

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144656.10 - phase: 0
(324 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
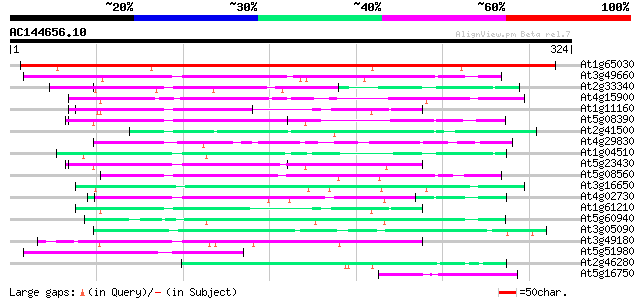
Score E
Sequences producing significant alignments: (bits) Value
At1g65030 Unknown protein (At1g65030) 407 e-114
At3g49660 putative WD-40 repeat - protein 63 2e-10
At2g33340 putative PRP19-like spliceosomal protein 59 3e-09
At4g15900 PRL1 protein 58 8e-09
At1g11160 hypothetical protein 56 3e-08
At5g08390 katanin p80 subunit - like protein 55 5e-08
At2g41500 putative U4/U6 small nuclear ribonucleoprotein 55 5e-08
At4g29830 unknown protein 55 7e-08
At1g04510 putative pre-mRNA splicing factor PRP19 54 1e-07
At5g23430 unknown protein 53 2e-07
At5g08560 WD-repeat protein-like 52 3e-07
At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2 52 4e-07
At4g02730 putative WD-repeat protein 51 7e-07
At1g61210 51 1e-06
At5g60940 cleavage stimulation factor 50K chain 50 2e-06
At3g05090 unknown protein 48 8e-06
At3g49180 unknown protein 47 1e-05
At5g51980 unknown protein 46 3e-05
At2g46280 eukaryotic translation initiation factor 3 delta subunit 46 3e-05
At5g16750 WD40-repeat protein 45 4e-05
>At1g65030 Unknown protein (At1g65030)
Length = 345
Score = 407 bits (1047), Expect = e-114
Identities = 209/322 (64%), Positives = 252/322 (77%), Gaps = 13/322 (4%)
Query: 7 ISLVAGSYERFIWGFNLNPN-------TQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSD 59
+S++AGSYERFIWGF L P T TL+P+FSYPSH+S I TVA S ASGGSD
Sbjct: 1 MSIIAGSYERFIWGFKLKPTKHDAESQTLTLSPLFSYPSHISPITTVACSGPAAASGGSD 60
Query: 60 DTIHLYDLSTSSSLGSLTDHS--STVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGF 117
DTIHLYDL ++SSLGSL DH+ +++TALSFY+P +L FPRNL+SA ADGS+AIFD D F
Sbjct: 61 DTIHLYDLPSASSLGSLLDHNHAASITALSFYTPSSLSFPRNLISAAADGSVAIFDTDPF 120
Query: 118 VHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFD 177
V LK+ HKK VNDLAIHPSGK+AL V RD FAM+NLVRG+RSFCCRL EAS+V+FD
Sbjct: 121 VLLKSFRPHKKAVNDLAIHPSGKLALAVYRDEFFAMLNLVRGKRSFCCRLGHEASLVKFD 180
Query: 178 VSGDSFFMAVDEIVNVHQAEDARLLMELQCP--KRVLCAAPAKNGLLYTGGEDRNITAWD 235
SG+ F+M V V VHQ+EDA+LL+EL+ P KR+LCA P ++G L+TGGEDR ITAWD
Sbjct: 181 PSGERFYMVVSNKVGVHQSEDAKLLLELENPSRKRILCATPGESGTLFTGGEDRAITAWD 240
Query: 236 LKSGKVAYCIEEAHAARVKGIVVL--SDEATGDDEPYLVASASSDGTIRAWDVRMAATEK 293
SGK+AY IE+AH AR+KGIVVL +D ++PYL+ SASSDG IR WDVRMAA E
Sbjct: 241 TNSGKLAYSIEDAHPARIKGIVVLTRNDSDGSLEDPYLIGSASSDGIIRVWDVRMAAKEN 300
Query: 294 SEPLAECKTQSRLTCLAGSSLK 315
++PLAE T+SRLTCLAGS+LK
Sbjct: 301 TKPLAETNTKSRLTCLAGSALK 322
>At3g49660 putative WD-40 repeat - protein
Length = 317
Score = 63.2 bits (152), Expect = 2e-10
Identities = 70/290 (24%), Positives = 119/290 (40%), Gaps = 30/290 (10%)
Query: 9 LVAGSYERFIWGFNLNP-NTQTLTPVFSYPSHLSLIKTVAVSNSV--VASGGSDDTIHLY 65
L + S ++ I + +N N PV + H + I VA S+ + S D T+ L+
Sbjct: 39 LASASADKTIRTYTINTINDPIAEPVQEFTGHENGISDVAFSSDARFIVSASDDKTLKLW 98
Query: 66 DLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSV 125
D+ T S + +L H++ ++F N+ +VS D ++ I+D LK L
Sbjct: 99 DVETGSLIKTLIGHTNYAFCVNFNPQSNM-----IVSGSFDETVRIWDVTTGKCLKVLPA 153
Query: 126 HKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCR--LDKE---ASIVRFDVSG 180
H V + + G + ++ S D + + G C + +D E S VRF +G
Sbjct: 154 HSDPVTAVDFNRDGSLIVSSSYDGLCRIWDSGTGH---CVKTLIDDENPPVSFVRFSPNG 210
Query: 181 DSFFMA-VDEIVNVHQAEDARLLM----ELQCPKRVLCAAPAKNGL-LYTGGEDRNITAW 234
+ +D + + A+ L + + A NG + +G ED + W
Sbjct: 211 KFILVGTLDNTLRLWNISSAKFLKTYTGHVNAQYCISSAFSVTNGKRIVSGSEDNCVHMW 270
Query: 235 DLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAW 284
+L S K+ +E H V + E L+AS S D T+R W
Sbjct: 271 ELNSKKLLQKLE-GHTETVMNVACHPTEN-------LIASGSLDKTVRIW 312
>At2g33340 putative PRP19-like spliceosomal protein
Length = 565
Score = 59.3 bits (142), Expect = 3e-09
Identities = 63/250 (25%), Positives = 100/250 (39%), Gaps = 55/250 (22%)
Query: 49 SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGS 108
S V+A+GG D T L+D + L +LT HS VT++ F +L +++A AD +
Sbjct: 233 SKDVIATGGVDATAVLFDRPSGQILSTLTGHSKKVTSVKFVGDSDL-----VLTASADKT 287
Query: 109 LAIFDADG---FVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCC 165
+ I+ G + TL+ H V + +HP+ K ++ S D + +L
Sbjct: 288 VRIWRNPGDGNYACGYTLNDHSAEVRAVTVHPTNKYFVSASLDGTWCFYDL--------- 338
Query: 166 RLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGL-LYT 224
SG D+ NV AA +GL L T
Sbjct: 339 ------------SSGSCLAQVSDDSKNVDYT----------------AAAFHPDGLILGT 370
Query: 225 GGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAW 284
G + WD+KS +A+ A+ G + + Y +A+A+ DG +R W
Sbjct: 371 GTSQSVVKIWDVKS--------QANVAKFDGHTGEVTAISFSENGYFLATAAEDG-VRLW 421
Query: 285 DVRMAATEKS 294
D+R KS
Sbjct: 422 DLRKLRNFKS 431
Score = 42.4 bits (98), Expect = 3e-04
Identities = 44/173 (25%), Positives = 71/173 (40%), Gaps = 15/173 (8%)
Query: 24 NPNTQTLTPVFSYPSHLSLIKTVAV--SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSS 81
NP ++ H + ++ V V +N S D T YDLS+ S L ++D S
Sbjct: 293 NPGDGNYACGYTLNDHSAEVRAVTVHPTNKYFVSASLDGTWCFYDLSSGSCLAQVSDDSK 352
Query: 82 TV--TALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSG 139
V TA +F+ P L + + + I+D ++ H V ++ +G
Sbjct: 353 NVDYTAAAFH-----PDGLILGTGTSQSVVKIWDVKSQANVAKFDGHTGEVTAISFSENG 407
Query: 140 KIALTVSRDSCFAMVNL--VRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEI 190
T + D V L +R R+F L +A+ V FD SG +A +I
Sbjct: 408 YFLATAAEDG----VRLWDLRKLRNFKSFLSADANSVEFDPSGSYLGIAASDI 456
>At4g15900 PRL1 protein
Length = 486
Score = 57.8 bits (138), Expect = 8e-09
Identities = 63/270 (23%), Positives = 113/270 (41%), Gaps = 56/270 (20%)
Query: 35 SYPSHLSLIKTVAVSNS--VVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPP 92
SY HLS + +A+ + V+ +GG D ++D+ T + +L+ H +TV S ++ P
Sbjct: 255 SYHGHLSGVYCLALHPTLDVLLTGGRDSVCRVWDIRTKMQIFALSGHDNTVC--SVFTRP 312
Query: 93 NLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFA 152
P +V+ D ++ +D + TL+ HKK V + +HP + S D+
Sbjct: 313 TDP---QVVTGSHDTTIKFWDLRYGKTMSTLTHHKKSVRAMTLHPKENAFASASADNT-K 368
Query: 153 MVNLVRGRRSFCCR-LDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRV 211
+L +G FC L ++ +I+ MAV+E
Sbjct: 369 KFSLPKGE--FCHNMLSQQKTIINA--------MAVNE---------------------- 396
Query: 212 LCAAPAKNGLLYTGGEDRNITAWDLKSG----KVAYCIEEAHAARVKGIVVLSDEATGDD 267
+G++ TGG++ +I WD KSG + ++ GI + TG
Sbjct: 397 -------DGVMVTGGDNGSIWFWDWKSGHSFQQSETIVQPGSLESEAGIYAACYDNTGS- 448
Query: 268 EPYLVASASSDGTIRAWDVRMAATEKSEPL 297
+ + +D TI+ W AT ++ P+
Sbjct: 449 ---RLVTCEADKTIKMWKEDENATPETHPI 475
Score = 37.7 bits (86), Expect = 0.009
Identities = 30/119 (25%), Positives = 50/119 (41%), Gaps = 16/119 (13%)
Query: 21 FNLNPNTQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHS 80
F L+ + T+ VF+ P+ + V +G D TI +DL ++ +LT H
Sbjct: 296 FALSGHDNTVCSVFTRPT-----------DPQVVTGSHDTTIKFWDLRYGKTMSTLTHHK 344
Query: 81 STVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSG 139
+V A++ + N SA AD + G LS K +N +A++ G
Sbjct: 345 KSVRAMTLHPKENA-----FASASADNTKKFSLPKGEFCHNMLSQQKTIINAMAVNEDG 398
Score = 30.8 bits (68), Expect = 1.0
Identities = 17/63 (26%), Positives = 29/63 (45%), Gaps = 8/63 (12%)
Query: 224 TGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRA 283
TG DR I WD+ +G + + H +V+G+ V + + SA D ++
Sbjct: 193 TGSADRTIKIWDVATGVLKLTL-TGHIEQVRGLAV-------SNRHTYMFSAGDDKQVKC 244
Query: 284 WDV 286
WD+
Sbjct: 245 WDL 247
>At1g11160 hypothetical protein
Length = 974
Score = 55.8 bits (133), Expect = 3e-08
Identities = 53/205 (25%), Positives = 84/205 (40%), Gaps = 51/205 (24%)
Query: 39 HLSLIKTVAVSNS--VVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPF 96
H S + +VA ++ +V +G S I L+DL S + + T H S +A+ F+ PF
Sbjct: 6 HTSPVDSVAFNSEEVLVLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVEFH-----PF 60
Query: 97 PRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNL 156
L S +D +L ++D ++T H +G++ + P G
Sbjct: 61 GEFLASGSSDTNLRVWDTRKKGCIQTYKGHTRGISTIEFSPDG----------------- 103
Query: 157 VRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQC---PKRVLC 213
R+ VSG +D +V V +LL E +C P R L
Sbjct: 104 ------------------RWVVSG-----GLDNVVKVWDLTAGKLLHEFKCHEGPIRSLD 140
Query: 214 AAPAKNGLLYTGGEDRNITAWDLKS 238
P + LL TG DR + WDL++
Sbjct: 141 FHPLEF-LLATGSADRTVKFWDLET 164
Score = 45.1 bits (105), Expect = 5e-05
Identities = 29/108 (26%), Positives = 47/108 (42%), Gaps = 7/108 (6%)
Query: 35 SYPSHLSLIKTVAVS--NSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPP 92
+Y H I T+ S V SGG D+ + ++DL+ L H + +L F+
Sbjct: 86 TYKGHTRGISTIEFSPDGRWVVSGGLDNVVKVWDLTAGKLLHEFKCHEGPIRSLDFH--- 142
Query: 93 NLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGK 140
P L + AD ++ +D + F + T GV +A HP G+
Sbjct: 143 --PLEFLLATGSADRTVKFWDLETFELIGTTRPEATGVRAIAFHPDGQ 188
Score = 32.7 bits (73), Expect = 0.27
Identities = 40/214 (18%), Positives = 86/214 (39%), Gaps = 55/214 (25%)
Query: 75 SLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLA 134
SL H+S V +++F S L +++ + G + ++D + ++ + H+ + +
Sbjct: 2 SLCGHTSPVDSVAFNSEEVL-----VLAGASSGVIKLWDLEESKMVRAFTGHRSNCSAVE 56
Query: 135 IHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVH 194
HP G+ + S D+ + + R+ C + K + I +
Sbjct: 57 FHPFGEFLASGSSDTNLRVWDT---RKKGCIQTYKGHT---------------RGISTIE 98
Query: 195 QAEDARLLMELQCPKRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVK 254
+ D R ++ +GG D + WDL +GK+ + + H ++
Sbjct: 99 FSPDGRWVV--------------------SGGLDNVVKVWDLTAGKLLHEF-KCHEGPIR 137
Query: 255 GIVVLSDEATGDDEP--YLVASASSDGTIRAWDV 286
+ D P +L+A+ S+D T++ WD+
Sbjct: 138 SL---------DFHPLEFLLATGSADRTVKFWDL 162
>At5g08390 katanin p80 subunit - like protein
Length = 823
Score = 55.1 bits (131), Expect = 5e-08
Identities = 49/256 (19%), Positives = 111/256 (43%), Gaps = 59/256 (23%)
Query: 35 SYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNL 94
++ + ++ +K S+ V+ +GG D ++L+ + +++ SL HSS + +++F + L
Sbjct: 107 AHSAAVNCLKIGRKSSRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEGL 166
Query: 95 PFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMV 154
+ + A G++ ++D + ++TL+ H+ + HP G+ + S D+ +
Sbjct: 167 -----VAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFHPFGEFFASGSLDTNLKIW 221
Query: 155 NLVRGRRSFCCRLDK----EASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKR 210
++ R+ C K +++RF G
Sbjct: 222 DI---RKKGCIHTYKGHTRGVNVLRFTPDG------------------------------ 248
Query: 211 VLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPY 270
+ +GGED + WDL +GK+ + ++H +++ + E +
Sbjct: 249 ---------RWIVSGGEDNVVKVWDLTAGKLLHEF-KSHEGKIQSLDFHPHE-------F 291
Query: 271 LVASASSDGTIRAWDV 286
L+A+ S+D T++ WD+
Sbjct: 292 LLATGSADKTVKFWDL 307
Score = 52.4 bits (124), Expect = 3e-07
Identities = 37/130 (28%), Positives = 61/130 (46%), Gaps = 7/130 (5%)
Query: 33 VFSYPSHLSLIKTVA--VSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYS 90
+ S H S I +V S +VA+G + TI L+DL + + +LT H S +++F+
Sbjct: 145 ILSLYGHSSGIDSVTFDASEGLVAAGAASGTIKLWDLEEAKVVRTLTGHRSNCVSVNFH- 203
Query: 91 PPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSC 150
PF S D +L I+D + T H +GVN L P G+ ++ D+
Sbjct: 204 ----PFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWIVSGGEDNV 259
Query: 151 FAMVNLVRGR 160
+ +L G+
Sbjct: 260 VKVWDLTAGK 269
Score = 34.7 bits (78), Expect = 0.072
Identities = 20/69 (28%), Positives = 32/69 (45%), Gaps = 8/69 (11%)
Query: 221 LLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGT 280
+L TGGED + W + + H++ + D T D LVA+ ++ GT
Sbjct: 124 VLVTGGEDHKVNLWAIGKPNAILSLY-GHSSGI-------DSVTFDASEGLVAAGAASGT 175
Query: 281 IRAWDVRMA 289
I+ WD+ A
Sbjct: 176 IKLWDLEEA 184
Score = 28.1 bits (61), Expect = 6.8
Identities = 19/71 (26%), Positives = 28/71 (38%), Gaps = 8/71 (11%)
Query: 217 AKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASAS 276
A GL+ G I WDL+ KV + V ++ G+ AS S
Sbjct: 162 ASEGLVAAGAASGTIKLWDLEEAKVVRTL----TGHRSNCVSVNFHPFGE----FFASGS 213
Query: 277 SDGTIRAWDVR 287
D ++ WD+R
Sbjct: 214 LDTNLKIWDIR 224
>At2g41500 putative U4/U6 small nuclear ribonucleoprotein
Length = 554
Score = 55.1 bits (131), Expect = 5e-08
Identities = 64/240 (26%), Positives = 96/240 (39%), Gaps = 19/240 (7%)
Query: 70 SSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKG 129
++++ L DH T + F P L +A AD + ++ DG + L+T H
Sbjct: 288 TNTIAVLKDHKERATDVVFS-----PVDDCLATASADRTAKLWKTDGTL-LQTFEGHLDR 341
Query: 130 VNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSFFMA--- 186
+ +A HPSGK T S D + + ++ G + S+ D A
Sbjct: 342 LARVAFHPSGKYLGTTSYDKTWRLWDINTGAE-LLLQEGHSRSVYGIAFQQDGALAASCG 400
Query: 187 VDEIVNVHQAEDARLLMELQCP-KRVLCAAPAKNGL-LYTGGEDRNITAWDLKSGKVAYC 244
+D + V R ++ Q K V + NG L +GGED WDL+ K Y
Sbjct: 401 LDSLARVWDLRTGRSILVFQGHIKPVFSVNFSPNGYHLASGGEDNQCRIWDLRMRKSLYI 460
Query: 245 IEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDVRMAATEKSEPLAECKTQS 304
I AHA ++S E Y +A+AS D + W R + KS E K S
Sbjct: 461 I-PAHAN------LVSQVKYEPQEGYFLATASYDMKVNIWSGRDFSLVKSLAGHESKVAS 513
Score = 38.5 bits (88), Expect = 0.005
Identities = 29/96 (30%), Positives = 45/96 (46%), Gaps = 4/96 (4%)
Query: 53 VASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIF 112
+ASGG D+ ++DL SL + H++ V+ + Y P F L +A D + I+
Sbjct: 438 LASGGEDNQCRIWDLRMRKSLYIIPAHANLVSQVK-YEPQEGYF---LATASYDMKVNIW 493
Query: 113 DADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRD 148
F +K+L+ H+ V L I TVS D
Sbjct: 494 SGRDFSLVKSLAGHESKVASLDITADSSCIATVSHD 529
>At4g29830 unknown protein
Length = 321
Score = 54.7 bits (130), Expect = 7e-08
Identities = 64/245 (26%), Positives = 106/245 (43%), Gaps = 42/245 (17%)
Query: 49 SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGS 108
S + AS D + ++D+ T++++ L S V + F P+ + A A GS
Sbjct: 71 SGIIAASSSIDSFVRVFDVDTNATIAVLEAPPSEVWGMQFE-------PKGTILAVAGGS 123
Query: 109 LA---IFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCC 165
A ++D + + TLS+ + A PS K T S+ ++ G+R C
Sbjct: 124 SASVKLWDTASWRLISTLSIPRPD----APKPSDK---TSSKKFVLSVAWSPNGKRLACG 176
Query: 166 RLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTG 225
+D +I FDV D +HQ E + P R L +P +L++G
Sbjct: 177 SMD--GTICVFDV---------DRSKLLHQLEGHNM------PVRSLVFSPVDPRVLFSG 219
Query: 226 GEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWD 285
+D ++ D + GK H + V LS +A+ D +A+ SSD T+R WD
Sbjct: 220 SDDGHVNMHDAE-GKTLLGSMSGHTSWV-----LSVDASPDGGA--IATGSSDRTVRLWD 271
Query: 286 VRMAA 290
++M A
Sbjct: 272 LKMRA 276
Score = 37.7 bits (86), Expect = 0.009
Identities = 22/82 (26%), Positives = 43/82 (51%), Gaps = 3/82 (3%)
Query: 35 SYPSHLSLIKTVAVS--NSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPP 92
S H S + +V S +A+G SD T+ L+DL +++ ++++H+ V +++F P
Sbjct: 238 SMSGHTSWVLSVDASPDGGAIATGSSDRTVRLWDLKMRAAIQTMSNHNDQVWSVAFRPPG 297
Query: 93 NLPF-PRNLVSADADGSLAIFD 113
L S D S++++D
Sbjct: 298 GTGVRAGRLASVSDDKSVSLYD 319
Score = 35.4 bits (80), Expect = 0.042
Identities = 23/100 (23%), Positives = 46/100 (46%), Gaps = 5/100 (5%)
Query: 48 VSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADG 107
V V+ SG D ++++D + LGS++ H+S V ++ P + + +D
Sbjct: 211 VDPRVLFSGSDDGHVNMHDAEGKTLLGSMSGHTSWVLSVD-----ASPDGGAIATGSSDR 265
Query: 108 SLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSR 147
++ ++D ++T+S H V +A P G + R
Sbjct: 266 TVRLWDLKMRAAIQTMSNHNDQVWSVAFRPPGGTGVRAGR 305
>At1g04510 putative pre-mRNA splicing factor PRP19
Length = 523
Score = 53.9 bits (128), Expect = 1e-07
Identities = 63/268 (23%), Positives = 108/268 (39%), Gaps = 57/268 (21%)
Query: 28 QTLTPVFSYPSHLS-----LIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSST 82
+ T + S+P H + + S V+A+GG D T L+D + L +LT HS
Sbjct: 207 EKFTQLSSHPLHKTNKPGIFSMDILHSKDVIATGGIDTTAVLFDRPSGQILSTLTGHSKK 266
Query: 83 VTALSFYSPPNLPFPRNLVSADADGSLAIF--DADG-FVHLKTLSVHKKGVNDLAIHPSG 139
VT++ F +L +++A +D ++ I+ DG + TL H V + +H +
Sbjct: 267 VTSIKFVGDTDL-----VLTASSDKTVRIWGCSEDGNYTSRHTLKDHSAEVRAVTVHATN 321
Query: 140 KIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDA 199
K ++ S DS + +L G C +AS DV+ + D ++
Sbjct: 322 KYFVSASLDSTWCFYDLSSG---LCLAQVTDAS--ENDVNYTAAAFHPDGLI-------- 368
Query: 200 RLLMELQCPKRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVL 259
L TG + WD+KS +A+ A+ G
Sbjct: 369 ----------------------LGTGTAQSIVKIWDVKS--------QANVAKFGGHNGE 398
Query: 260 SDEATGDDEPYLVASASSDGTIRAWDVR 287
+ + Y +A+A+ DG +R WD+R
Sbjct: 399 ITSISFSENGYFLATAALDG-VRLWDLR 425
Score = 36.6 bits (83), Expect = 0.019
Identities = 45/188 (23%), Positives = 76/188 (39%), Gaps = 16/188 (8%)
Query: 18 IWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLT 77
IWG + + N + + + + + + TV +N S D T YDLS+ L +T
Sbjct: 290 IWGCSEDGNYTSRHTLKDHSAEVRAV-TVHATNKYFVSASLDSTWCFYDLSSGLCLAQVT 348
Query: 78 DHSS---TVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLA 134
D S TA +F+ P L + A + I+D ++ H + ++
Sbjct: 349 DASENDVNYTAAAFH-----PDGLILGTGTAQSIVKIWDVKSQANVAKFGGHNGEITSIS 403
Query: 135 IHPSGKIALTVSRDSCFAMVNL--VRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVN 192
+G T + D V L +R ++F +A+ V FD SG +A +I
Sbjct: 404 FSENGYFLATAALDG----VRLWDLRKLKNFRTFDFPDANSVEFDHSGSYLGIAASDI-R 458
Query: 193 VHQAEDAR 200
V QA +
Sbjct: 459 VFQAASVK 466
>At5g23430 unknown protein
Length = 837
Score = 53.1 bits (126), Expect = 2e-07
Identities = 47/211 (22%), Positives = 98/211 (46%), Gaps = 15/211 (7%)
Query: 35 SYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNL 94
++ + ++ +K S+ V+ +GG D ++L+ + +++ SL HSS + +++F + L
Sbjct: 14 AHSAAVNCLKIGRKSSRVLVTGGEDHKVNLWAIGKPNAILSLYGHSSGIDSVTFDASEVL 73
Query: 95 PFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMV 154
+ + A G++ ++D + ++TL+ H+ + HP G+ + S D+ +
Sbjct: 74 -----VAAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFHPFGEFFASGSLDTNLKIW 128
Query: 155 NLVRGRRSFCCRLDK----EASIVRFDVSGDSFFM-AVDEIVNVHQAEDARLLMELQCPK 209
++ R+ C K +++RF G D IV V +LL E + +
Sbjct: 129 DI---RKKGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNIVKVWDLTAGKLLTEFKSHE 185
Query: 210 RVLCAAP--AKNGLLYTGGEDRNITAWDLKS 238
+ + LL TG DR + WDL++
Sbjct: 186 GQIQSLDFHPHEFLLATGSADRTVKFWDLET 216
Score = 51.2 bits (121), Expect = 7e-07
Identities = 37/130 (28%), Positives = 60/130 (45%), Gaps = 7/130 (5%)
Query: 33 VFSYPSHLSLIKTVA--VSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYS 90
+ S H S I +V S +VA+G + TI L+DL + + +LT H S ++ F+
Sbjct: 52 ILSLYGHSSGIDSVTFDASEVLVAAGAASGTIKLWDLEEAKIVRTLTGHRSNCISVDFH- 110
Query: 91 PPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSC 150
PF S D +L I+D + T H +GVN L P G+ ++ D+
Sbjct: 111 ----PFGEFFASGSLDTNLKIWDIRKKGCIHTYKGHTRGVNVLRFTPDGRWVVSGGEDNI 166
Query: 151 FAMVNLVRGR 160
+ +L G+
Sbjct: 167 VKVWDLTAGK 176
Score = 35.4 bits (80), Expect = 0.042
Identities = 20/69 (28%), Positives = 32/69 (45%), Gaps = 8/69 (11%)
Query: 221 LLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGT 280
+L TGGED + W + + H++ + D T D LVA+ ++ GT
Sbjct: 31 VLVTGGEDHKVNLWAIGKPNAILSLY-GHSSGI-------DSVTFDASEVLVAAGAASGT 82
Query: 281 IRAWDVRMA 289
I+ WD+ A
Sbjct: 83 IKLWDLEEA 91
Score = 27.7 bits (60), Expect = 8.8
Identities = 13/37 (35%), Positives = 22/37 (59%)
Query: 52 VVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSF 88
++A+G +D T+ +DL T +GS ++ V LSF
Sbjct: 199 LLATGSADRTVKFWDLETFELIGSGGPETAGVRCLSF 235
>At5g08560 WD-repeat protein-like
Length = 589
Score = 52.4 bits (124), Expect = 3e-07
Identities = 54/242 (22%), Positives = 106/242 (43%), Gaps = 26/242 (10%)
Query: 53 VASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIF 112
V + G+++ I +D+ + + + + +Y P + +++ D S+ ++
Sbjct: 334 VLTCGAEEVIRRWDVDSGDCVHMYEKGGISPISCGWY-----PDGQGIIAGMTDRSICMW 388
Query: 113 DADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRL-DKEA 171
D DG + V+D+A+ GK ++V +DS ++ + R + RL ++E
Sbjct: 389 DLDGREKECWKGQRTQKVSDIAMTDDGKWLVSVCKDSVISLFD----REATVERLIEEED 444
Query: 172 SIVRFDVSGDSFFMAVD----EIVNVHQAEDARLLMELQCPKRVL-----CAAPAKNGLL 222
I F +S D+ ++ V+ EI + D +++ + KR C K +
Sbjct: 445 MITSFSLSNDNKYILVNLLNQEIRLWNIEGDPKIVSRYKGHKRSRFIIRSCFGGYKQAFI 504
Query: 223 YTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIR 282
+G ED + W +GK+ + HA V + + +++ASAS DGTIR
Sbjct: 505 ASGSEDSQVYIWHRSTGKLIVELP-GHAGAVNCV------SWSPTNLHMLASASDDGTIR 557
Query: 283 AW 284
W
Sbjct: 558 IW 559
>At3g16650 PP1/PP2A phosphatases pleiotropic regulator PRL2
Length = 479
Score = 52.0 bits (123), Expect = 4e-07
Identities = 70/309 (22%), Positives = 118/309 (37%), Gaps = 59/309 (19%)
Query: 39 HLSLIKTVAV--SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPF 96
HL +++VA SN +G +D TI ++D++T +LT H V L+ + F
Sbjct: 169 HLGWVRSVAFDPSNEWFCTGSADRTIKIWDVATGVLKLTLTGHIGQVRGLAVSNRHTYMF 228
Query: 97 PRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNL 156
SA D + +D + +++ H GV LA+HP+ + LT RDS + ++
Sbjct: 229 -----SAGDDKQVKCWDLEQNKVIRSYHGHLHGVYCLALHPTLDVVLTGGRDSVCRVWDI 283
Query: 157 VRGRRSFCCRLDKEA------------------SIVRF-DVS-GDSF-----------FM 185
+ F D + S ++F D+ G S M
Sbjct: 284 RTKMQIFVLPHDSDVFSVLARPTDPQVITGSHDSTIKFWDLRYGKSMATITNHKKTVRAM 343
Query: 186 AVDEIVNVHQAEDARLLMELQCPKRVLC-------------AAPAKNGLLYTGGEDRNIT 232
A+ N + A + + PK C A ++G++ TGG+ +
Sbjct: 344 ALHPKENDFVSASADNIKKFSLPKGEFCHNMLSLQRDIINAVAVNEDGVMVTGGDKGGLW 403
Query: 233 AWDLKSG----KVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDVRM 288
WD KSG + ++ GI + TG + + D TI+ W
Sbjct: 404 FWDWKSGHNFQRAETIVQPGSLESEAGIYAACYDQTGS----RLVTCEGDKTIKMWKEDE 459
Query: 289 AATEKSEPL 297
AT ++ PL
Sbjct: 460 DATPETHPL 468
Score = 40.4 bits (93), Expect = 0.001
Identities = 29/113 (25%), Positives = 51/113 (44%), Gaps = 6/113 (5%)
Query: 28 QTLTPVFSYPSHLSLIKTVA-VSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTAL 86
+T +F P + +A ++ V +G D TI +DL S+ ++T+H TV A+
Sbjct: 284 RTKMQIFVLPHDSDVFSVLARPTDPQVITGSHDSTIKFWDLRYGKSMATITNHKKTVRAM 343
Query: 87 SFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSG 139
+ + P + VSA AD G LS+ + +N +A++ G
Sbjct: 344 ALH-----PKENDFVSASADNIKKFSLPKGEFCHNMLSLQRDIINAVAVNEDG 391
>At4g02730 putative WD-repeat protein
Length = 333
Score = 51.2 bits (121), Expect = 7e-07
Identities = 48/200 (24%), Positives = 85/200 (42%), Gaps = 25/200 (12%)
Query: 50 NSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSL 109
++++ SG D+TI ++++ T + + HS ++++ F +L +VSA DGS
Sbjct: 140 SNLIVSGSFDETIRIWEVKTGKCVRMIKAHSMPISSVHFNRDGSL-----IVSASHDGSC 194
Query: 110 AIFDADGFVHLKTLSVHKK-GVNDLAIHPSGKIALTVSRDSCFAMVNLVRGR-------- 160
I+DA LKTL K V+ P+GK L + DS + N G+
Sbjct: 195 KIWDAKEGTCLKTLIDDKSPAVSFAKFSPNGKFILVATLDSTLKLSNYATGKFLKVYTGH 254
Query: 161 --RSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAA--- 215
+ FC + ++ VSG D V + + +L L+ + +
Sbjct: 255 TNKVFCITSAFSVTNGKYIVSGSE-----DNCVYLWDLQARNILQRLEGHTDAVISVSCH 309
Query: 216 PAKNGLLYTGGE-DRNITAW 234
P +N + +G D+ I W
Sbjct: 310 PVQNEISSSGNHLDKTIRIW 329
Score = 41.2 bits (95), Expect = 8e-04
Identities = 52/251 (20%), Positives = 93/251 (36%), Gaps = 71/251 (28%)
Query: 46 VAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADA 105
VA +NS + G+ + +Y L +L H++ ++ + F + NL L SA
Sbjct: 12 VANANST-GNAGTSGNVPIY--KPYRHLKTLEGHTAAISCVKFSNDGNL-----LASASV 63
Query: 106 DGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRD---------SCFAMVNL 156
D ++ ++ A + + H G++DLA + S D S + + +
Sbjct: 64 DKTMILWSATNYSLIHRYEGHSSGISDLAWSSDSHYTCSASDDCTLRIWDARSPYECLKV 123
Query: 157 VRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAP 216
+RG +F V F+ P
Sbjct: 124 LRGHTNF-------VFCVNFN--------------------------------------P 138
Query: 217 AKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASAS 276
N L+ +G D I W++K+GK I +AH+ + + D + L+ SAS
Sbjct: 139 PSN-LIVSGSFDETIRIWEVKTGKCVRMI-KAHSMPISSVHFNRDGS-------LIVSAS 189
Query: 277 SDGTIRAWDVR 287
DG+ + WD +
Sbjct: 190 HDGSCKIWDAK 200
>At1g61210
Length = 282
Score = 50.8 bits (120), Expect = 1e-06
Identities = 51/205 (24%), Positives = 83/205 (39%), Gaps = 51/205 (24%)
Query: 39 HLSLIKTVAVSNS--VVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPF 96
H S + +VA ++ +V +G S I L+D+ + + + T H S +A+ F+ PF
Sbjct: 87 HTSAVDSVAFDSAEVLVLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEFH-----PF 141
Query: 97 PRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNL 156
L S +D +L I+D ++T H +G++ + P G
Sbjct: 142 GEFLASGSSDANLKIWDIRKKGCIQTYKGHSRGISTIRFTPDG----------------- 184
Query: 157 VRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQC---PKRVLC 213
R+ VSG +D +V V +LL E + P R L
Sbjct: 185 ------------------RWVVSG-----GLDNVVKVWDLTAGKLLHEFKFHEGPIRSLD 221
Query: 214 AAPAKNGLLYTGGEDRNITAWDLKS 238
P + LL TG DR + WDL++
Sbjct: 222 FHPLEF-LLATGSADRTVKFWDLET 245
Score = 40.8 bits (94), Expect = 0.001
Identities = 49/272 (18%), Positives = 112/272 (41%), Gaps = 65/272 (23%)
Query: 27 TQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSL----------GSL 76
T++L ++ ++++ + ++ + +GG D ++L+ + +SL SL
Sbjct: 25 TESLGEFLAHSANVNCLSIGKKTSRLFITGGDDYKVNLWAIGKPTSLMKNDAIAFYWQSL 84
Query: 77 TDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIH 136
H+S V +++F S L +++ + G + ++D + ++ + H+ + + H
Sbjct: 85 CGHTSAVDSVAFDSAEVL-----VLAGASSGVIKLWDVEEAKMVRAFTGHRSNCSAVEFH 139
Query: 137 PSGKIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVNVHQA 196
P G+ + S D+ + ++ R+ C + K S I +
Sbjct: 140 PFGEFLASGSSDANLKIWDI---RKKGCIQTYKGHS---------------RGISTIRFT 181
Query: 197 EDARLLMELQCPKRVLCAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGI 256
D R ++ +GG D + WDL +GK+ + + H ++ +
Sbjct: 182 PDGRWVV--------------------SGGLDNVVKVWDLTAGKLLHEF-KFHEGPIRSL 220
Query: 257 VVLSDEATGDDEP--YLVASASSDGTIRAWDV 286
D P +L+A+ S+D T++ WD+
Sbjct: 221 ---------DFHPLEFLLATGSADRTVKFWDL 243
Score = 32.0 bits (71), Expect = 0.47
Identities = 21/85 (24%), Positives = 37/85 (42%), Gaps = 7/85 (8%)
Query: 35 SYPSHLSLIKTVAVS--NSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPP 92
+Y H I T+ + V SGG D+ + ++DL+ L H + +L F+
Sbjct: 167 TYKGHSRGISTIRFTPDGRWVVSGGLDNVVKVWDLTAGKLLHEFKFHEGPIRSLDFH--- 223
Query: 93 NLPFPRNLVSADADGSLAIFDADGF 117
P L + AD ++ +D + F
Sbjct: 224 --PLEFLLATGSADRTVKFWDLETF 246
>At5g60940 cleavage stimulation factor 50K chain
Length = 429
Score = 49.7 bits (117), Expect = 2e-06
Identities = 59/265 (22%), Positives = 104/265 (38%), Gaps = 40/265 (15%)
Query: 44 KTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSA 103
+T ++ SV + GS TI ++ T L++H S V F SP + F +
Sbjct: 93 RTASIDFSVNHAKGSSKTIPKHESKT------LSEHKSVVRCARF-SPDGMFF----ATG 141
Query: 104 DADGSLAIF-----------DADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFA 152
AD S+ +F D ++T H + +NDL HP I ++ ++D+C
Sbjct: 142 GADTSIKLFEVPKVKQMISGDTQARPLIRTFYDHAEPINDLDFHPRSTILISSAKDNCIK 201
Query: 153 MVNLVRGRRSFCCRLDKEASIVR---FDVSGDSFFMAVDE-IVNVHQAEDARLLMELQCP 208
+ + ++ ++ VR F SG+ D I +++ + + P
Sbjct: 202 FFDFSKTTAKRAFKVFQDTHNVRSISFHPSGEFLLAGTDHPIPHLYDVNTYQCFLPSNFP 261
Query: 209 KRVLCAA-----PAKNGLLY-TGGEDRNITAWDLKSGKVAYCIEEAHA-ARVKGIVVLSD 261
+ A + G +Y T +D I +D S K I AH + V V D
Sbjct: 262 DSGVSGAINQVRYSSTGSIYITASKDGAIRLFDGVSAKCVRSIGNAHGKSEVTSAVFTKD 321
Query: 262 EATGDDEPYLVASASSDGTIRAWDV 286
+ V S+ D T++ W++
Sbjct: 322 QR-------FVLSSGKDSTVKLWEI 339
Score = 40.8 bits (94), Expect = 0.001
Identities = 58/282 (20%), Positives = 103/282 (35%), Gaps = 47/282 (16%)
Query: 39 HLSLIKTVAVS--NSVVASGGSDDTIHLY-----------DLSTSSSLGSLTDHSSTVTA 85
H S+++ S A+GG+D +I L+ D + + DH+ +
Sbjct: 122 HKSVVRCARFSPDGMFFATGGADTSIKLFEVPKVKQMISGDTQARPLIRTFYDHAEPIND 181
Query: 86 LSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKK--GVNDLAIHPSGKIAL 143
L F+ P L+S+ D + FD + V + V ++ HPSG+ L
Sbjct: 182 LDFH-----PRSTILISSAKDNCIKFFDFSKTTAKRAFKVFQDTHNVRSISFHPSGEFLL 236
Query: 144 T-----------VSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGDSFFMAVDE--- 189
V+ CF N S VR+ +G + A +
Sbjct: 237 AGTDHPIPHLYDVNTYQCFLPSNFPDSGVSGAINQ------VRYSSTGSIYITASKDGAI 290
Query: 190 -IVNVHQAEDARLLMELQCPKRVLCAAPAKNG-LLYTGGEDRNITAWDLKSGKVAYCIEE 247
+ + A+ R + V A K+ + + G+D + W++ SG++ E
Sbjct: 291 RLFDGVSAKCVRSIGNAHGKSEVTSAVFTKDQRFVLSSGKDSTVKLWEIGSGRMVK--EY 348
Query: 248 AHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDVRMA 289
A RVK + S D E ++++ + + WD R A
Sbjct: 349 LGAKRVK---LRSQAIFNDTEEFVISIDEASNEVVTWDARTA 387
>At3g05090 unknown protein
Length = 753
Score = 47.8 bits (112), Expect = 8e-06
Identities = 61/279 (21%), Positives = 105/279 (36%), Gaps = 36/279 (12%)
Query: 49 SNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGS 108
+ +++ SGG++ + ++D T S L H+ V L S R +S +D
Sbjct: 224 TGTMLVSGGTEKVLRVWDPRTGSKSMKLRGHTDNVRVLLLDSTG-----RFCLSGSSDSM 278
Query: 109 LAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRGRRSFCCRLD 168
+ ++D L T +VH V LA +PS + RD C + +L C
Sbjct: 279 IRLWDLGQQRCLHTYAVHTDSVWALACNPSFSHVYSGGRDQCLYLTDLATRESVLLC--T 336
Query: 169 KEASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQCPKRVLCAAPAKNGLLYTGGED 228
KE I + + +S ++A + + R E+Q PK V + G G
Sbjct: 337 KEHPIQQLALQDNSIWVATTD------SSVERWPAEVQSPKTVF----QRGGSFLAGNLS 386
Query: 229 RNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDV-- 286
N L+ A +E ++ E + L A+ G+++ WD+
Sbjct: 387 FNRARVSLEGLNPAPAYKEPSITVPGTHPIVQHEILNNKRQILTKDAA--GSVKLWDITR 444
Query: 287 --------RMAATEKSEPLAEC-------KTQSRLTCLA 310
+++ EK E L E +RL CL+
Sbjct: 445 GVVVEDYGKISFEEKKEELFEMVSIPSWFTVDTRLGCLS 483
Score = 28.1 bits (61), Expect = 6.8
Identities = 41/223 (18%), Positives = 96/223 (42%), Gaps = 20/223 (8%)
Query: 36 YPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLP 95
+ +++ + A +N+VVASGG + ++D+ + S + + ++ ++ + + P
Sbjct: 125 HSDYVTCLAVAAKNNNVVASGGLGGEVFIWDIEAALSPVTKPNDANEDSSSNGANGP--- 181
Query: 96 FPRNLVSADADGSLAIFDADGFVHLKTLSV-HKKGVNDLAIHPSGKIALTVSRDSCFAMV 154
+L + + ++++ + + T++ HK+ V LA++ +G + ++ + +
Sbjct: 182 -VTSLRTVGSSNNISVQSSPSHGYTPTIAKGHKESVYALAMNDTGTMLVSGGTEKVLRVW 240
Query: 155 NLVRGRRSFCCRLDKE-------ASIVRFDVSGDSFFMAVDEIVNVHQAEDARLLMELQC 207
+ G +S R + S RF +SG S D ++ + R L
Sbjct: 241 DPRTGSKSMKLRGHTDNVRVLLLDSTGRFCLSGSS-----DSMIRLWDLGQQRCLHTYAV 295
Query: 208 PKRVLCAAPAKNGL--LYTGGEDRNITAWDLKSGK-VAYCIEE 247
+ A +Y+GG D+ + DL + + V C +E
Sbjct: 296 HTDSVWALACNPSFSHVYSGGRDQCLYLTDLATRESVLLCTKE 338
>At3g49180 unknown protein
Length = 438
Score = 47.0 bits (110), Expect = 1e-05
Identities = 59/245 (24%), Positives = 99/245 (40%), Gaps = 35/245 (14%)
Query: 17 FIWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVSN--SVVASGGSDDTIHLYDLSTSSSLG 74
F W + T+ V SYP + IK +A +N + + GG I+L++++T L
Sbjct: 62 FYWSW-----TKPQAEVKSYP--VEPIKALAANNEGTYLVGGGISGDIYLWEVATGKLLK 114
Query: 75 SLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDA----DGF-------VHLKTL 123
H +VT L F +L LVS DGS+ ++ D F ++
Sbjct: 115 KWHGHYRSVTCLVFSGDDSL-----LVSGSQDGSIRVWSLIRLFDDFQRQQGNTLYEHNF 169
Query: 124 SVHKKGVNDLAIHPSG--KIALTVSRDSCFAMVNLVRGRRSFCCRLDKEASIVRFDVSGD 181
+ H V D+ I G + ++ S D + +L RG+ + + D G
Sbjct: 170 NEHTMSVTDIVIDYGGCNAVIISSSEDRTCKVWSLSRGKLLKNIIFPSVINALALDPGGC 229
Query: 182 SFFMAVDE------IVNVHQAEDARLLMEL-QCPKRVLCAAPAKNG-LLYTGGEDRNITA 233
F+ + +N ++L + + K + C A +G LL +G ED +
Sbjct: 230 VFYAGARDSKIYIGAINATSEYGTQVLGSVSEKGKAITCLAYCADGNLLISGSEDGVVCV 289
Query: 234 WDLKS 238
WD KS
Sbjct: 290 WDPKS 294
Score = 37.7 bits (86), Expect = 0.009
Identities = 26/96 (27%), Positives = 49/96 (50%), Gaps = 13/96 (13%)
Query: 41 SLIKTVAVS--NSVVASGGSDDTIHLYDLSTSSS-----LGSLTDHSSTVTALSFYSPPN 93
S+I +A+ V +G D I++ ++ +S LGS+++ +T L++ + N
Sbjct: 217 SVINALALDPGGCVFYAGARDSKIYIGAINATSEYGTQVLGSVSEKGKAITCLAYCADGN 276
Query: 94 LPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKKG 129
L L+S DG + ++D H++TL +H KG
Sbjct: 277 L-----LISGSEDGVVCVWDPKSLRHVRTL-IHAKG 306
Score = 29.3 bits (64), Expect = 3.0
Identities = 22/73 (30%), Positives = 37/73 (50%), Gaps = 8/73 (10%)
Query: 214 AAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVA 273
AA + L GG +I W++ +GK+ +++ H + + L +GDD L+
Sbjct: 85 AANNEGTYLVGGGISGDIYLWEVATGKL---LKKWHG-HYRSVTCLV--FSGDDS--LLV 136
Query: 274 SASSDGTIRAWDV 286
S S DG+IR W +
Sbjct: 137 SGSQDGSIRVWSL 149
>At5g51980 unknown protein
Length = 437
Score = 45.8 bits (107), Expect = 3e-05
Identities = 35/127 (27%), Positives = 58/127 (45%), Gaps = 7/127 (5%)
Query: 9 LVAGSYERFIWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVSNSVVASGGSDDTIHLYDLS 68
L AG+ + I + N T P S H + T+ V + + SG D TI ++ L
Sbjct: 243 LFAGTQDGSILAWRYNAATNCFEPSASLTGHTLAVVTLYVGANRLYSGSMDKTIKVWSLD 302
Query: 69 TSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSLAIFDADGFVHLKTLSVHKK 128
+ +LTDHSS V +L + + + L+S D ++ I+ A +L+ HK+
Sbjct: 303 NLQCIQTLTDHSSVVMSL-------ICWDQFLLSCSLDNTVKIWAAIEGGNLEVTYTHKE 355
Query: 129 GVNDLAI 135
LA+
Sbjct: 356 EHGVLAL 362
Score = 29.6 bits (65), Expect = 2.3
Identities = 15/48 (31%), Positives = 26/48 (53%), Gaps = 2/48 (4%)
Query: 195 QAEDARLLMELQCPKRVLC--AAPAKNGLLYTGGEDRNITAWDLKSGK 240
+ E LL +L ++++ A P+ + LYTG +D + WD SG+
Sbjct: 137 KGESFALLTQLDGHEKLVSGIALPSGSDKLYTGSKDETLRVWDCASGQ 184
>At2g46280 eukaryotic translation initiation factor 3 delta subunit
Length = 328
Score = 45.8 bits (107), Expect = 3e-05
Identities = 52/204 (25%), Positives = 82/204 (39%), Gaps = 21/204 (10%)
Query: 100 LVSADADGSLAIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVSRDSCFAMVNLVRG 159
L S D + ++ AD L T H V + +T S D + ++ G
Sbjct: 25 LFSCAKDHTPTLWFADNGERLGTYRGHNGAVWCCDVSRDSSRLITGSADQTAKLWDVKSG 84
Query: 160 RRSFCCRLDKEASIVRFDVSGDSFFMAVDEIVN----VH--------QAEDARLLMELQC 207
+ F + + V F V + D V+ +H + +DA ++ L C
Sbjct: 85 KELFTFKFNAPTRSVDFAVGDRLAVITTDHFVDRTAAIHVKRIAEDPEEQDAESVLVLHC 144
Query: 208 P---KRVLCAAPAK-NGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEA 263
P KR+ A N + +GGED+ I WD ++GK+ +E K I L A
Sbjct: 145 PDGKKRINRAVWGPLNQTIVSGGEDKVIRIWDAETGKLLKQSDE-EVGHKKDITSLCKAA 203
Query: 264 TGDDEPYLVASASSDGTIRAWDVR 287
DD +L + S D T + WD+R
Sbjct: 204 --DDSHFL--TGSLDKTAKLWDMR 223
>At5g16750 WD40-repeat protein
Length = 876
Score = 45.4 bits (106), Expect = 4e-05
Identities = 29/80 (36%), Positives = 36/80 (44%), Gaps = 6/80 (7%)
Query: 214 AAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEATGDDEPYLVA 273
A A GLL T G DR + WD+ G +C H R VV S D ++
Sbjct: 109 ACHASGGLLATAGADRKVLVWDVDGG---FC---THYFRGHKGVVSSILFHPDSNKNILI 162
Query: 274 SASSDGTIRAWDVRMAATEK 293
S S D T+R WD+ TEK
Sbjct: 163 SGSDDATVRVWDLNAKNTEK 182
Score = 39.7 bits (91), Expect = 0.002
Identities = 69/341 (20%), Positives = 128/341 (37%), Gaps = 53/341 (15%)
Query: 9 LVAGSYERFIWGFNLNPNTQTLTPVFSYPSHLSLIKTVAVSNS--VVASGGSDDTIHLYD 66
L++GS + + ++LN + H S + ++A+S + S G D ++L+D
Sbjct: 161 LISGSDDATVRVWDLNAKNTEKKCLAIMEKHFSAVTSIALSEDGLTLFSAGRDKVVNLWD 220
Query: 67 L---------STSSSLGSLTDHSSTVTALSFYSPPNLPFPRN---------LVSADADGS 108
L +T L ++T SS SF + + + ++ G
Sbjct: 221 LHDYSCKATVATYEVLEAVTTVSSGTPFASFVASLDQKKSKKKESDSQATYFITVGERGV 280
Query: 109 LAIFDADGFVHL---KTLSV--------HKKGVNDLAIHPSGKIALTVSRDSCFAMVNLV 157
+ I+ ++G + L K+ + K+G A+ PS L V+ D F ++V
Sbjct: 281 VRIWKSEGSICLYEQKSSDITVSSDDEESKRGFTAAAMLPSDHGLLCVTADQQFFFYSVV 340
Query: 158 RGRRSFCCRLDKEASIVRFDVSGDSF------FMAVD---EIVNVHQAEDARLLMELQCP 208
L K +++ F F+AV E V V+ L
Sbjct: 341 ENVEETELVLSKRLVGYNEEIADMKFLGDEEQFLAVATNLEEVRVYDVATMSCSYVLAGH 400
Query: 209 KRVL-----CAAPAKNGLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIVVLSDEA 263
K V+ C + + N L+ TG +D+ + W+ S K + H + +
Sbjct: 401 KEVVLSLDTCVSSSGNVLIVTGSKDKTVRLWNATS-KSCIGVGTGHNGDILAVAFAKKSF 459
Query: 264 TGDDEPYLVASASSDGTIRAWDVRMAATEKSEPLAECKTQS 304
+ S S D T++ W + + + EP+ KT+S
Sbjct: 460 S------FFVSGSGDRTLKVWSLDGISEDSEEPI-NLKTRS 493
Score = 38.5 bits (88), Expect = 0.005
Identities = 33/117 (28%), Positives = 56/117 (47%), Gaps = 16/117 (13%)
Query: 176 FDVSGDSFFMAV--DEIVNVHQAEDARLLMELQCPKRVLCAAPAK--NGLLYTGGEDRNI 231
F VS D F+A +++N+ + D+ + ++ L A + LL++ G R I
Sbjct: 25 FIVSSDGSFIACACGDVINIVDSTDSSVKSTIEGESDTLTALALSPDDKLLFSAGHSRQI 84
Query: 232 TAWDLKSGKVAYCIE--EAHAARVKGIVVLSDEATGDDEPYLVASASSDGTIRAWDV 286
WDL++ K CI + H V G ++ A+G L+A+A +D + WDV
Sbjct: 85 RVWDLETLK---CIRSWKGHEGPVMG---MACHASGG----LLATAGADRKVLVWDV 131
Score = 35.4 bits (80), Expect = 0.042
Identities = 23/97 (23%), Positives = 41/97 (41%), Gaps = 5/97 (5%)
Query: 50 NSVVASGGSDDTIHLYDLSTSSSLGSLTDHSSTVTALSFYSPPNLPFPRNLVSADADGSL 109
+ ++ S G I ++DL T + S H V ++ ++ L L +A AD +
Sbjct: 72 DKLLFSAGHSRQIRVWDLETLKCIRSWKGHEGPVMGMACHASGGL-----LATAGADRKV 126
Query: 110 AIFDADGFVHLKTLSVHKKGVNDLAIHPSGKIALTVS 146
++D DG HK V+ + HP + +S
Sbjct: 127 LVWDVDGGFCTHYFRGHKGVVSSILFHPDSNKNILIS 163
Score = 27.7 bits (60), Expect = 8.8
Identities = 24/81 (29%), Positives = 37/81 (45%), Gaps = 13/81 (16%)
Query: 209 KRVLCAAPAKN-GLLYTGGEDRNITAWDLKSGKVAYCIEEAHAARVKGIV--VLSDEATG 265
K + A A+N L+ TG EDR + W L + H +KG + S E +
Sbjct: 500 KDINSVAVARNDSLVCTGSEDRTASIWRLP--------DLVHVVTLKGHKRRIFSVEFST 551
Query: 266 DDEPYLVASASSDGTIRAWDV 286
D+ V +AS D T++ W +
Sbjct: 552 VDQ--CVMTASGDKTVKIWAI 570
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.132 0.385
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,112,423
Number of Sequences: 26719
Number of extensions: 289183
Number of successful extensions: 1619
Number of sequences better than 10.0: 165
Number of HSP's better than 10.0 without gapping: 65
Number of HSP's successfully gapped in prelim test: 100
Number of HSP's that attempted gapping in prelim test: 1077
Number of HSP's gapped (non-prelim): 501
length of query: 324
length of database: 11,318,596
effective HSP length: 99
effective length of query: 225
effective length of database: 8,673,415
effective search space: 1951518375
effective search space used: 1951518375
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144656.10


