

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144657.4 + phase: 0
(776 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
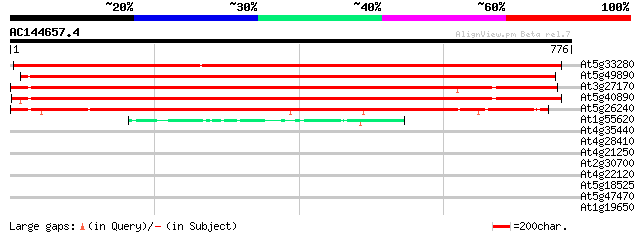
Score E
Sequences producing significant alignments: (bits) Value
At5g33280 chloride channel-like protein 1118 0.0
At5g49890 chloride channel (emb|CAA70310.1) 924 0.0
At3g27170 CLC-b chloride channel protein 823 0.0
At5g40890 anion channel protein (gb|AAC05742.1) 811 0.0
At5g26240 CLC-d chloride channel protein 614 e-176
At1g55620 CLC-f chloride channel protein 49 1e-05
At4g35440 CLC-e chloride channel protein 38 0.019
At4g28410 unknown protein 36 0.094
At4g21250 hypothetical protein 32 1.8
At2g30700 unknown protein 31 2.3
At4g22120 unknown protein 31 3.0
At5g18525 unknown protein 30 5.2
At5g47470 nodulin-like protein 29 8.8
At1g19650 sec14 cytosolic factor- like protein 29 8.8
>At5g33280 chloride channel-like protein
Length = 763
Score = 1118 bits (2891), Expect = 0.0
Identities = 549/758 (72%), Positives = 642/758 (84%), Gaps = 2/758 (0%)
Query: 6 LSNGESEPLLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGV 65
+ N +E + PLL S R NSTSQVAIVGANVCPIESLDYEI EN+FFKQDWR R
Sbjct: 1 MPNSTTEDSVAVPLLPSLRRATNSTSQVAIVGANVCPIESLDYEIAENDFFKQDWRGRSK 60
Query: 66 VQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFF 125
V+I QY+ MKWLLCF IG+IV IGF NNLAVENLAG+KFV TSNMM+ RF F +F
Sbjct: 61 VEIFQYVFMKWLLCFCIGIIVSLIGFANNLAVENLAGVKFVVTSNMMIAGRFAMGFVVFS 120
Query: 126 ASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSG 185
+NL LTLFAS+ITAF+AP AAGSGI EVKAYLNGVDAP IF++RTL +KIIG+I+AVS
Sbjct: 121 VTNLILTLFASVITAFVAPAAAGSGIPEVKAYLNGVDAPEIFSLRTLIIKIIGNISAVSA 180
Query: 186 SLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAA 245
SL+IGKAGPMVHTGACVA++LGQGGSKRY +TWRWLRFFKNDRDRRDL+ CG+AAGIAA+
Sbjct: 181 SLLIGKAGPMVHTGACVASILGQGGSKRYRLTWRWLRFFKNDRDRRDLVTCGAAAGIAAS 240
Query: 246 FRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIM 305
FRAPVGGVLFALEEM+SW +ALLWR FF+TA VAI LRA+IDVCLS KCGLFGKGGLIM
Sbjct: 241 FRAPVGGVLFALEEMSSW--SALLWRIFFSTAVVAIVLRALIDVCLSGKCGLFGKGGLIM 298
Query: 306 FDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACL 365
FD YS + SYHL DV PV +L VVGG+LGSL+NF+ +KVLR YN I EKG ++ LAC
Sbjct: 299 FDVYSENASYHLGDVLPVLLLGVVGGILGSLYNFLLDKVLRAYNYIYEKGVTWKILLACA 358
Query: 366 ISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTND 425
ISIFTSCLLFGLP+LA C+PCP DA+E CPTIGRSG +KK+QCPP HYN LASLIFNTND
Sbjct: 359 ISIFTSCLLFGLPFLASCQPCPVDALEECPTIGRSGNFKKYQCPPGHYNDLASLIFNTND 418
Query: 426 DAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGI 485
DAI+NLFS +TD EF S+LVFF+ C FLSIFS GIVAPAGLFVP+IVTGASYGR VG+
Sbjct: 419 DAIKNLFSKNTDFEFHYFSVLVFFVTCFFLSIFSYGIVAPAGLFVPVIVTGASYGRFVGM 478
Query: 486 LVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVA 545
L+G +NL++GL+AVLGAAS LGG+MR TVS CVI+LELTNNLLLLP++M+VL++SK+VA
Sbjct: 479 LLGSNSNLNHGLFAVLGAASFLGGTMRMTVSTCVILLELTNNLLLLPMMMVVLLISKTVA 538
Query: 546 NVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTT 605
+ FNAN+Y+LIMK KG PYL +HAEPYMRQL VGDVVTGPLQ+FNGIEKV IV +L+TT
Sbjct: 539 DGFNANIYNLIMKLKGFPYLYSHAEPYMRQLLVGDVVTGPLQVFNGIEKVETIVHVLKTT 598
Query: 606 AHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKK 665
HNGFPV+D PP + AP+L G+ILR H+ TLLKK+ F+PSPVA + + +F +++FAKK
Sbjct: 599 NHNGFPVVDGPPLAAAPVLHGLILRAHILTLLKKRVFMPSPVACDSNTLSQFKAEEFAKK 658
Query: 666 YSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIP 725
S KIED++L+EEE+ M++DLHPF+NASPYTVVETMSLAKALILFREVG+RHLLVIP
Sbjct: 659 GSGRSDKIEDVELSEEELNMYLDLHPFSNASPYTVVETMSLAKALILFREVGIRHLLVIP 718
Query: 726 KIPGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLR 763
K R PVVGILTRHDF PEHILG+HP + +S+WKRLR
Sbjct: 719 KTSNRPPVVGILTRHDFMPEHILGLHPSVSRSKWKRLR 756
>At5g49890 chloride channel (emb|CAA70310.1)
Length = 779
Score = 924 bits (2387), Expect = 0.0
Identities = 452/740 (61%), Positives = 557/740 (75%), Gaps = 3/740 (0%)
Query: 16 RRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDWRSRGVVQILQYICMK 75
R+PLL+ R N+TSQ+AIVGAN CPIESLDYEIFEN+FFKQDWRSR ++ILQY +K
Sbjct: 38 RQPLLARNRK--NTTSQIAIVGANTCPIESLDYEIFENDFFKQDWRSRKKIEILQYTFLK 95
Query: 76 WLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFAFFIFFASNLSLTLFA 135
W L F+IGL G +GF NNL VEN+AG K + N+ML+ ++ AFF F NL L A
Sbjct: 96 WALAFLIGLATGLVGFLNNLGVENIAGFKLLLIGNLMLKEKYFQAFFAFAGCNLILATAA 155
Query: 136 SIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPM 195
+ + AFIAP AAGSGI EVKAYLNG+DA I TL VKI GSI V+ V+GK GPM
Sbjct: 156 ASLCAFIAPAAAGSGIPEVKAYLNGIDAYSILAPSTLFVKIFGSIFGVAAGFVVGKEGPM 215
Query: 196 VHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLF 255
VHTGAC+A LLGQGGSK+Y +TW+WLRFFKNDRDRRDLI CG+AAG+AAAFRAPVGGVLF
Sbjct: 216 VHTGACIANLLGQGGSKKYRLTWKWLRFFKNDRDRRDLITCGAAAGVAAAFRAPVGGVLF 275
Query: 256 ALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASISY 315
ALEE ASWWR ALLWR FFTTA VA+ LR++I+ C S +CGLFGKGGLIMFD S + Y
Sbjct: 276 ALEEAASWWRNALLWRTFFTTAVVAVVLRSLIEFCRSGRCGLFGKGGLIMFDVNSGPVLY 335
Query: 316 HLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLF 375
D+ + L V+GG+LGSL+N++ +KVLR Y++INEKG ++ L +SI +SC F
Sbjct: 336 STPDLLAIVFLGVIGGVLGSLYNYLVDKVLRTYSIINEKGPRFKIMLVMAVSILSSCCAF 395
Query: 376 GLPWLAPCRPCPPDAVE-PCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSM 434
GLPWL+ C PCP E CP++GRS IYK FQCPPNHYN L+SL+ NTNDDAIRNLF+
Sbjct: 396 GLPWLSQCTPCPIGIEEGKCPSVGRSSIYKSFQCPPNHYNDLSSLLLNTNDDAIRNLFTS 455
Query: 435 HTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYGRLVGILVGERTNLS 494
++NEF +S++ +FF+ L I + GI P+GLF+P+I+ GASYGRLVG L+G + L
Sbjct: 456 RSENEFHISTLAIFFVAVYCLGIITYGIAIPSGLFIPVILAGASYGRLVGRLLGPVSQLD 515
Query: 495 NGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNANVYD 554
GL+++LGAAS LGG+MR TVSLCVI+LELTNNLL+LPL+M+VL++SK+VA+ FN VYD
Sbjct: 516 VGLFSLLGAASFLGGTMRMTVSLCVILLELTNNLLMLPLVMLVLLISKTVADCFNRGVYD 575
Query: 555 LIMKAKGLPYLETHAEPYMRQLTVGDVVTGPLQMFNGIEKVRNIVFILRTTAHNGFPVID 614
I+ KGLPY+E HAEPYMR L DVV+G L F+ +EKV I L+ T HNGFPVID
Sbjct: 576 QIVTMKGLPYMEDHAEPYMRNLVAKDVVSGALISFSRVEKVGVIWQALKMTRHNGFPVID 635
Query: 615 EPPGSEAPILFGIILRHHLTTLLKKKAFLPSPVANSYDVVRKFSSDDFAKKYSVERVKIE 674
EPP +EA L GI LR HL LL+ K F ++R + DF K + +KIE
Sbjct: 636 EPPFTEASELCGIALRSHLLVLLQGKKFSKQRTTFGSQILRSCKARDFGKAGLGKGLKIE 695
Query: 675 DIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILFREVGLRHLLVIPKIPGRSPVV 734
D+ L+EEEM M+VDLHP TN SPYTV+ET+SLAKA ILFR++GLRHL V+PK PGR P+V
Sbjct: 696 DLDLSEEEMEMYVDLHPITNTSPYTVLETLSLAKAAILFRQLGLRHLCVVPKTPGRPPIV 755
Query: 735 GILTRHDFTPEHILGMHPFL 754
GILTRHDF PEH+LG++P +
Sbjct: 756 GILTRHDFMPEHVLGLYPHI 775
>At3g27170 CLC-b chloride channel protein
Length = 780
Score = 823 bits (2126), Expect = 0.0
Identities = 408/769 (53%), Positives = 547/769 (71%), Gaps = 18/769 (2%)
Query: 2 SNNHLSNGESEP-LLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFFKQDW 60
SN + G+ E L +PL+ + R++ S++ +A+VGA V IESLDYEI EN+ FK DW
Sbjct: 13 SNYNGEGGDPESNTLNQPLVKANRTL--SSTPLALVGAKVSHIESLDYEINENDLFKHDW 70
Query: 61 RSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERRFMFA 120
R R Q+LQY+ +KW L ++GL G I NLAVEN+AG K + + + + R++
Sbjct: 71 RKRSKAQVLQYVFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGHFLTQERYVTG 130
Query: 121 FFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSI 180
+ +NL LTL AS++ APTAAG GI E+KAYLNGVD P +F T+ VKI+GSI
Sbjct: 131 LMVLVGANLGLTLVASVLCVCFAPTAAGPGIPEIKAYLNGVDTPNMFGATTMIVKIVGSI 190
Query: 181 TAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAA 240
AV+ L +GK GP+VH G+C+A+LLGQGG+ + I WRWLR+F NDRDRRDLI CGSAA
Sbjct: 191 GAVAAGLDLGKEGPLVHIGSCIASLLGQGGTDNHRIKWRWLRYFNNDRDRRDLITCGSAA 250
Query: 241 GIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGK 300
G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA V + LR I++C S KCGLFGK
Sbjct: 251 GVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLREFIEICNSGKCGLFGK 310
Query: 301 GGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRL 360
GGLIMFD + +YH+ D+ PV ++ V+GG+LGSL+N + +KVLR+YN+INEKG I ++
Sbjct: 311 GGLIMFDVSHVTYTYHVTDIIPVMLIGVIGGILGSLYNHLLHKVLRLYNLINEKGKIHKV 370
Query: 361 FLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLI 420
L+ +S+FTS L+GLP+LA C+PC P E CPT GRSG +K+F CP +YN LA+L+
Sbjct: 371 LLSLTVSLFTSVCLYGLPFLAKCKPCDPSIDEICPTNGRSGNFKQFHCPKGYYNDLATLL 430
Query: 421 FNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTGASYG 480
TNDDA+RNLFS +T NEF + S+ +FF++ L +F+ GI P+GLF+PII+ GA+YG
Sbjct: 431 LTTNDDAVRNLFSSNTPNEFGMGSLWIFFVLYCILGLFTFGIATPSGLFLPIILMGAAYG 490
Query: 481 RLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVV 540
R++G +G T++ GLYAVLGAA+L+ GSMR TVSLCVI LELTNNLLLLP+ M+VL++
Sbjct: 491 RMLGAAMGSYTSIDQGLYAVLGAAALMAGSMRMTVSLCVIFLELTNNLLLLPITMIVLLI 550
Query: 541 SKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEKVRNI 598
+K+V + FN ++YD+I+ KGLP+LE + EP+MR LTVG++ P+ G+EKV NI
Sbjct: 551 AKTVGDSFNPSIYDIILHLKGLPFLEANPEPWMRNLTVGELGDAKPPVVTLQGVEKVSNI 610
Query: 599 VFILRTTAHNGFPVIDEPP------GSEAPILFGIILRHHLTTLLKKKAFLPSP-VANSY 651
V +L+ T HN FPV+DE + A L G+ILR HL +LKK+ FL +
Sbjct: 611 VDVLKNTTHNAFPVLDEAEVPQVGLATGATELHGLILRAHLVKVLKKRWFLTEKRRTEEW 670
Query: 652 DVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALI 711
+V KF D+ A++ +D+ +T EM M+VDLHP TN +PYTV+E MS+AKAL+
Sbjct: 671 EVREKFPWDELAERED----NFDDVAITSAEMEMYVDLHPLTNTTPYTVMENMSVAKALV 726
Query: 712 LFREVGLRHLLVIPKI--PGRSPVVGILTRHDFTPEHILGMHPFLVKSR 758
LFR+VGLRHLL++PKI G PVVGILTR D +IL P L KS+
Sbjct: 727 LFRQVGLRHLLIVPKIQASGMCPVVGILTRQDLRAYNILQAFPLLEKSK 775
>At5g40890 anion channel protein (gb|AAC05742.1)
Length = 775
Score = 811 bits (2096), Expect = 0.0
Identities = 406/772 (52%), Positives = 540/772 (69%), Gaps = 17/772 (2%)
Query: 3 NNHLSNGESEP------LLRRPLLSSQRSIINSTSQVAIVGANVCPIESLDYEIFENEFF 56
+N NGE E L +PLL R++ S++ +A+VGA V IESLDYEI EN+ F
Sbjct: 10 SNSNYNGEEEGEDPENNTLNQPLLKRHRTL--SSTPLALVGAKVSHIESLDYEINENDLF 67
Query: 57 KQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERR 116
K DWRSR Q+ QYI +KW L ++GL G I NLAVEN+AG K + + + R
Sbjct: 68 KHDWRSRSKAQVFQYIFLKWTLACLVGLFTGLIATLINLAVENIAGYKLLAVGYYIAQDR 127
Query: 117 FMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKI 176
F +F +NL LTL A+++ + APTAAG GI E+KAYLNG+D P +F T+ VKI
Sbjct: 128 FWTGLMVFTGANLGLTLVATVLVVYFAPTAAGPGIPEIKAYLNGIDTPNMFGFTTMMVKI 187
Query: 177 IGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIIC 236
+GSI AV+ L +GK GP+VH G+C+A+LLGQGG + I WRWLR+F NDRDRRDLI C
Sbjct: 188 VGSIGAVAAGLDLGKEGPLVHIGSCIASLLGQGGPDNHRIKWRWLRYFNNDRDRRDLITC 247
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCG 296
GSA+G+ AAFR+PVGGVLFALEE+A+WWR+ALLWR FF+TA V + LRA I++C S KCG
Sbjct: 248 GSASGVCAAFRSPVGGVLFALEEVATWWRSALLWRTFFSTAVVVVVLRAFIEICNSGKCG 307
Query: 297 LFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLRIYNVINEKGT 356
LFG GGLIMFD + YH D+ PV ++ V GG+LGSL+N + +KVLR+YN+IN+KG
Sbjct: 308 LFGSGGLIMFDVSHVEVRYHAADIIPVTLIGVFGGILGSLYNHLLHKVLRLYNLINQKGK 367
Query: 357 ICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGL 416
I ++ L+ +S+FTS LFGLP+LA C+PC P E CPT GRSG +K+F CP +YN L
Sbjct: 368 IHKVLLSLGVSLFTSVCLFGLPFLAECKPCDPSIDEICPTNGRSGNFKQFNCPNGYYNDL 427
Query: 417 ASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFVPIIVTG 476
++L+ TNDDA+RN+FS +T NEF + S+ +FF + L + + GI P+GLF+PII+ G
Sbjct: 428 STLLLTTNDDAVRNIFSSNTPNEFGMVSLWIFFGLYCILGLITFGIATPSGLFLPIILMG 487
Query: 477 ASYGRLVGILVGERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMM 536
++YGR++G +G TN+ GLYAVLGAASL+ GSMR TVSLCVI LELTNNLLLLP+ M
Sbjct: 488 SAYGRMLGTAMGSYTNIDQGLYAVLGAASLMAGSMRMTVSLCVIFLELTNNLLLLPITMF 547
Query: 537 VLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDV--VTGPLQMFNGIEK 594
VL+++K+V + FN ++Y++I+ KGLP+LE + EP+MR LTVG++ P+ NG+EK
Sbjct: 548 VLLIAKTVGDSFNLSIYEIILHLKGLPFLEANPEPWMRNLTVGELNDAKPPVVTLNGVEK 607
Query: 595 VRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKAFL-PSPVANSYDV 653
V NIV +LR T HN FPV+D + L G+ILR HL +LKK+ FL ++V
Sbjct: 608 VANIVDVLRNTTHNAFPVLDGADQNTGTELHGLILRAHLVKVLKKRWFLNEKRRTEEWEV 667
Query: 654 VRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVETMSLAKALILF 713
KF+ + A++ +D+ +T EM ++VDLHP TN +PYTVV++MS+AKAL+LF
Sbjct: 668 REKFTPVELAERED----NFDDVAITSSEMQLYVDLHPLTNTTPYTVVQSMSVAKALVLF 723
Query: 714 REVGLRHLLVIPKI--PGRSPVVGILTRHDFTPEHILGMHPFLVKSRWKRLR 763
R VGLRHLLV+PKI G SPV+GILTR D +IL P L K + + R
Sbjct: 724 RSVGLRHLLVVPKIQASGMSPVIGILTRQDLRAYNILQAFPHLDKHKSGKAR 775
>At5g26240 CLC-d chloride channel protein
Length = 792
Score = 614 bits (1583), Expect = e-176
Identities = 344/763 (45%), Positives = 479/763 (62%), Gaps = 28/763 (3%)
Query: 1 MSNNHLSNG-ESEPLLRRPLLSSQRSIINSTSQVAIVGANVCP---IESLDYEIFENEFF 56
M +NHL NG ES+ LL + S + ST + ++ ++ + SLDYE+ EN +
Sbjct: 1 MLSNHLQNGIESDNLLWSRVPESDDT---STDDITLLNSHRDGDGGVNSLDYEVIENYAY 57
Query: 57 KQDWRSRGVVQILQYICMKWLLCFMIGLIVGFIGFCNNLAVENLAGIKFVTTSNMMLERR 116
+++ RG + + Y+ +KW +IG+ G NL+VEN AG KF T ++++
Sbjct: 58 REEQAHRGKLYVGYYVAVKWFFSLLIGIGTGLAAVFINLSVENFAGWKFALTF-AIIQKS 116
Query: 117 FMFAFFIFFASNLSLTLFASIITAFIAPTAAGSGISEVKAYLNGVDAPGIFTVRTLCVKI 176
+ F ++ NL L ++ I AP AAGSGI E+K YLNG+D PG RTL KI
Sbjct: 117 YFAGFIVYLLINLVLVFSSAYIITQFAPAAAGSGIPEIKGYLNGIDIPGTLLFRTLIGKI 176
Query: 177 IGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFFKNDRDRRDLIIC 236
GSI +V G L +GK GP+VHTGAC+A+LLGQGGS +Y + RW + FK+DRDRRDL+ C
Sbjct: 177 FGSIGSVGGGLALGKEGPLVHTGACIASLLGQGGSTKYHLNSRWPQLFKSDRDRRDLVTC 236
Query: 237 GSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFLRAMIDVCLSDKCG 296
G AAG+AAAFRAPVGGVLFALEE+ SWWR+ L+WR FFT+A VA+ +R + C S CG
Sbjct: 237 GCAAGVAAAFRAPVGGVLFALEEVTSWWRSQLMWRVFFTSAIVAVVVRTAMGWCKSGICG 296
Query: 297 LFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNKVLR-IYNVINEKG 355
FG GG I++D Y+ ++ P+ ++ V+GGLLG+LFN +T + N +++KG
Sbjct: 297 HFGGGGFIIWDVSDGQDDYYFKELLPMAVIGVIGGLLGALFNQLTLYMTSWRRNSLHKKG 356
Query: 356 TICRLFLACLISIFTSCLLFGLPWLAPCRPCP---PDAVEPCP-TIGRSGIYKKFQC-PP 410
++ AC+IS TS + FGLP L C PCP PD+ CP G G Y F C
Sbjct: 357 NRVKIIEACIISCITSAISFGLPLLRKCSPCPESVPDSGIECPRPPGMYGNYVNFFCKTD 416
Query: 411 NHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIVAPAGLFV 470
N YN LA++ FNT DDAIRNLFS T EF S+L F + L++ + G PAG FV
Sbjct: 417 NEYNDLATIFFNTQDDAIRNLFSAKTMREFSAQSLLTFLAMFYTLAVVTFGTAVPAGQFV 476
Query: 471 PIIVTGASYGRLVGILV---GERTNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNN 527
P I+ G++YGRLVG+ V ++ N+ G YA+LGAAS LGGSMR TVSLCVIM+E+TNN
Sbjct: 477 PGIMIGSTYGRLVGMFVVRFYKKLNIEEGTYALLGAASFLGGSMRMTVSLCVIMVEITNN 536
Query: 528 LLLLPLIMMVLVVSKSVANVFNANVYDLIMKAKGLPYLETHAEPYMRQLTVGDVV-TGPL 586
L LLPLIM+VL++SK+V + FN +Y++ + KG+P LE+ + +MRQ+ + + +
Sbjct: 537 LKLLPLIMLVLLISKAVGDAFNEGLYEVQARLKGIPLLESRPKYHMRQMIAKEACQSQKV 596
Query: 587 QMFNGIEKVRNIVFILRTTAHNGFPVIDEPPGSEAPILFGIILRHHLTTLLKKKA-FLPS 645
+ +V ++ IL + HNGFPVID E ++ G++LR HL LL+ K F S
Sbjct: 597 ISLPRVIRVADVASILGSNKHNGFPVIDHTRSGET-LVIGLVLRSHLLVLLQSKVDFQHS 655
Query: 646 PV---ANSYDVVRKFSSDDFAKKYSVERVKIEDIQLTEEEMGMFVDLHPFTNASPYTVVE 702
P+ ++ ++ FS +FAK S + + IEDI LT +++ M++DL PF N SPY V E
Sbjct: 656 PLPCDPSARNIRHSFS--EFAKPVSSKGLCIEDIHLTSDDLEMYIDLAPFLNPSPYVVPE 713
Query: 703 TMSLAKALILFREVGLRHLLVIPKIPGRSPVVGILTRHDFTPE 745
MSL K LFR++GLRHL V+P+ P R V+G++TR D E
Sbjct: 714 DMSLTKVYNLFRQLGLRHLFVVPR-PSR--VIGLITRKDLLIE 753
>At1g55620 CLC-f chloride channel protein
Length = 781
Score = 48.9 bits (115), Expect = 1e-05
Identities = 89/395 (22%), Positives = 151/395 (37%), Gaps = 79/395 (20%)
Query: 165 GIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGGSKRYGITWRWLRFF 224
GI+ V +K I + + +G GP V G A +
Sbjct: 224 GIYPV----IKAIQAAVTLGTGCSLGPEGPSVDIGKSCAN--------------GFALMM 265
Query: 225 KNDRDRR-DLIICGSAAGIAAAFRAPVGGVLFALEEMASWWRTALLWRAFFTTATVAIFL 283
+N+R+RR L G+A+GIA+ F A V G FA+E + R FTTA I L
Sbjct: 266 ENNRERRIALTAAGAASGIASGFNAAVAGCFFAIETVLRPLRAEN--SPPFTTA--MIIL 321
Query: 284 RAMIDVCLSDKCGLFGKGGLIMFDAYSASISYHLVDVPPVFILAVVGGLLGSLFNFMTNK 343
++I +S+ L G +Y + ++P IL ++ G + +F+ +
Sbjct: 322 ASVISSTVSN--ALLGTQSAFTVPSYDLKSA---AELPLYLILGMLCGAVSVVFSRLVTW 376
Query: 344 VLRIYNVINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRPCPPDAVEPCPTIGRSGIY 403
+ ++ I +K FGLP + CP +G G
Sbjct: 377 FTKSFDFIKDK--------------------FGLPAIV------------CPALGGLGA- 403
Query: 404 KKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSMLVFFIICLFLSIFSCGIV 463
Y G+ F ++ + S + L+ + ++ L S G+V
Sbjct: 404 ---GIIALKYPGILYWGFTNVEEILHTGKSASAPGIWLLAQLAAAKVVATALCKGS-GLV 459
Query: 464 APAGLFVPIIVTGASYGRLVG---------ILVGERTNLSNGLYAVLGAASLLGGSMRTT 514
GL+ P ++ GA+ G + G + G YA++G A+ L
Sbjct: 460 --GGLYAPSLMIGAAVGAVFGGSAAEIINRAIPGNAAVAQPQAYALVGMAATLASMCSVP 517
Query: 515 VSLCVIMLELTNNL-LLLPLIMMV--LVVSKSVAN 546
++ +++ ELT + +LLPL+ V + SVAN
Sbjct: 518 LTSVLLLFELTKDYRILLPLMGAVGLAIWVPSVAN 552
>At4g35440 CLC-e chloride channel protein
Length = 710
Score = 38.1 bits (87), Expect = 0.019
Identities = 73/402 (18%), Positives = 149/402 (36%), Gaps = 76/402 (18%)
Query: 146 AAGSGISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAAL 205
+AG + + L+ V A +R + +T +G+ +G GP V GA +A
Sbjct: 149 SAGKSTGDSHSSLDRVKA----VLRPFLKTVAACVTLGTGNS-LGPEGPSVEIGASIAKG 203
Query: 206 LGQGGSKRYGITWRWLRFFKNDRDRRDLIICGSAAGIAAAFRAPVGGVLFALEEMASWWR 265
+ F K+ + L+ GSAAGI++ F A V G FA+E + W
Sbjct: 204 VNS-------------LFNKSPQTGFSLLAAGSAAGISSGFNAAVAGCFFAVESVL--WP 248
Query: 266 TALLWRAFFTTATVAIFLRAMIDVCLSDKCGLFGKGGLIMFDAYSASISYHLVDVPPVFI 325
++ + T ++ + + + + + GL + + D S ++P +
Sbjct: 249 SSSTDSSTSLPNTTSMVILSAVTASVVSEIGLGSEPAFKVPDYDFRSPG----ELPLYLL 304
Query: 326 LAVVGGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGLPWLAPCRP 385
L + GL+ + T+ + + +N+ I + + + + P +
Sbjct: 305 LGALCGLVSLALSRCTSSMTSAVDSLNKDAGIPKAVFPVMGGLSVGIIALVYPEV----- 359
Query: 386 CPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHTDNEFELSSM 445
+Y FQ N D + +L
Sbjct: 360 ----------------LYWGFQ----------------NVDILLEKRPFVKGLSADLLLQ 387
Query: 446 LVFFIICLFLSIFSCGIVAPAGLFVPII----VTGASYGRLVGILVGERTNLSNGL---- 497
LV I + G+V G + P + G +YG+ +G+ + + + + +
Sbjct: 388 LVAVKIAATAWCRASGLV--GGYYAPSLFIGGAAGMAYGKFIGLALAQNPDFNLSILEVA 445
Query: 498 ----YAVLGAASLLGGSMRTTVSLCVIMLELTNNL-LLLPLI 534
Y ++G A+ L G + ++ +++ ELT + ++LPL+
Sbjct: 446 SPQAYGLVGMAATLAGVCQVPLTAVLLLFELTQDYRIVLPLL 487
>At4g28410 unknown protein
Length = 447
Score = 35.8 bits (81), Expect = 0.094
Identities = 25/74 (33%), Positives = 35/74 (46%), Gaps = 2/74 (2%)
Query: 330 GGLLGSLFNFMTNKVLRIYNVINEKGTICRLFLACLISIFTSCLLFGL--PWLAPCRPCP 387
GG+ GS++ F NK + ++ KGT+ RLF C + + L G P + PC
Sbjct: 28 GGIGGSVWRFKGNKAAKEAASVSMKGTLARLFDCCSKDVKKTILPLGHGDPSVYPCFQTS 87
Query: 388 PDAVEPCPTIGRSG 401
DA E RSG
Sbjct: 88 VDAEEAVVESLRSG 101
>At4g21250 hypothetical protein
Length = 449
Score = 31.6 bits (70), Expect = 1.8
Identities = 27/93 (29%), Positives = 40/93 (42%), Gaps = 20/93 (21%)
Query: 439 EFELSSMLVFFIICLFLS--IFSCGIVAPAGLFVPII------------------VTGAS 478
E +LSS ++ + FL+ I S G + GLF+PI+ VTG S
Sbjct: 51 ELKLSSAIIMAGVLCFLAALISSAGGIGGGGLFIPIMTIVAGVDLKTASSFSAFMVTGGS 110
Query: 479 YGRLVGILVGERTNLSNGLYAVLGAASLLGGSM 511
++ L G + L L +L LLG S+
Sbjct: 111 IANVISNLFGGKALLDYDLALLLEPCMLLGVSI 143
>At2g30700 unknown protein
Length = 480
Score = 31.2 bits (69), Expect = 2.3
Identities = 53/233 (22%), Positives = 87/233 (36%), Gaps = 47/233 (20%)
Query: 377 LPWLAPCRPCPPDAVEPCPTIGRSGIYKKFQCPPNHYNGLASLIFNTNDDAIRNLFSMHT 436
LPW P P PD EP T +CP + + ++S+I D +
Sbjct: 83 LPWTPPMYPTFPDTYEPKLT---------GKCPTD-FQAISSVIDTAASDCSQPF----- 127
Query: 437 DNEFELSSMLVFFIICL--FLSIFSC----GIVAPAGLFVPIIVTGASYGRLVGILVGER 490
+ LV +IC F+S+ V L +P V + +V ILV R
Sbjct: 128 -------AALVGNVICCPQFVSLLHIFQGQHNVKSNKLVLPDAVATDCFSDIVSILVSRR 180
Query: 491 TNLSNGLYAVLGAASLLGGSMRTTVSLCVIMLELTNNLLLLPLIMMVLVVSKSVANVFNA 550
N++ + +++L GGS V+ ++ N+ LL V + + +
Sbjct: 181 ANMTIPALCSVTSSNLTGGS--CPVTDVTTFEKVVNSSKLLDACRTVDPLKECCRPICQP 238
Query: 551 NVYDLIMKAKGLPYLETHAEPYMRQLTVGDVVTGPL---QMFNGIEKVRNIVF 600
+ + + G Q+TVGD + PL N I +N+VF
Sbjct: 239 AIMEAALIISG------------HQMTVGDKI--PLAGSNNVNAINDCKNVVF 277
>At4g22120 unknown protein
Length = 771
Score = 30.8 bits (68), Expect = 3.0
Identities = 24/87 (27%), Positives = 39/87 (44%), Gaps = 18/87 (20%)
Query: 80 FMIGLIVGFI-GFCNNLAVENLAGIKFVT------TSNMMLERRFMFAFFIFFASNLSLT 132
FM +I GF+ G L + L I + TS LERR F ++IF
Sbjct: 420 FMKSVIQGFLPGIALKLFLAFLPSILMIMSKFEGFTSISSLERRAAFRYYIF-------- 471
Query: 133 LFASIITAFIAPTAAGSGISEVKAYLN 159
+++ F+A AG+ ++ ++LN
Sbjct: 472 ---NLVNVFLASVIAGAAFEQLNSFLN 495
>At5g18525 unknown protein
Length = 909
Score = 30.0 bits (66), Expect = 5.2
Identities = 17/44 (38%), Positives = 22/44 (49%), Gaps = 10/44 (22%)
Query: 728 PGRSPVVGILTRHD----------FTPEHILGMHPFLVKSRWKR 761
P SPV+G+LT D +T E+IL P +KS W R
Sbjct: 218 PNLSPVLGLLTSSDCLVSVLPKAPYTLENILYYSPSAIKSEWHR 261
>At5g47470 nodulin-like protein
Length = 364
Score = 29.3 bits (64), Expect = 8.8
Identities = 26/95 (27%), Positives = 42/95 (43%), Gaps = 10/95 (10%)
Query: 290 CLSDKCGLFGK-----GGLIMFDAYSASISYHLVDVPPVFIL---AVVG--GLLGSLFNF 339
C+ K + G G L M +S SIS+ D P+F+ VVG LLG++F
Sbjct: 151 CVYSKLKILGTLLCVFGALAMSVMHSTSISHKEEDDTPIFVFDRDKVVGCIYLLGAVFVL 210
Query: 340 MTNKVLRIYNVINEKGTICRLFLACLISIFTSCLL 374
TN VL+ + I + L+ + + ++
Sbjct: 211 STNVVLQASTLAEFPAPISLSAITALLGVLITTVV 245
>At1g19650 sec14 cytosolic factor- like protein
Length = 608
Score = 29.3 bits (64), Expect = 8.8
Identities = 20/74 (27%), Positives = 33/74 (44%), Gaps = 5/74 (6%)
Query: 151 ISEVKAYLNGVDAPGIFTVRTLCVKIIGSITAVSGSLVIGKAGPMVHTGACVAALLGQGG 210
++ V Y NG +P + + CV ++ + V+ L + P G + LG+ G
Sbjct: 402 LTPVSEYANGNISPTVLSEYEECVPMVDKVVDVAWQL---QEMPNASEGPQYTSSLGKIG 458
Query: 211 SKRYGITWRWLRFF 224
S R+ W WL F
Sbjct: 459 SVRH--IWSWLTAF 470
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.328 0.143 0.436
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 17,082,429
Number of Sequences: 26719
Number of extensions: 736040
Number of successful extensions: 2021
Number of sequences better than 10.0: 14
Number of HSP's better than 10.0 without gapping: 8
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 1988
Number of HSP's gapped (non-prelim): 17
length of query: 776
length of database: 11,318,596
effective HSP length: 107
effective length of query: 669
effective length of database: 8,459,663
effective search space: 5659514547
effective search space used: 5659514547
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144657.4


