

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.13 + phase: 0 /pseudo/partial
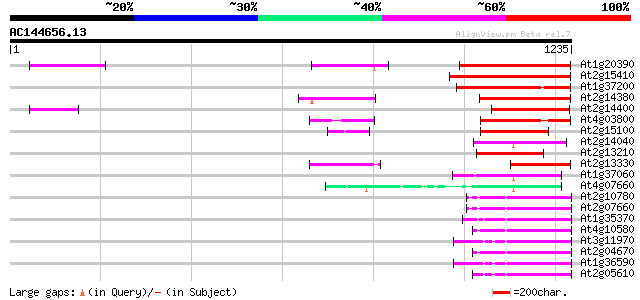
(1235 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At1g20390 hypothetical protein 246 6e-65
At2g15410 putative retroelement pol polyprotein 237 4e-62
At1g37200 hypothetical protein 218 2e-56
At2g14380 putative retroelement pol polyprotein 206 7e-53
At2g14400 putative retroelement pol polyprotein 190 4e-48
At4g03800 172 8e-43
At2g15100 putative retroelement pol polyprotein 162 8e-40
At2g14040 putative retroelement pol polyprotein 155 1e-37
At2g13210 pseudogene 151 2e-36
At2g13330 F14O4.9 151 3e-36
At1g37060 Athila retroelment ORF 1, putative 150 3e-36
At4g07660 putative athila transposon protein 149 7e-36
At2g10780 pseudogene 136 8e-32
At2g07660 putative retroelement pol polyprotein 136 8e-32
At1g35370 hypothetical protein 130 4e-30
At4g10580 putative reverse-transcriptase -like protein 128 2e-29
At3g11970 hypothetical protein 127 5e-29
At2g04670 putative retroelement pol polyprotein 127 5e-29
At1g36590 hypothetical protein 127 5e-29
At2g05610 putative retroelement pol polyprotein 124 3e-28
>At1g20390 hypothetical protein
Length = 1791
Score = 246 bits (628), Expect = 6e-65
Identities = 122/244 (50%), Positives = 164/244 (67%)
Query: 991 EFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVEHRIPT 1050
E +NI + R + VGA + ++ ++I LL+ FAWS EDM G+DP I H +
Sbjct: 712 EMVNIDESDPTRCVGVGAEISPSIRLELIALLKRNSKTFAWSIEDMKGIDPAITAHELNV 771
Query: 1051 KPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMC 1110
P PV+QK R+ P+ A + EV+K + AG ++ V+YPEW+AN V V KK+GK R+C
Sbjct: 772 DPTFKPVKQKRRKLGPERARAVNEEVEKLLKAGQIIEVKYPEWLANPVVVKKKNGKWRVC 831
Query: 1111 VDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITP 1170
VD+ DLNKA PKD++PL HID LV+ + + + FMD FSGYNQI M +D+EKTSF+T
Sbjct: 832 VDYTDLNKACPKDSYPLPHIDRLVEATSGNGLLSFMDAFSGYNQILMHKDDQEKTSFVTD 891
Query: 1171 WGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKM 1230
GT+CYKVM GL NAGATYQR + + D I + VEVY+DDM+VKS E HVE+L+K
Sbjct: 892 RGTYCYKVMSFGLKNAGATYQRFVNKMLADQIGRTVEVYIDDMLVKSLKPEDHVEHLSKC 951
Query: 1231 FERL 1234
F+ L
Sbjct: 952 FDVL 955
Score = 71.2 bits (173), Expect = 3e-12
Identities = 46/176 (26%), Positives = 82/176 (46%), Gaps = 7/176 (3%)
Query: 665 FSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRIS 724
F++ +L HN L + + ++ VL+DTGSS++++ K L ++ + ++
Sbjct: 555 FTDVDLEGLDTPHNDPLVVELIISDSRVTRVLIDTGSSVDLIFKDVLTAMNITDRQIKPV 614
Query: 725 TFLVKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTST 784
+ + FDG +G I LP+ +G + F V+ A Y+ +LG PWIH A+ ST
Sbjct: 615 SKPLAGFDGDFVMTIGTIKLPIFVGGLIAWVKFVVIGKPAVYNVILGTPWIHQMQAIPST 674
Query: 785 LHQNFKFVKNGKLVTVHG-------EEAYLVSQLSSFSCIEAGSAEGTAFQGLTIE 833
HQ KF + + T+ +Y S+L + ++ T G+ E
Sbjct: 675 YHQCVKFPTHNGIFTLRAPKEAKTPSRSYEESELCRTEMVNIDESDPTRCVGVGAE 730
Score = 45.4 bits (106), Expect = 2e-04
Identities = 36/172 (20%), Positives = 71/172 (40%), Gaps = 4/172 (2%)
Query: 43 EFDRYNGLTCPHNHIIKY---VRKMGNYSDN-DSLMIHCFQDSLMEDAAEWYTSLSKDDV 98
+ + YNG P + + + + DN D+ F + L A W++ L + +
Sbjct: 173 KLESYNGRNDPKEFLTSFNVAINRAELTIDNFDAGRCQIFIEHLTGPAHNWFSRLKPNSI 232
Query: 99 HTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKDESFREYAQRWRGAAARITPALDEEE 158
+F +L ++F HY + + L S+SQ ES R + R++ IT +
Sbjct: 233 DSFHQLTSSFLKHYAPLIENQTSNADLWSISQGAKESLRSFVDRFKLVVTNITVPDEAAI 292
Query: 159 MTQRFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVREGIIVFEKVESS 210
+ R + + + + AP+ + + +R E E +I+ K S+
Sbjct: 293 VALRNAVWYDSRFRDDITLHAPSTLEDALHRASRFIELEEEKLILARKHNST 344
>At2g15410 putative retroelement pol polyprotein
Length = 1787
Score = 237 bits (604), Expect = 4e-62
Identities = 118/267 (44%), Positives = 168/267 (62%), Gaps = 1/267 (0%)
Query: 968 IPYEITRLLEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPD 1027
I Y + + Q+++ P + + +NI + R + + L ++ +++ LR+
Sbjct: 695 IIYNVILVPTQDERP-NPQKGTVVQVNIDESDPSRCVGIRIDLPSELQNELVNFLRQNAA 753
Query: 1028 IFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMT 1087
FAWS EDMPG+D + H + P P++QK R+ PD + EV+K +DAG ++
Sbjct: 754 TFAWSVEDMPGIDSAVTCHELNVDPTYKPLKQKRRKLGPDRTKDVNEEVKKLLDAGSIVE 813
Query: 1088 VEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMD 1147
V YP+W+ N V V KK+GK R+C+DF DLNKA PKD+FPL HID LV+ A +++ FMD
Sbjct: 814 VRYPDWLRNPVVVKKKNGKWRVCIDFTDLNKACPKDSFPLPHIDRLVEATAGNELLSFMD 873
Query: 1148 GFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVE 1207
FSGYNQI M DREKT FIT GT+CYKVMP GL NAGATY R + +F D + +E
Sbjct: 874 AFSGYNQILMHQNDREKTVFITDQGTYCYKVMPFGLKNAGATYPRLVNQMFTDQLDHSME 933
Query: 1208 VYVDDMIVKSKDEEQHVEYLTKMFERL 1234
VY+DDM+VKS E+H+ +L + F+ L
Sbjct: 934 VYIDDMLVKSLRAEEHITHLRQCFQVL 960
Score = 31.6 bits (70), Expect = 2.9
Identities = 24/102 (23%), Positives = 42/102 (40%), Gaps = 4/102 (3%)
Query: 47 YNGLTCPHNHIIKYVRKMG--NYSDNDSLMIHC--FQDSLMEDAAEWYTSLSKDDVHTFD 102
Y GL P + +G ++SD D C F L A W++ L + + +
Sbjct: 258 YEGLVDPRPFLTSVSIAIGRAHFSDEDRDAGSCQLFVKHLSGAALTWFSRLEANSIDSVH 317
Query: 103 ELAAAFKSHYGFNTRLKPNREFLRSLSQKKDESFREYAQRWR 144
L +F +YG + L +++Q ES R + R++
Sbjct: 318 ALTTSFLKNYGVFMEKGASNVDLWTMAQTAKESLRSFIGRFK 359
>At1g37200 hypothetical protein
Length = 1564
Score = 218 bits (555), Expect = 2e-56
Identities = 109/250 (43%), Positives = 158/250 (62%), Gaps = 4/250 (1%)
Query: 985 PHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIV 1044
P + I + I + K+ + VG L+ +++ I L+E D FAWS ++ G+ ++
Sbjct: 614 PQKSSITQVYIDESDPKQCVGVGQDLDPAIREDFITFLKENKDSFAWSSANLQGISLEVT 673
Query: 1045 EHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKD 1104
H + P P++QK R+ P+ A +++EV + + G + V+YP+W+AN V V K+
Sbjct: 674 SHELNVDPTYRPIKQKRRKLGPERAKAVQDEVDRLLKIGSIREVKYPDWLANPVVVKNKN 733
Query: 1105 GKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREK 1164
GK R+C+DF DLNKA PKD+FPL HID LV A ++ FMD FSGYNQI M P+D+EK
Sbjct: 734 GKWRVCIDFMDLNKACPKDSFPLPHIDRLVKATAGHELLSFMDAFSGYNQILMRPDDQEK 793
Query: 1165 TSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHV 1224
T+FIT CYKVMP GL N GATYQR + +F D + K +E+Y+DDM+VKS E+ H+
Sbjct: 794 TAFITD----CYKVMPFGLKNTGATYQRLVNRMFADQLGKTMELYIDDMLVKSAHEKDHL 849
Query: 1225 EYLTKMFERL 1234
L + F+ L
Sbjct: 850 PQLRECFKIL 859
Score = 40.4 bits (93), Expect = 0.006
Identities = 21/53 (39%), Positives = 28/53 (52%), Gaps = 6/53 (11%)
Query: 764 ASYSCLLGRPWIHDAGAVTSTLHQNFKFVKNGKLVTVHGEEAYLVSQLSSFSC 816
A ++ +LGRPW+H AV ST HQ KF + ++G SQ SS C
Sbjct: 530 APFNAILGRPWLHAMKAVPSTYHQCIKFPSEKGIAVIYG------SQRSSRRC 576
Score = 30.8 bits (68), Expect = 5.0
Identities = 16/45 (35%), Positives = 27/45 (59%), Gaps = 2/45 (4%)
Query: 663 LWFSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGSSLNVMP 707
LW E E + K H+ A+ I ++ + LS +++DTGSS++ P
Sbjct: 489 LW--ESETTDLDKPHDDAIVIRIDVGNYKLSRIMIDTGSSVDKAP 531
>At2g14380 putative retroelement pol polyprotein
Length = 764
Score = 206 bits (524), Expect = 7e-53
Identities = 99/200 (49%), Positives = 135/200 (67%)
Query: 1035 DMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWV 1094
DM G+DP++ H + P V+QK R+ P+ + + +EV K +DAG ++ V+YPEW+
Sbjct: 476 DMVGIDPEVACHELNVDPTFKLVKQKRRKLGPERSKAVNDEVDKLLDAGSIVEVKYPEWL 535
Query: 1095 ANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQ 1154
AN V V KK+ K R+C+DF DLNKA PKD+FPL HID +V+ +++ FMD FSGYNQ
Sbjct: 536 ANPVVVKKKNDKWRVCIDFTDLNKACPKDSFPLPHIDRMVEATTGNELLSFMDAFSGYNQ 595
Query: 1155 IKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMI 1214
I M +D+EKTSFI GT+CYKVMP GL N GA YQR + +F + K +EVY+DDM+
Sbjct: 596 IPMHKDDQEKTSFIIDRGTYCYKVMPFGLKNVGARYQRLVNQMFAPQLGKTMEVYIDDML 655
Query: 1215 VKSKDEEQHVEYLTKMFERL 1234
VKS H+++L FE L
Sbjct: 656 VKSTRSADHIDHLKACFETL 675
Score = 80.5 bits (197), Expect = 6e-15
Identities = 54/195 (27%), Positives = 88/195 (44%), Gaps = 27/195 (13%)
Query: 637 EQAYVDHEVTMDRFGSIVGNITACNN---------------LW------------FSEDE 669
+Q + D V R I+G +TAC + LW F+ ++
Sbjct: 277 DQQHNDAPVPRRRVNMIMGGLTACRDSFRSIKEYIKSGAATLWSSPATKEMTPLTFTSED 336
Query: 670 LPEAGKYHNLALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSLRISTFLVK 729
L HN L I ++ ++ +L+DTGSS+NV+ K L ++ + ++ S +
Sbjct: 337 LFGVDLPHNDPLVIELHIGESEVTRILIDTGSSVNVVFKDVLQKMKVHDRHIKPSVRPLT 396
Query: 730 AFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGAVTSTLHQNF 789
FDG+ G I LP+ +G F V+D Y+ +LG PWIHD A+ S+ HQ
Sbjct: 397 GFDGNTMMTNGTIKLPIYLGGAATWHKFVVVDKPTIYNIILGTPWIHDMQAIPSSYHQCI 456
Query: 790 KFVKNGKLVTVHGEE 804
K + + T+ G +
Sbjct: 457 KIPTSIGIETIRGNQ 471
>At2g14400 putative retroelement pol polyprotein
Length = 1466
Score = 190 bits (483), Expect = 4e-48
Identities = 90/170 (52%), Positives = 123/170 (71%)
Query: 1062 RRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASP 1121
R+ + A + +EV K + G + V+YP+W+AN V V KK+GK R+C+DF DLNKA P
Sbjct: 470 RKLGVERAKAVNDEVDKLLKIGSIREVQYPDWLANTVVVKKKNGKDRVCIDFTDLNKACP 529
Query: 1122 KDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPI 1181
KD+FPL HID LV++ A +++ FMD FSGYNQI M+PED+EKT FIT G +CYKVMP
Sbjct: 530 KDSFPLPHIDRLVESTAGNELLSFMDAFSGYNQIMMNPEDQEKTLFITDRGIYCYKVMPF 589
Query: 1182 GLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMF 1231
GL NAGATY R + +F + + K +EVY+DDM++KS +E HV++L + F
Sbjct: 590 GLRNAGATYPRLVNKMFSEHVGKTMEVYIDDMLIKSLKKEDHVKHLEECF 639
Score = 44.7 bits (104), Expect = 3e-04
Identities = 26/111 (23%), Positives = 50/111 (44%), Gaps = 4/111 (3%)
Query: 45 DRYNGLTCPHNHIIKYVRKMG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHT 100
+ YNGL P ++ ++ G N +D D F ++L A W+T L +
Sbjct: 163 ESYNGLEDPKGYLAAFLIAAGRVDLNEADEDVRYCKLFSENLCGQALMWFTQLEPGSISN 222
Query: 101 FDELAAAFKSHYGFNTRLKPNREFLRSLSQKKDESFREYAQRWRGAAARIT 151
F+EL+ F Y + L +LSQ +E+ R + +++ ++++
Sbjct: 223 FNELSVVFLKQYSILMDKSISDTDLWNLSQGPNETLRAFITKFKYVLSKLS 273
Score = 34.7 bits (78), Expect = 0.35
Identities = 23/88 (26%), Positives = 46/88 (52%), Gaps = 4/88 (4%)
Query: 662 NLWFSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETSL 721
++ F E E + H+ AL ++++ + +S +L+DTGSS++++ STL+++ +
Sbjct: 408 SILFDEKETQHLERSHDDALVVTLDVANFEVSRILIDTGSSVDLIFLSTLERMGISRADV 467
Query: 722 RISTFLVKAFDGSRKNVLGEIDLPMTIG 749
V+ K V E+D + IG
Sbjct: 468 NRRKLGVE----RAKAVNDEVDKLLKIG 491
>At4g03800
Length = 637
Score = 172 bits (437), Expect = 8e-43
Identities = 88/199 (44%), Positives = 126/199 (63%), Gaps = 15/199 (7%)
Query: 1036 MPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVA 1095
MP +DP I+ H + P P++QK R+ + A + ++ K + G + V+YP+WVA
Sbjct: 321 MPDIDPSIICHELNVDPRFKPLKQKRRKLGVERAKAVNGKIDKLLKIGSIREVQYPDWVA 380
Query: 1096 NIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQI 1155
V V KK+GK R+C+DF DLNKA PKD+FPL HID LV++ A +++ FMD F GYNQI
Sbjct: 381 ITVVVKKKNGKDRVCIDFTDLNKACPKDSFPLPHIDRLVESTAGNELLTFMDAFLGYNQI 440
Query: 1156 KMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIV 1215
M+PED+EKTSFIT + GATYQ + +F++ + K +EV +DD +V
Sbjct: 441 MMNPEDQEKTSFIT---------------DRGATYQWLVNKMFNEHLRKTMEVSIDDTLV 485
Query: 1216 KSKDEEQHVEYLTKMFERL 1234
KS +E HV++L + FE L
Sbjct: 486 KSLKKEDHVKHLGECFEIL 504
Score = 70.5 bits (171), Expect = 6e-12
Identities = 47/147 (31%), Positives = 74/147 (49%), Gaps = 20/147 (13%)
Query: 660 CNNLWFSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGSSLNVM---PKSTLDQLSY 716
C+++ F E+E + H+ AL I+++ + +S +LVDTGSS++++ P S L
Sbjct: 182 CSSVSFDEEETRHIERPHDDALIITLDVANFKISRILVDTGSSVDLIFLGPPSPL----- 236
Query: 717 RETSLRISTFLVKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIH 776
AF LG I LP+ G + ++ F V D A+Y+ +LG PWI
Sbjct: 237 ------------VAFTSESAMSLGTIKLPVLAGKMSKIVDFVVFDKPATYNIILGTPWIF 284
Query: 777 DAGAVTSTLHQNFKFVKNGKLVTVHGE 803
AV ST HQ KF + + T+ G+
Sbjct: 285 QMKAVPSTYHQWLKFPTSNGVETIWGD 311
>At2g15100 putative retroelement pol polyprotein
Length = 1329
Score = 162 bits (411), Expect = 8e-40
Identities = 80/151 (52%), Positives = 103/151 (67%)
Query: 1036 MPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVA 1095
M G+ +++ H + P PV+QK R+ PD A + EV + ++ G + V+YPEW+A
Sbjct: 412 MTGISTEVISHELNVDPTFKPVKQKRRKLGPDRAQAVNIEVVRLLEVGRIREVKYPEWLA 471
Query: 1096 NIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQI 1155
N V V KK+GK R+CVDF DLNKA KD FPL HID LV++ ++ FMD FSGYNQI
Sbjct: 472 NPVVVKKKNGKWRVCVDFTDLNKACSKDFFPLPHIDRLVESTTGHEMLSFMDAFSGYNQI 531
Query: 1156 KMSPEDREKTSFITPWGTFCYKVMPIGLINA 1186
M+PED+EKTSFIT GT+ YKVMP GL NA
Sbjct: 532 LMNPEDQEKTSFITECGTYYYKVMPFGLKNA 562
Score = 45.8 bits (107), Expect = 2e-04
Identities = 30/94 (31%), Positives = 45/94 (46%), Gaps = 5/94 (5%)
Query: 701 SSLNVMPKSTLDQLSYRETSLR---ISTFLVKAFDGSRKNVLGEIDLPMTIGPETFLITF 757
S+ N PK+ + + T LR T + S LG + LP+T ++ F
Sbjct: 314 SASNAAPKAPQPATTKKPTELRKPAAGTCTTMLCETSMS--LGTVVLPVTAQGVVKMVEF 371
Query: 758 QVMDINASYSCLLGRPWIHDAGAVTSTLHQNFKF 791
V D A+Y+ +LG PW+++ V ST HQ KF
Sbjct: 372 TVFDRPAAYNVILGTPWLYEMKVVPSTYHQCVKF 405
>At2g14040 putative retroelement pol polyprotein
Length = 841
Score = 155 bits (392), Expect = 1e-37
Identities = 79/221 (35%), Positives = 125/221 (55%), Gaps = 18/221 (8%)
Query: 1022 LREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQID 1081
LR+Y +S +D+ G+ P + H I + E + RR + ++ +K E+ K +D
Sbjct: 340 LRKYRKALGYSLDDITGISPTLCMHMIHLEGESITSVEHQRRLNSNLRDVVKKEIMKLLD 399
Query: 1082 AGFLMTVEYPEWVANIVPVPKKDGKV------------------RMCVDFRDLNKASPKD 1123
AG + + WV+ + VPKK G + RMC+D+R LN A+ KD
Sbjct: 400 AGIIYPISDSTWVSPVHVVPKKGGVIVIKNEKNELIPTRTVTGHRMCIDYRKLNSATRKD 459
Query: 1124 NFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGL 1183
NFPL ID +++ + + F+DG+ G+ QI + P+D+EKT+F P+GTF Y+ MP GL
Sbjct: 460 NFPLSFIDQMLERLSNQPYYCFLDGYLGFFQILIHPDDQEKTTFTCPYGTFAYRRMPFGL 519
Query: 1184 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHV 1224
NA AT+Q M +F DMI +EV++DD +V + E++H+
Sbjct: 520 CNAPATFQHCMKYIFSDMIEDFMEVFIDDFLVLQRCEDKHL 560
>At2g13210 pseudogene
Length = 152
Score = 151 bits (382), Expect = 2e-36
Identities = 72/147 (48%), Positives = 98/147 (65%)
Query: 1029 FAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTV 1088
F WS ED+PG+ +IV H + P PV+QK R+ + A +K EV+K + G +
Sbjct: 6 FVWSPEDLPGVSVEIVAHELNVDPTFKPVKQKRRKLGRERAEAVKAEVEKLLRIGSITKA 65
Query: 1089 EYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDG 1148
+YPEW+AN V V KK+GK R+C+DF DLNKA PKD+FP+ HI LV++ + K+ FMD
Sbjct: 66 KYPEWLANPVVVKKKNGKWRVCIDFTDLNKACPKDSFPMSHIGRLVESTSGDKLLSFMDA 125
Query: 1149 FSGYNQIKMSPEDREKTSFITPWGTFC 1175
F+GYNQI M+ ED+EKT+F T G C
Sbjct: 126 FAGYNQIMMNSEDQEKTTFYTEQGIVC 152
>At2g13330 F14O4.9
Length = 889
Score = 151 bits (381), Expect = 3e-36
Identities = 74/133 (55%), Positives = 95/133 (70%)
Query: 1102 KKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPED 1161
+K+GK R+C+DFRDLNKA PKD+FPL HID LV+ + + FMD F GYNQI M +
Sbjct: 289 EKNGKWRVCIDFRDLNKACPKDSFPLPHIDRLVEATVEHEKLSFMDAFYGYNQILMRRDG 348
Query: 1162 REKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEE 1221
+EKT+FIT GT+ YKVMP GL NAG TYQR + +F D + K +EVY+DDM+VKS E+
Sbjct: 349 QEKTAFITDRGTYYYKVMPFGLKNAGTTYQRLVNRMFVDQLGKTIEVYIDDMLVKSAHEK 408
Query: 1222 QHVEYLTKMFERL 1234
HV L + F+ L
Sbjct: 409 DHVPQLRECFKIL 421
Score = 75.1 bits (183), Expect = 2e-13
Identities = 50/156 (32%), Positives = 77/156 (49%), Gaps = 6/156 (3%)
Query: 661 NNLWFSEDELPEAGKYHNLALHISVNCKSDMLSNVLVDTGSSLNVMPKSTLDQLSYRETS 720
+ + F E E+ + K H+ AL I ++ + LS ++VDTGSS++V+ + + ++
Sbjct: 45 DGITFWESEITDLDKPHDDALVIRIDVGNYELSCIMVDTGSSVDVLFYDAFKRTGHLDSK 104
Query: 721 LRISTFLVKAFDGSRKNVLGEIDLPMTIGPETFLITFQVMDINASYSCLLGRPWIHDAGA 780
L+ + F G +G I LP L F V+D A ++ +LGRPW+H A
Sbjct: 105 LQGRKTPLTGFAGDTTFSIGTIQLPTIARGVRQLTNFLVVDKKAPFNAILGRPWLHVMKA 164
Query: 781 VTSTLHQNFKFVKNGKLVTVHGEEAYLVSQLSSFSC 816
V ST HQ KF + V+G SQ SS C
Sbjct: 165 VPSTYHQCIKFPSYKGIAVVYG------SQRSSRKC 194
>At1g37060 Athila retroelment ORF 1, putative
Length = 1734
Score = 150 bits (380), Expect = 3e-36
Identities = 85/258 (32%), Positives = 138/258 (52%), Gaps = 19/258 (7%)
Query: 976 LEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYED 1035
++ K ++P + + ++ +G I + V I +L++ Y +S +D
Sbjct: 820 IKAPKVDLKPLPKGLRYVFLGLNSTYPVIVNDELTADQVNLLITELMK-YRKAIGYSLDD 878
Query: 1036 MPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVA 1095
+ G+ P + HRI + E + RR +P++ +K E+ K +DAG + + WV+
Sbjct: 879 IKGISPTLCTHRIHLENESYSSIEPQRRLNPNLKEVVKKEILKLLDAGVIYPISDSTWVS 938
Query: 1096 NIVPVPKKDGKV------------------RMCVDFRDLNKASPKDNFPLLHIDVLVDNN 1137
+ VPKK G RMC+++R LN AS K++FPL ID +++
Sbjct: 939 PVHCVPKKGGMTVVKNSKDELIPTRTITGHRMCIEYRKLNVASRKEHFPLPFIDHMLERL 998
Query: 1138 AQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTL 1197
A + F+D +SG+ QI + P D+ KT+F P+GTF YK MP GL NA AT+QR MT++
Sbjct: 999 ANHPYYCFLDSYSGFFQIPIHPNDQGKTTFTCPYGTFAYKRMPFGLCNAPATFQRCMTSI 1058
Query: 1198 FHDMIHKEVEVYVDDMIV 1215
F D+I + VEV++DD V
Sbjct: 1059 FSDLIEEMVEVFMDDFSV 1076
Score = 39.7 bits (91), Expect = 0.011
Identities = 33/146 (22%), Positives = 67/146 (45%), Gaps = 15/146 (10%)
Query: 15 NRGNGDSVKTHDLC--LVPKVDVPKKFKVP-------EFDRYNGLTC--PHNHIIKYVRK 63
N G GD+ + H+ +VP F++ + ++++GL P +H+ ++ R
Sbjct: 13 NIGAGDAPRNHNQRNGIVPPPVQNNNFEIKSGLIAMVQSNKFHGLPMEDPLDHLDEFDRL 72
Query: 64 MG----NYSDNDSLMIHCFQDSLMEDAAEWYTSLSKDDVHTFDELAAAFKSHYGFNTRLK 119
N D + F SL + A +W SL + + ++++ AF + + N+R
Sbjct: 73 CSLTKINRVSEDGFKLRLFPFSLGDKAHQWEKSLPQGSITSWNDCKKAFLAKFFSNSRTA 132
Query: 120 PNREFLRSLSQKKDESFREYAQRWRG 145
R + +Q +E+F E +R++G
Sbjct: 133 RLRNDISGFTQTNNETFYEAWERFKG 158
>At4g07660 putative athila transposon protein
Length = 724
Score = 149 bits (377), Expect = 7e-36
Identities = 145/558 (25%), Positives = 225/558 (39%), Gaps = 104/558 (18%)
Query: 696 LVDTGSSLNVMPKSTLDQLSYRE-TSLRISTFLVKAFDGSRKNVLGEIDLPMTIGPETFL 754
L D G+ +++MP S +L + + S IS L ++ +LP+ IG
Sbjct: 35 LCDLGALVSLMPLSVAKRLGFTQYKSCNISLILADRSVRIPHSLFE--NLPIRIGAVDIP 92
Query: 755 ITFQV--MDINASYSCLLGRPWIHDAGAVTST------------LHQNFKFVKNGKLVTV 800
F V MD +LGRP++ AGA+ F K T+
Sbjct: 93 TDFVVLEMDEEPKDPLILGRPFLATAGAMNDVKKGKIDLNLGKYCRMTFDVKDAMKKPTI 152
Query: 801 HGEEAYL--VSQLSSFSCIEAGSAEGTAFQGLTIEGME--LKKTGTAMASLKDAQKAVQE 856
G+ ++ + QL+ +E + E + LT G + L L D+ KA++E
Sbjct: 153 KGQLFWIEEIDQLAD-ELLEERAEEDHLYSALTKRGEDGFLHLETLGYQKLLDSHKAMEE 211
Query: 857 GQAAGRGKLIQLRENKHKEGLGFSPTSGVSTGTFHSAGF-VNAITEEATGFGPRPVFVIP 915
+ + E + S HS + N T + G +IP
Sbjct: 212 SEP------FEELNGPETEVMVMSEEGSTQVQPAHSRTYSTNNSTSTISNSGE---LIIP 262
Query: 916 GGIARDWDAIDIPSIMHVSELNHNKPVGHSNPTFPPNFEFPMYEAEAEEGDDIPYEITRL 975
+ DW + P + S P G F PN +P+
Sbjct: 263 --TSDDWSELKAPKVDLKS-----LPKGLRYVFFGPNSTYPVI----------------- 298
Query: 976 LEQEKKAIQPHQEEIEFINIGTEENKREIKVGAALEEWVKKKIIQLLREYPDIFAWSYED 1035
I E N E+ + ++ LR+Y +S D
Sbjct: 299 -------------------INVELNDNEVNL-----------LLSELRKYKRAIGYSLSD 328
Query: 1036 MPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVA 1095
+ + P + HRI + E + RR + ++ +K E+ K +DAG + + WV
Sbjct: 329 IKRISPSLCNHRIHLENESYSSIEPQRRLNLNLKEVVKKEILKLLDAGVIYPISDSTWVF 388
Query: 1096 NIVPVPKKDGKV------------------RMCVDFRDLNKASPKDNFPLLHIDVLVDNN 1137
+ VPKK G R+C+D+R LN AS KD+FPL + +++
Sbjct: 389 PVHCVPKKGGMTVVKNEKDELIPTRTITGHRVCIDYRKLNAASRKDHFPLPFTNQMLEGL 448
Query: 1138 AQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTL 1197
A F+DG+SG+ QI + P D+EKT+F P+GTF YK MP GL NA T+QR MT++
Sbjct: 449 ANHLYNCFLDGYSGFFQIPIHPNDQEKTTFTCPYGTFAYKRMPFGLCNAPTTFQRCMTSI 508
Query: 1198 FHDMIHKEVEVYVDDMIV 1215
F D+I K VEV++DD V
Sbjct: 509 FSDLIEKMVEVFMDDFSV 526
>At2g10780 pseudogene
Length = 1611
Score = 136 bits (342), Expect = 8e-32
Identities = 82/232 (35%), Positives = 131/232 (56%), Gaps = 10/232 (4%)
Query: 1005 KVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVE-HRIPTKPECPPVRQKLRR 1063
+VGA+ E K I ++ E+ D+FA + G+ P + I +P P+ + R
Sbjct: 615 EVGASAE----LKDIPIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYR 666
Query: 1064 THPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKD 1123
P K+K ++++ +D GF+ P W A ++ V KKDG R+C+D+R LNK + K+
Sbjct: 667 MAPAEMAKLKKQLEELLDKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKN 725
Query: 1124 NFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGL 1183
+PL ID L+D ++ F +D SGY+QI + P D KT+F T + F + VMP GL
Sbjct: 726 KYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYDHFEFVVMPFGL 785
Query: 1184 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
NA A + + M +F D + + V ++++D++V SK E H E+L + ERLR
Sbjct: 786 TNAPAAFMKMMNGVFRDFLDEFVIIFINDILVYSKSWEAHQEHLRAVLERLR 837
Score = 35.4 bits (80), Expect = 0.20
Identities = 27/119 (22%), Positives = 52/119 (43%), Gaps = 5/119 (4%)
Query: 85 DAAEWYTSLSK---DDVHTFDELAAAFKSHYGFNTRLKPNREFLRSLSQKKDESFREYAQ 141
DA W+ ++ K D+V +F + F Y F E + + + REY +
Sbjct: 223 DAHNWWLTVEKRRGDEVRSFADFEDEFNKKY-FPPEAWDRLECAYLDLVQGNRTVREYDE 281
Query: 142 RWRGAAARITPALDEEE-MTQRFLKTLKKDYVERMIIAAPNNFSEMVTMGTRLEEAVRE 199
+ + L+EE+ +RF++ L+ + ++ N+ SE+V +EE + E
Sbjct: 282 EFNRLRRYVGRELEEEQAQLRRFIRGLRIEIRNHCLVRTFNSVSELVERAAMIEEGIEE 340
>At2g07660 putative retroelement pol polyprotein
Length = 949
Score = 136 bits (342), Expect = 8e-32
Identities = 82/232 (35%), Positives = 131/232 (56%), Gaps = 10/232 (4%)
Query: 1005 KVGAALEEWVKKKIIQLLREYPDIFAWSYEDMPGLDPKIVE-HRIPTKPECPPVRQKLRR 1063
+VGA+ E K I ++ E+ D+FA + G+ P + I +P P+ + R
Sbjct: 110 EVGASAE----LKDILIVNEFSDVFA----AVSGVPPDRSDPFTIELEPGTTPISKAPYR 161
Query: 1064 THPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKD 1123
P ++K ++++ + GF+ P W A ++ V KKDG R+C+D+R LNK + K+
Sbjct: 162 MAPAEMAELKKQLEELLAKGFIRPSSSP-WGAPVLFVKKKDGSFRLCIDYRGLNKVTVKN 220
Query: 1124 NFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGL 1183
+PL ID L+D ++ F +D SGY+QI + P D KT+F T +G F + VMP GL
Sbjct: 221 KYPLPRIDELMDQLGGAQWFSKIDLASGYHQIPIEPTDVRKTAFRTRYGHFEFVVMPFGL 280
Query: 1184 INAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
NA A + + M +F D + + V +++DD++V SK E H E+L + ERLR
Sbjct: 281 TNAPAAFMKMMNGVFRDFLDEFVIIFIDDILVHSKSWEAHQEHLRAVLERLR 332
>At1g35370 hypothetical protein
Length = 1447
Score = 130 bits (328), Expect = 4e-30
Identities = 84/240 (35%), Positives = 134/240 (55%), Gaps = 4/240 (1%)
Query: 997 TEENKREIKVGAALEEWVKKKIIQ-LLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECP 1055
++E + + A + V++ ++Q ++ E+PD+FA D+P K +H+I
Sbjct: 522 SDEEQEIGSISALTSDVVEESVVQNIVEEFPDVFA-EPTDLPPFREKH-DHKIKLLEGAN 579
Query: 1056 PVRQKLRRTHPDMALKIKNEVQKQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRD 1115
PV Q+ R +I VQ I +G + P + + +V V KKDG R+CVD+ +
Sbjct: 580 PVNQRPYRYVVHQKDEIDKIVQDMIKSGTIQVSSSP-FASPVVLVKKKDGTWRLCVDYTE 638
Query: 1116 LNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFC 1175
LN + KD F + I+ L+D S VF +D +GY+Q++M P+D +KT+F T G F
Sbjct: 639 LNGMTVKDRFLIPLIEDLMDELGGSVVFSKIDLRAGYHQVRMDPDDIQKTAFKTHNGHFE 698
Query: 1176 YKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
Y VM GL NA AT+Q M ++F D + K V V+ DD+++ S E+H E+L +FE +R
Sbjct: 699 YLVMLFGLTNAPATFQSLMNSVFRDFLRKFVLVFFDDILIYSSSIEEHKEHLRLVFEVMR 758
>At4g10580 putative reverse-transcriptase -like protein
Length = 1240
Score = 128 bits (322), Expect = 2e-29
Identities = 72/218 (33%), Positives = 126/218 (57%), Gaps = 6/218 (2%)
Query: 1019 IQLLREYPDIFAWSYEDMPGLDPKIVE-HRIPTKPECPPVRQKLRRTHPDMALKIKNEVQ 1077
I++++E+ D+F + + GL P + I +P P+ + R P ++K +++
Sbjct: 457 IRVVQEFQDVF----QSLQGLPPSQSDPFTIELEPGTAPLSKAPYRMAPAEMAELKKQLK 512
Query: 1078 KQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNN 1137
+ GF+ P W A ++ V KKDG R+C+D+R+LN+ + K+ +PL ID L+D
Sbjct: 513 DLLGKGFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRELNRVTVKNRYPLPRIDELLDQL 571
Query: 1138 AQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTL 1197
+ F +D SGY+QI ++ D KT+F T +G F + VMP GL NA A + R M ++
Sbjct: 572 RGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFGLTNAPAVFMRLMNSV 631
Query: 1198 FHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
F + + + V +++DD++V SK E+ +L ++ E+LR
Sbjct: 632 FQEFLDEFVIIFIDDILVYSKSPEEQEVHLRRVMEKLR 669
>At3g11970 hypothetical protein
Length = 1499
Score = 127 bits (318), Expect = 5e-29
Identities = 87/266 (32%), Positives = 146/266 (54%), Gaps = 15/266 (5%)
Query: 978 QEKKAIQPHQEEIEFINIG--TEENKREI-KVGAALEEWVKKKIIQ-LLREYPDIFAWSY 1033
Q+ + +Q Q ++ + + +E + E+ + A E ++ +++ +L EYPDIF
Sbjct: 521 QKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVLNEYPDIFIEPT 580
Query: 1034 EDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDA----GFLMTVE 1089
P + H+I PV Q+ R ++ KNE+ K ++ G +
Sbjct: 581 ALPPFREKH--NHKIKLLEGSNPVNQRPYR----YSIHQKNEIDKLVEDLLTNGTVQASS 634
Query: 1090 YPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGF 1149
P + + +V V KKDG R+CVD+R+LN + KD+FP+ I+ L+D + +F +D
Sbjct: 635 SP-YASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLR 693
Query: 1150 SGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVY 1209
+GY+Q++M P+D +KT+F T G F Y VMP GL NA AT+Q M +F + K V V+
Sbjct: 694 AGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVF 753
Query: 1210 VDDMIVKSKDEEQHVEYLTKMFERLR 1235
DD++V S E+H ++L ++FE +R
Sbjct: 754 FDDILVYSSSLEEHRQHLKQVFEVMR 779
>At2g04670 putative retroelement pol polyprotein
Length = 1411
Score = 127 bits (318), Expect = 5e-29
Identities = 72/218 (33%), Positives = 124/218 (56%), Gaps = 6/218 (2%)
Query: 1019 IQLLREYPDIFAWSYEDMPGLDPKIVE-HRIPTKPECPPVRQKLRRTHPDMALKIKNEVQ 1077
I++++E+ D+F + + GL P + I +P P+ + R P ++K +++
Sbjct: 483 IRVIQEFEDVF----QSLQGLPPSRSDPFTIELEPGTAPLSKAPYRMAPAEMTELKKQLE 538
Query: 1078 KQIDAGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNN 1137
+ GF+ P W A ++ V KKDG R+C+D+R LN + K+ +PL ID L+D
Sbjct: 539 DLLGKGFIRPSTSP-WGAPVLFVKKKDGSFRLCIDYRGLNWVTVKNKYPLPRIDELLDQL 597
Query: 1138 AQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTL 1197
+ F +D SGY+QI ++ D KT+F T +G F + VMP L NA A + R M ++
Sbjct: 598 RGATCFSKIDLTSGYHQIPIAEADVRKTAFRTRYGHFEFVVMPFALTNAPAAFMRLMNSV 657
Query: 1198 FHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
F + + + V +++DD++V SK E+H +L ++ E+LR
Sbjct: 658 FQEFLDEFVIIFIDDILVYSKSPEEHEVHLRRVMEKLR 695
>At1g36590 hypothetical protein
Length = 1499
Score = 127 bits (318), Expect = 5e-29
Identities = 87/266 (32%), Positives = 146/266 (54%), Gaps = 15/266 (5%)
Query: 978 QEKKAIQPHQEEIEFINIG--TEENKREI-KVGAALEEWVKKKIIQ-LLREYPDIFAWSY 1033
Q+ + +Q Q ++ + + +E + E+ + A E ++ +++ +L EYPDIF
Sbjct: 521 QKLQKLQEDQVQLAMLCVQEVSESTEGELCTINALTSELGEESVVEEVLNEYPDIFIEPT 580
Query: 1034 EDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQIDA----GFLMTVE 1089
P + H+I PV Q+ R ++ KNE+ K ++ G +
Sbjct: 581 ALPPFREKH--NHKIKLLEGSNPVNQRPYR----YSIHQKNEIDKLVEDLLTNGTVQASS 634
Query: 1090 YPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVDNNAQSKVFFFMDGF 1149
P + + +V V KKDG R+CVD+R+LN + KD+FP+ I+ L+D + +F +D
Sbjct: 635 SP-YASPVVLVKKKDGTWRLCVDYRELNGMTVKDSFPIPLIEDLMDELGGAVIFSKIDLR 693
Query: 1150 SGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMTTLFHDMIHKEVEVY 1209
+GY+Q++M P+D +KT+F T G F Y VMP GL NA AT+Q M +F + K V V+
Sbjct: 694 AGYHQVRMDPDDIQKTAFKTHSGHFEYLVMPFGLTNAPATFQGLMNFIFKPFLRKFVLVF 753
Query: 1210 VDDMIVKSKDEEQHVEYLTKMFERLR 1235
DD++V S E+H ++L ++FE +R
Sbjct: 754 FDDILVYSSSLEEHRQHLKQVFEVMR 779
>At2g05610 putative retroelement pol polyprotein
Length = 780
Score = 124 bits (312), Expect = 3e-28
Identities = 78/220 (35%), Positives = 123/220 (55%), Gaps = 11/220 (5%)
Query: 1020 QLLREYPDIFAWSYEDMPGLDPKIVEHRIPTKPECPPVRQKLRRTHPDMALKIKNEVQKQ 1079
+++ E+PDIF + P P +H+I + PV Q+ R + KNE+ K
Sbjct: 9 KVVTEFPDIFVEPTDLPPFRAPH--DHKIELLEDSNPVNQRPYR----YVVHQKNEIDKI 62
Query: 1080 ID----AGFLMTVEYPEWVANIVPVPKKDGKVRMCVDFRDLNKASPKDNFPLLHIDVLVD 1135
+D +G + P + + +V V KKDG R+CVD+R+LN + KD FP+ I+ L+D
Sbjct: 63 VDDMLASGTIQASSSP-YASPVVLVKKKDGTWRLCVDYRELNGMTVKDRFPIPLIEDLMD 121
Query: 1136 NNAQSKVFFFMDGFSGYNQIKMSPEDREKTSFITPWGTFCYKVMPIGLINAGATYQRGMT 1195
S V+ +D +GY+Q++M P D KT+F T G + Y VMP GL NA A++Q M
Sbjct: 122 ELGGSNVYSKIDLRAGYHQVRMDPLDIHKTAFKTHNGHYEYLVMPFGLTNAPASFQSLMN 181
Query: 1196 TLFHDMIHKEVEVYVDDMIVKSKDEEQHVEYLTKMFERLR 1235
+ F + K V V+ DD+++ S E+H ++L +FE +R
Sbjct: 182 SFFKPFLRKFVLVFFDDILIYSTSMEEHKKHLEAVFEVMR 221
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.136 0.403
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 28,883,285
Number of Sequences: 26719
Number of extensions: 1342616
Number of successful extensions: 4552
Number of sequences better than 10.0: 135
Number of HSP's better than 10.0 without gapping: 70
Number of HSP's successfully gapped in prelim test: 66
Number of HSP's that attempted gapping in prelim test: 4235
Number of HSP's gapped (non-prelim): 313
length of query: 1235
length of database: 11,318,596
effective HSP length: 111
effective length of query: 1124
effective length of database: 8,352,787
effective search space: 9388532588
effective search space used: 9388532588
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 66 (30.0 bits)
Medicago: description of AC144656.13


