550 receptor protein kinase like protein
Length = 895
Score = 229 bits (585), Expect = 2e-60
Identities = 127/289 (43%), Positives = 180/289 (61%), Gaps = 8/289 (2%)
Query: 155 HLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLV-VAVKRAKREHFESLRTEFSSE 213
H + +++ AT+NF E+ +G GGFG VY+ +D G VA+KR + + EF +E
Sbjct: 523 HFSFAEIKAATKNFDESRVLGVGGFGKVYRGEIDGGTTKVAIKRGNPMSEQGVH-EFQTE 581
Query: 214 VELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAID 273
+E+L+K+ HR+LV L+GY ++ E IL+ +++A+GT+REHL + L + QRLEI I
Sbjct: 582 IEMLSKLRHRHLVSLIGYCEENCEMILVYDYMAHGTMREHLYKTQNPSLPWKQRLEICIG 641
Query: 274 VAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKG 333
A GL YLH A+ IIHRDVK++NILL E AKV+DFG +K GP DHTH+ST VKG
Sbjct: 642 AARGLHYLHTGAKHTIIHRDVKTTNILLDEKWVAKVSDFGLSKTGP-TLDHTHVSTVVKG 700
Query: 334 TVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTL-RWAFRKYNE 392
+ GYLDPEY + LT KSDVYSFG++L E L RP A+E+V+L WA Y +
Sbjct: 701 SFGYLDPEYFRRQQLTEKSDVYSFGVVLFEALCA-RPALNPTLAKEQVSLAEWAPYCYKK 759
Query: 393 GSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
G + ++DP ++ + + K + A C +RP M G+ LW
Sbjct: 760 GMLDQIVDPYLKGKITPECFKKFAETAMKCVLDQGIERPSM---GDVLW 805
>At2g39360 putative protein kinase
Length = 815
Score = 229 bits (585), Expect = 2e-60
Identities = 126/285 (44%), Positives = 179/285 (62%), Gaps = 6/285 (2%)
Query: 158 LSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELL 217
L+ + +AT +F E+L IG GGFG VYK L D VAVKR + + L EF +EVE+L
Sbjct: 477 LALIKEATDDFDESLVIGVGGFGKVYKGVLRDKTEVAVKRGAPQSRQGL-AEFKTEVEML 535
Query: 218 AKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKI-LDFNQRLEIAIDVAH 276
+ HR+LV L+GY D+ +E I++ E++ GTL++HL L K L + QRLEI + A
Sbjct: 536 TQFRHRHLVSLIGYCDENSEMIIVYEYMEKGTLKDHLYDLDDKPRLSWRQRLEICVGAAR 595
Query: 277 GLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVG 336
GL YLH + + IIHRDVKS+NILL ++ AKVADFG +K GP + D TH+ST VKG+ G
Sbjct: 596 GLHYLHTGSTRAIIHRDVKSANILLDDNFMAKVADFGLSKTGP-DLDQTHVSTAVKGSFG 654
Query: 337 YLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVV 396
YLDPEY+ LT KSDVYSFG+++LE++ GR ++ E+ + WA + +G +
Sbjct: 655 YLDPEYLTRQQLTEKSDVYSFGVVMLEVVCGRPVIDPSLPREKVNLIEWAMKLVKKGKLE 714
Query: 397 ALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
++DP + VK + K ++ C + +RP M G+ LW
Sbjct: 715 DIIDPFLVGKVKLEEVKKYCEVTEKCLSQNGIERPAM---GDLLW 756
>At4g39110 putative receptor-like protein kinase
Length = 878
Score = 229 bits (584), Expect = 2e-60
Identities = 131/305 (42%), Positives = 184/305 (59%), Gaps = 7/305 (2%)
Query: 155 HLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEV 214
+ +LS++ +AT+NF + IG GGFG VY LDDG VAVKR + + + TEF +E+
Sbjct: 513 YFSLSELQEATKNFEASQIIGVGGFGNVYIGTLDDGTKVAVKRGNPQSEQGI-TEFQTEI 571
Query: 215 ELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDV 274
++L+K+ HR+LV L+GY D+ +E IL+ EF++NG R+HL G L + QRLEI I
Sbjct: 572 QMLSKLRHRHLVSLIGYCDENSEMILVYEFMSNGPFRDHLYGKNLAPLTWKQRLEICIGS 631
Query: 275 AHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGT 334
A GL YLH + IIHRDVKS+NILL E++ AKVADFG +K V H+ST VKG+
Sbjct: 632 ARGLHYLHTGTAQGIIHRDVKSTNILLDEALVAKVADFGLSK--DVAFGQNHVSTAVKGS 689
Query: 335 VGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGS 394
GYLDPEY + LT KSDVYSFG++LLE L R + + E+ WA + +G
Sbjct: 690 FGYLDPEYFRRQQLTDKSDVYSFGVVLLEALCARPAINPQLPREQVNLAEWAMQWKRKGL 749
Query: 395 VVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAIR-ADYLKSSST 453
+ ++DP + + + K + A C DRP T+G+ LW + A L+ + T
Sbjct: 750 LEKIIDPHLAGTINPESMKKFAEAAEKCLEDYGVDRP---TMGDVLWNLEYALQLQEAFT 806
Query: 454 TARRE 458
+ E
Sbjct: 807 QGKAE 811
>At3g46350 receptor-like protein kinase homolog
Length = 871
Score = 227 bits (578), Expect = 1e-59
Identities = 145/381 (38%), Positives = 205/381 (53%), Gaps = 27/381 (7%)
Query: 61 STSQGDPKQVSSSQVGIFAGGALLVCCAVVCPCFYGKRRKATSHAVLEKDPNSMELGSSF 120
STS K+ S V I A + V V F+G R+K TS V P+
Sbjct: 479 STSCNPKKKFSVMIVAIVASTVVFVL-VVSLALFFGLRKKKTSSHVKAIPPS-------- 529
Query: 121 EPSVSDKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVSKATRNFSETLQIGEGGFG 180
P +PL S S S ++ R + + S+V K T NF L GEGGFG
Sbjct: 530 --------PTTPLENVMSTSISETSIEMKRKK---FSYSEVMKMTNNFQRAL--GEGGFG 576
Query: 181 TVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDHRNLVKLLGYIDKGNERIL 240
TVY LD VAVK + + + EF +EV+LL ++ H NL+ L+GY D+ + L
Sbjct: 577 TVYHGDLDSSQQVAVKLLSQSSTQGYK-EFKAEVDLLLRVHHINLLNLVGYCDERDHLAL 635
Query: 241 ITEFVANGTLREHLDGLRG-KILDFNQRLEIAIDVAHGLTYLHLYAEKQIIHRDVKSSNI 299
I E+++NG L+ HL G G +L +N RL IA+D A GL YLH+ ++HRDVKS+NI
Sbjct: 636 IYEYMSNGDLKHHLSGEHGGSVLSWNIRLRIAVDAALGLEYLHIGCRPSMVHRDVKSTNI 695
Query: 300 LLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDPEYMKTYHLTPKSDVYSFGI 359
LL E+ AK+ADFG ++ + +H+ST V G++GYLDPEY +T L SDVYSFGI
Sbjct: 696 LLDENFMAKIADFGLSR-SFILGGESHVSTVVAGSLGYLDPEYYRTSRLAEMSDVYSFGI 754
Query: 360 LLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALLDPLMQEAVKTDVAVKMFDLA 419
+LLEI+T +R ++ K+ E+ W N G + ++DP + + + +LA
Sbjct: 755 VLLEIITNQRVID--KTREKPHITEWTAFMLNRGDITRIMDPNLNGDYNSHSVWRALELA 812
Query: 420 FNCAAPVRSDRPDMKTVGEQL 440
+CA P +RP M V +L
Sbjct: 813 MSCANPSSENRPSMSQVVAEL 833
>At2g21480 putative protein kinase
Length = 871
Score = 226 bits (577), Expect = 2e-59
Identities = 126/287 (43%), Positives = 175/287 (60%), Gaps = 6/287 (2%)
Query: 155 HLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEV 214
+ +LS++ + T+NF + IG GGFG VY +DDG VA+KR + + + TEF +E+
Sbjct: 512 YFSLSELQEVTKNFDASEIIGVGGFGNVYIGTIDDGTQVAIKRGNPQSEQGI-TEFHTEI 570
Query: 215 ELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDV 274
++L+K+ HR+LV L+GY D+ E IL+ E+++NG R+HL G L + QRLEI I
Sbjct: 571 QMLSKLRHRHLVSLIGYCDENAEMILVYEYMSNGPFRDHLYGKNLSPLTWKQRLEICIGA 630
Query: 275 AHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGT 334
A GL YLH + IIHRDVKS+NILL E++ AKVADFG +K V H+ST VKG+
Sbjct: 631 ARGLHYLHTGTAQGIIHRDVKSTNILLDEALVAKVADFGLSK--DVAFGQNHVSTAVKGS 688
Query: 335 VGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGS 394
GYLDPEY + LT KSDVYSFG++LLE L R + + E+ WA +G
Sbjct: 689 FGYLDPEYFRRQQLTDKSDVYSFGVVLLEALCARPAINPQLPREQVNLAEWAMLWKQKGL 748
Query: 395 VVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
+ ++DP + AV + K + A C A DRP T+G+ LW
Sbjct: 749 LEKIIDPHLVGAVNPESMKKFAEAAEKCLADYGVDRP---TMGDVLW 792
>At5g49770 receptor protein kinase-like
Length = 946
Score = 224 bits (570), Expect = 1e-58
Identities = 123/285 (43%), Positives = 182/285 (63%), Gaps = 4/285 (1%)
Query: 160 QVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAK 219
++SK T NFS+ +G GG+G VYK L +G V+A+KRA++ + EF +E+ELL++
Sbjct: 626 ELSKCTNNFSDANDVGGGGYGQVYKGTLPNGQVIAIKRAQQGSMQGA-FEFKTEIELLSR 684
Query: 220 IDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGLT 279
+ H+N+VKLLG+ E++L+ E++ NG+LR+ L G G LD+ +RL+IA+ GL
Sbjct: 685 VHHKNVVKLLGFCFDQKEQMLVYEYIPNGSLRDGLSGKNGVKLDWTRRLKIALGSGKGLA 744
Query: 280 YLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLD 339
YLH A+ IIHRDVKS+NILL E + AKVADFG +KL + + H++T+VKGT+GYLD
Sbjct: 745 YLHELADPPIIHRDVKSNNILLDEHLTAKVADFGLSKL-VGDPEKAHVTTQVKGTMGYLD 803
Query: 340 PEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGSVVALL 399
PEY T LT KSDVY FG+++LE+LTG+ P++ + V + + N + LL
Sbjct: 804 PEYYMTNQLTEKSDVYGFGVVMLELLTGKSPIDRGSYVVKEVKKKMD-KSRNLYDLQELL 862
Query: 400 D-PLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAI 443
D ++Q + K D+A C P +RP M V ++L +I
Sbjct: 863 DTTIIQNSGNLKGFEKYVDVALQCVEPEGVNRPTMSEVVQELESI 907
>At1g06840 receptor protein kinase, putative
Length = 939
Score = 223 bits (569), Expect = 1e-58
Identities = 123/286 (43%), Positives = 175/286 (61%), Gaps = 11/286 (3%)
Query: 159 SQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLA 218
++++ AT NF+ + QIG+GG+G VYK L G VVA+KRA+ + + EF +E+ELL+
Sbjct: 602 AELALATDNFNSSTQIGQGGYGKVYKGTLGSGTVVAIKRAQEGSLQGEK-EFLTEIELLS 660
Query: 219 KIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRLEIAIDVAHGL 278
++ HRNLV LLG+ D+ E++L+ E++ NGTLR+++ + LDF RL IA+ A G+
Sbjct: 661 RLHHRNLVSLLGFCDEEGEQMLVYEYMENGTLRDNISVKLKEPLDFAMRLRIALGSAKGI 720
Query: 279 TYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNN----DHTHISTKVKGT 334
YLH A I HRD+K+SNILL AKVADFG ++L PV + H+ST VKGT
Sbjct: 721 LYLHTEANPPIFHRDIKASNILLDSRFTAKVADFGLSRLAPVPDMEGISPQHVSTVVKGT 780
Query: 335 VGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFRKYNEGS 394
GYLDPEY T+ LT KSDVYS G++LLE+ TG +P+ K+ + + Y GS
Sbjct: 781 PGYLDPEYFLTHQLTDKSDVYSLGVVLLELFTGMQPITHGKNIVREINI-----AYESGS 835
Query: 395 VVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQL 440
+++ +D M +V + K LA C RP M V +L
Sbjct: 836 ILSTVDKRM-SSVPDECLEKFATLALRCCREETDARPSMAEVVREL 880
>At5g56890 unknown protein
Length = 1113
Score = 223 bits (568), Expect = 2e-58
Identities = 148/402 (36%), Positives = 217/402 (53%), Gaps = 32/402 (7%)
Query: 50 RKLLQKDLNNNSTSQGDPKQVSSSQVGIFAGGALLVCCAVVCPCFYGKRRKATSHAVLEK 109
RK +K+LN S + V + + A + C V+ F RR+ + ++
Sbjct: 617 RKPRKKELNGGSIA-----------VIVLSAAAFIGLCFVIV-WFLVFRRQRDRRRLSKR 664
Query: 110 DPNSM-ELGSSFEPSVS------DKIPASPLRVPPSPSRFSMSPKLSRLQSLHLNLSQVS 162
P + L S +PS S + ++ L S + F++S K S++
Sbjct: 665 TPLARPSLPSLSKPSGSARSLTGSRFSSTSLSFESSIAPFTLSAKT-------FTASEIM 717
Query: 163 KATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRTEFSSEVELLAKIDH 222
KAT NF E+ +GEGGFG VY+ DDG VAVK KR+ + R EF +EVE+L+++ H
Sbjct: 718 KATNNFDESRVLGEGGFGRVYEGVFDDGTKVAVKVLKRDDQQGSR-EFLAEVEMLSRLHH 776
Query: 223 RNLVKLLGYIDKGNERILITEFVANGTLREHLDGL--RGKILDFNQRLEIAIDVAHGLTY 280
RNLV L+G + R L+ E + NG++ HL G+ LD++ RL+IA+ A GL Y
Sbjct: 777 RNLVNLIGICIEDRNRSLVYELIPNGSVESHLHGIDKASSPLDWDARLKIALGAARGLAY 836
Query: 281 LHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVKGTVGYLDP 340
LH + ++IHRD KSSNILL KV+DFG A+ + D+ HIST+V GT GY+ P
Sbjct: 837 LHEDSSPRVIHRDFKSSNILLENDFTPKVSDFGLARNALDDEDNRHISTRVMGTFGYVAP 896
Query: 341 EYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWA--FRKYNEGSVVAL 398
EY T HL KSDVYS+G++LLE+LTGR+PV++ + + + W F EG + A+
Sbjct: 897 EYAMTGHLLVKSDVYSYGVVLLELLTGRKPVDMSQPPGQENLVSWTRPFLTSAEG-LAAI 955
Query: 399 LDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQL 440
+D + + D K+ +A C P S RP M V + L
Sbjct: 956 IDQSLGPEISFDSIAKVAAIASMCVQPEVSHRPFMGEVVQAL 997
>At5g24010 receptor-protein kinase-like protein
Length = 824
Score = 223 bits (567), Expect = 2e-58
Identities = 124/293 (42%), Positives = 173/293 (58%), Gaps = 5/293 (1%)
Query: 149 SRLQSLHLNLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGLVVAVKRAKREHFESLRT 208
S +L ++ +++ T NF +L IG GGFG V++ L D VAVKR + L
Sbjct: 470 SGYHTLRISFAELQSGTNNFDRSLVIGVGGFGMVFRGSLKDNTKVAVKRGSPGSRQGL-P 528
Query: 209 EFSSEVELLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHLDGLRGKILDFNQRL 268
EF SE+ +L+KI HR+LV L+GY ++ +E IL+ E++ G L+ HL G L + QRL
Sbjct: 529 EFLSEITILSKIRHRHLVSLVGYCEEQSEMILVYEYMDKGPLKSHLYGSTNPPLSWKQRL 588
Query: 269 EIAIDVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHIS 328
E+ I A GL YLH + + IIHRD+KS+NILL + AKVADFG ++ GP D TH+S
Sbjct: 589 EVCIGAARGLHYLHTGSSQGIIHRDIKSTNILLDNNYVAKVADFGLSRSGPC-IDETHVS 647
Query: 329 TKVKGTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTLRWAFR 388
T VKG+ GYLDPEY + LT KSDVYSFG++L E+L R V+ E+ WA
Sbjct: 648 TGVKGSFGYLDPEYFRRQQLTDKSDVYSFGVVLFEVLCARPAVDPLLVREQVNLAEWAIE 707
Query: 389 KYNEGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLW 441
+G + ++DP + + +K K + A C A DRP T+G+ LW
Sbjct: 708 WQRKGMLDQIVDPNIADEIKPCSLKKFAETAEKCCADYGVDRP---TIGDVLW 757
>At5g39000 protein kinase - like protein
Length = 873
Score = 221 bits (563), Expect = 6e-58
Identities = 122/292 (41%), Positives = 181/292 (61%), Gaps = 10/292 (3%)
Query: 157 NLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGL-VVAVKRAKREHFESLRTEFSSEVE 215
++ ++ AT +F + L IG GGFG+VYK +D G +VAVKR + + + EF +E+E
Sbjct: 507 SIFEIKSATNDFEDKLIIGVGGFGSVYKGQIDGGATLVAVKRLEITSNQGAK-EFETELE 565
Query: 216 LLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHL---DGLRGKILDFNQRLEIAI 272
+L+K+ H +LV L+GY D+ NE +L+ E++ +GTL++HL D L + +RLEI I
Sbjct: 566 MLSKLRHVHLVSLIGYCDEDNEMVLVYEYMPHGTLKDHLFRRDKTSDPPLSWKRRLEICI 625
Query: 273 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVK 332
A GL YLH A+ IIHRD+K++NILL E+ KV+DFG +++GP + TH+ST VK
Sbjct: 626 GAARGLQYLHTGAKYTIIHRDIKTTNILLDENFVTKVSDFGLSRVGPTSASQTHVSTVVK 685
Query: 333 GTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTL-RWAFRKYN 391
GT GYLDPEY + LT KSDVYSFG++LLE+L RP+ ++ E+ L RW Y
Sbjct: 686 GTFGYLDPEYYRRQVLTEKSDVYSFGVVLLEVLC-CRPIRMQSVPPEQADLIRWVKSNYR 744
Query: 392 EGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAI 443
G+V ++D + + + K ++A C +RP M V +WA+
Sbjct: 745 RGTVDQIIDSDLSADITSTSLEKFCEIAVRCVQDRGMERPPMNDV---VWAL 793
>At5g38990 protein kinase - like protein
Length = 880
Score = 221 bits (563), Expect = 6e-58
Identities = 123/292 (42%), Positives = 182/292 (62%), Gaps = 10/292 (3%)
Query: 157 NLSQVSKATRNFSETLQIGEGGFGTVYKAHLDDGL-VVAVKRAKREHFESLRTEFSSEVE 215
++ ++ AT +F E L IG GGFG+VYK +D G +VAVKR + + + EF +E+E
Sbjct: 514 SIYEIKSATNDFEEKLIIGVGGFGSVYKGRIDGGATLVAVKRLEITSNQGAK-EFDTELE 572
Query: 216 LLAKIDHRNLVKLLGYIDKGNERILITEFVANGTLREHL---DGLRGKILDFNQRLEIAI 272
+L+K+ H +LV L+GY D NE +L+ E++ +GTL++HL D L + +RLEI I
Sbjct: 573 MLSKLRHVHLVSLIGYCDDDNEMVLVYEYMPHGTLKDHLFRRDKASDPPLSWKRRLEICI 632
Query: 273 DVAHGLTYLHLYAEKQIIHRDVKSSNILLTESMRAKVADFGFAKLGPVNNDHTHISTKVK 332
A GL YLH A+ IIHRD+K++NILL E+ AKV+DFG +++GP + TH+ST VK
Sbjct: 633 GAARGLQYLHTGAKYTIIHRDIKTTNILLDENFVAKVSDFGLSRVGPTSASQTHVSTVVK 692
Query: 333 GTVGYLDPEYMKTYHLTPKSDVYSFGILLLEILTGRRPVELKKSAEERVTL-RWAFRKYN 391
GT GYLDPEY + LT KSDVYSFG++LLE+L RP+ ++ E+ L RW +N
Sbjct: 693 GTFGYLDPEYYRRQILTEKSDVYSFGVVLLEVLC-CRPIRMQSVPPEQADLIRWVKSNFN 751
Query: 392 EGSVVALLDPLMQEAVKTDVAVKMFDLAFNCAAPVRSDRPDMKTVGEQLWAI 443
+ +V ++D + + + K ++A C +RP M V +WA+
Sbjct: 752 KRTVDQIIDSDLTADITSTSMEKFCEIAIRCVQDRGMERPPMNDV---VWAL 800
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.134 0.381
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 9,722,571
Number of Sequences: 26719
Number of extensions: 409190
Number of successful extensions: 4695
Number of sequences better than 10.0: 1013
Number of HSP's better than 10.0 without gapping: 863
Number of HSP's successfully gapped in prelim test: 151
Number of HSP's that attempted gapping in prelim test: 1299
Number of HSP's gapped (non-prelim): 1152
length of query: 458
length of database: 11,318,596
effective HSP length: 103
effective length of query: 355
effective length of database: 8,566,539
effective search space: 3041121345
effective search space used: 3041121345
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 62 (28.5 bits)
Medicago: description of AC144657.5


