

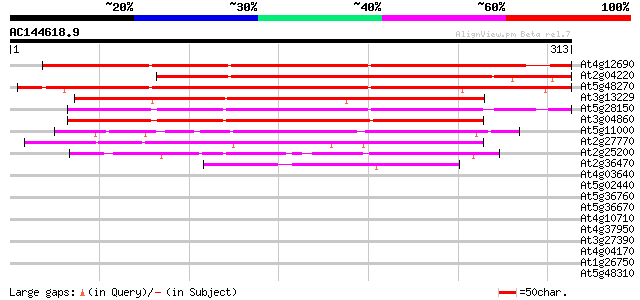
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144618.9 + phase: 0
(313 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At4g12690 unknown protein 353 5e-98
At2g04220 hypothetical protein 320 5e-88
At5g48270 putative protein 308 3e-84
At3g13229 putative protein 213 1e-55
At5g28150 unknown protein 199 2e-51
At3g04860 unknown protein 189 2e-48
At5g11000 unknown protein 126 1e-29
At2g27770 unknown protein 125 3e-29
At2g25200 hypothetical protein 107 8e-24
At2g36470 unknown protein 72 3e-13
At4g03640 hypothetical protein 34 0.090
At5g02440 putative protein 31 0.99
At5g36760 hypothetical protein 29 2.9
At5g36670 putative protein 29 2.9
At4g10710 putative transcriptional regulator 29 3.8
At4g37950 hypothetical protein 28 4.9
At3g27390 unknown protein 28 4.9
At4g04170 putative protein 28 6.4
At1g26750 unknown protein 28 6.4
At5g48310 putative protein 28 8.4
>At4g12690 unknown protein
Length = 285
Score = 353 bits (907), Expect = 5e-98
Identities = 173/295 (58%), Positives = 221/295 (74%), Gaps = 16/295 (5%)
Query: 19 SLMSQRITEEPLPTKTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQ 78
S +++ITE+P+ KTAQS+VTC+YQ + G WRN+ VLW KNL+NH+L + V S +G+
Sbjct: 7 SSTAEKITEDPVTYKTAQSSVTCIYQAHMVGFWRNVRVLWSKNLMNHSLTVMVTSVQGDM 66
Query: 79 FHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDE 138
+ CK+++KPW FW +KGY+SF+V+GNQ+DVYWD RSAKF G PEP SD+YV LVS+E
Sbjct: 67 -NYCCKVDLKPWHFWYKKGYKSFEVEGNQVDVYWDFRSAKFNGG-PEPSSDFYVALVSEE 124
Query: 139 EVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVES 198
EVVLLLGD+KKKA+KR K R +LV+A L K+ENVF KK FST+AKF ++++E +IVVES
Sbjct: 125 EVVLLLGDHKKKAFKRTKSRPSLVDAALFYKKENVFGKKIFSTRAKFHDRKREHEIVVES 184
Query: 199 STAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGP 258
ST G K+PEMWIS+DGIVL+ ++NLQWKFRGNQTV+V+K+PV VFWDVYDWLFS G+G
Sbjct: 185 ST-GAKEPEMWISVDGIVLVQVRNLQWKFRGNQTVLVDKEPVQVFWDVYDWLFSTPGTGH 243
Query: 259 GLFIFKPGPTEVESDKEEKGLEGESDDGSATSGYYSTKSYTPFESCLVLCAYKLE 313
GLFIFKP E E+ E K S S+ E CL L A+KLE
Sbjct: 244 GLFIFKPESGESETSNETKNCSASSSSSSS-------------EFCLFLYAWKLE 285
>At2g04220 hypothetical protein
Length = 246
Score = 320 bits (821), Expect = 5e-88
Identities = 159/236 (67%), Positives = 189/236 (79%), Gaps = 7/236 (2%)
Query: 83 CKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVL 142
CK+++KPW FWN+KGY+SFDV+GN ++VYWD RSAKF + PEP SD+YV LVS+EEVVL
Sbjct: 13 CKVDLKPWHFWNKKGYKSFDVEGNPVEVYWDFRSAKFTSS-PEPSSDFYVALVSEEEVVL 71
Query: 143 LLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAG 202
L+GDYKKKA+KR K R ALVEA L K+ENVF KK F+T+AKF +++KE +I+VESST+G
Sbjct: 72 LVGDYKKKAFKRTKSRPALVEAALFYKKENVFGKKCFTTRAKFYDRKKEHEIIVESSTSG 131
Query: 203 NKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGPGLFI 262
K+PEMWISIDGIVLI +KNLQWKFRGNQTV+V+KQPV VFWDVYDWLFS+ G+G GLFI
Sbjct: 132 PKEPEMWISIDGIVLIQVKNLQWKFRGNQTVLVDKQPVQVFWDVYDWLFSMPGTGHGLFI 191
Query: 263 FKPGPTEVESDKEEKGL---EGESDDGSATSGYYSTKSYTPF--ESCLVLCAYKLE 313
FKPG TE +SD E G G D S S YYSTKS P+ E CL L AYKLE
Sbjct: 192 FKPGTTE-DSDMEGSGHGGGGGGESDTSTGSRYYSTKSSNPWPPEFCLFLHAYKLE 246
>At5g48270 putative protein
Length = 322
Score = 308 bits (788), Expect = 3e-84
Identities = 168/325 (51%), Positives = 225/325 (68%), Gaps = 20/325 (6%)
Query: 5 LSFRSQSFSKQNLASLMSQRITEEP--------LPTKTAQSTVTCLYQTSIGGQWRNISV 56
LSFR S Q A + +IT+ P + + STVTC YQ + G +RN++V
Sbjct: 2 LSFRMPSIGSQPTA--FAAKITDNPKTVAQLKSAASPSPHSTVTCGYQAHVAGFFRNVTV 59
Query: 57 LWCKNLINHTLNLKVDSARGEQFHQNCKIE-VKPWCFWNRKGYRSFDVDGNQIDVYWDLR 115
LW KNL+NH+L + V S + + CKI+ VKPW FW+++G +SFDV+GN ++V+WDLR
Sbjct: 60 LWSKNLMNHSLTVMVSSLDNDM-NYCCKIDLVKPWQFWSKRGSKSFDVEGNFVEVFWDLR 118
Query: 116 SAKFAGN-CPEPCSDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVF 174
SAK AGN PEP SDYYV +VSDEEVVLLLGD K+KAYKR K R ALVE + K+E++F
Sbjct: 119 SAKLAGNGSPEPVSDYYVAVVSDEEVVLLLGDLKQKAYKRTKSRPALVEGFIYFKKESIF 178
Query: 175 AKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVM 234
KK+FST+A+FDE+RKE ++VVESS G +PEMWIS+DGIV++++KNLQWKFRGNQ VM
Sbjct: 179 GKKTFSTRARFDEQRKEHEVVVESSN-GAAEPEMWISVDGIVVVNVKNLQWKFRGNQMVM 237
Query: 235 VNKQPVHVFWDVYDWLF--SVSGSGPGLFIFKPGPTEVESDKE-EKGLEGESDDGSATSG 291
V++ PV V++DV+DWLF S + + GLF+FKP P D+ EG+S GS+
Sbjct: 238 VDRTPVMVYYDVHDWLFASSETAASSGLFLFKPVPVGAMVDESFSDAEEGDSGGGSSPLS 297
Query: 292 YYSTKS--YTPF-ESCLVLCAYKLE 313
Y++ S + P + CL L A+KLE
Sbjct: 298 RYNSASSGFGPLHDFCLFLYAWKLE 322
>At3g13229 putative protein
Length = 289
Score = 213 bits (541), Expect = 1e-55
Identities = 103/234 (44%), Positives = 151/234 (64%), Gaps = 6/234 (2%)
Query: 37 STVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQ--FHQNCKIEVKPWCFWN 94
S+V+ +Y I +N+ V W K +H+L +K+++ + EQ HQ KI++ FW
Sbjct: 6 SSVSLIYVVEIAKTPQNVDVTWSKTTSSHSLTIKIENVKDEQQNHHQPVKIDLSGSSFWA 65
Query: 95 RKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAYKR 154
+KG +S + +G ++DVYWD R AKF+ N PEP S +YV LVS VL +GD + +A KR
Sbjct: 66 KKGLKSLEANGTRVDVYWDFRQAKFS-NFPEPSSGFYVSLVSQNATVLTIGDLRNEALKR 124
Query: 155 MKMRQALVEATLLVKRENVFAKKSFSTKAKFD--EKRKESDIVVESSTAGNKDPEMWISI 212
K + EA L+ K+E+V K+ F T+ F E R+E+++V+E+S +G DPEMWI++
Sbjct: 125 TKKNPSATEAALVSKQEHVHGKRVFYTRTAFGGGESRRENEVVIETSLSGPSDPEMWITV 184
Query: 213 DGIVLIHIKNLQWKFRGNQTVMVNK-QPVHVFWDVYDWLFSVSGSGPGLFIFKP 265
DG+ I I NL W+FRGN+ V V+ + +FWDV+DWLF SGS GLF+FKP
Sbjct: 185 DGVPAIRIMNLNWRFRGNEVVTVSDGVSLEIFWDVHDWLFEPSGSSSGLFVFKP 238
>At5g28150 unknown protein
Length = 289
Score = 199 bits (506), Expect = 2e-51
Identities = 111/281 (39%), Positives = 163/281 (57%), Gaps = 17/281 (6%)
Query: 33 KTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKIEVKPWCF 92
K AQ+ VTC+YQ I G+ I+V W KNL+ ++ + VD + + CK+E+KPW F
Sbjct: 26 KNAQNLVTCIYQCRIRGRNCLITVTWTKNLMGQSVTVGVDDSCNQSL---CKVEIKPWLF 82
Query: 93 WNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAY 152
RKG +S + IDV+WDL SAKF G+ PE +YV +V D+E+VLLLGD KK+A+
Sbjct: 83 TKRKGSKSLEAYSCNIDVFWDLSSAKF-GSGPEALGGFYVGVVVDKEMVLLLGDMKKEAF 141
Query: 153 KRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWISI 212
K+ + + A + K+E+VF K+ F+TKA+ K D+++E T DP + + +
Sbjct: 142 KKTNASPSSLGAVFIAKKEHVFGKRVFATKAQLFADGKFHDLLIECDT-NVTDPCLVVRV 200
Query: 213 DGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGPGLFIFKPGPTEVES 272
DG L+ +K L+WKFRGN T++VNK V V WDV+ WLF + +G +F+F+
Sbjct: 201 DGKTLLQVKRLKWKFRGNDTIVVNKMTVEVLWDVHSWLFGLPTTGNAVFMFR------TC 254
Query: 273 DKEEKGLEGESDDGSATSGYYSTKSYTPFESCLVLCAYKLE 313
EK L D + S +S F L+L A+K E
Sbjct: 255 QSTEKSLSFSQDVTTTNSKSHS------FGFSLILYAWKSE 289
>At3g04860 unknown protein
Length = 289
Score = 189 bits (479), Expect = 2e-48
Identities = 96/234 (41%), Positives = 147/234 (62%), Gaps = 7/234 (2%)
Query: 33 KTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNCKIEVKPWCF 92
+ AQ+ V C+Y+ I G+ I+V W KNL+ + + VD + CK+E+KPW F
Sbjct: 26 RNAQNLVICIYRCRIRGRTCLITVTWTKNLMGQCVTVGVDDSCNRSL---CKVEIKPWLF 82
Query: 93 WNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAY 152
RKG ++ + IDV+WDL SAKF G+ PEP +YV +V D+E+VLLLGD KK+A+
Sbjct: 83 TKRKGSKTLEAYACNIDVFWDLSSAKF-GSSPEPLGGFYVGVVVDKEMVLLLGDMKKEAF 141
Query: 153 KRMKMR-QALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWIS 211
K+ + + A + K+E+VF K++F+TKA+F K D+V+E T+ DP + +
Sbjct: 142 KKTNAAPSSSLGAVFIAKKEHVFGKRTFATKAQFSGDGKTHDLVIECDTS-LSDPCLIVR 200
Query: 212 IDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSV-SGSGPGLFIFK 264
+DG +L+ ++ L WKFRGN T++VN+ V V WDV+ W F + S G +F+F+
Sbjct: 201 VDGKILMQVQRLHWKFRGNDTIIVNRISVEVLWDVHSWFFGLPSSPGNAVFMFR 254
>At5g11000 unknown protein
Length = 389
Score = 126 bits (317), Expect = 1e-29
Identities = 94/287 (32%), Positives = 139/287 (47%), Gaps = 42/287 (14%)
Query: 26 TEEPLPTKTAQSTVT-CLYQTS------------IGGQWRNISVLWCKNLINHTLNLKVD 72
++ P P + +T CLYQT +GG N+ L ++ NH+ L
Sbjct: 21 SQSPSPLSSGDPNLTTCLYQTDHGVFYLTWSRTFLGGHSVNL-FLHSQDYYNHSSPLSFS 79
Query: 73 SA-----RGEQFHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPC 127
SA FH N + FW ++G R +I V+WDL AKF EP
Sbjct: 80 SADLSLSSAVSFHLN----LNTLAFWKKRGSRFVSP---KIQVFWDLSKAKFDSGS-EPR 131
Query: 128 SDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEA-TLLVKRENVFAKKSFSTKAKFD 186
S +Y+ +V D E+ LL+GD K+AY R K + LL+++E+VF + F+TKA+F
Sbjct: 132 SGFYIAVVVDGEMGLLVGDSVKEAYARAKSAKPPTNPQALLLRKEHVFGARVFTTKARFG 191
Query: 187 EKRKESDIVVESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDV 246
K +E I ++D ++ S+D ++ IK L+WKFRGN+ V ++ V + WDV
Sbjct: 192 GKNREISI----DCRVDEDAKLCFSVDSKQVLQIKRLRWKFRGNEKVEIDGVHVQISWDV 247
Query: 247 YDWLFSVSGSGPG---------LFIFKPGPTEVESDKEEKGLEGESD 284
Y+WLF SG G +F F+ P E E E K E E +
Sbjct: 248 YNWLFQSKSSGDGGGGGGHAVFMFRFESDP-EAEEVCETKRKEEEDE 293
>At2g27770 unknown protein
Length = 320
Score = 125 bits (314), Expect = 3e-29
Identities = 82/267 (30%), Positives = 137/267 (50%), Gaps = 13/267 (4%)
Query: 9 SQSFSKQNLASLMSQRITEEPLPTKTAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLN 68
S S S + + + + P + Q+++T +Y+ ++ + I V WC N+ L+
Sbjct: 20 SISSSSSSCSKYSTNNVCISPSLIPSIQTSITSIYRITLS-KHLIIKVTWCNPHNNNGLS 78
Query: 69 LKVDSARGEQFHQNCKIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNC--PEP 126
+ V SA + K+ F +KG +S D D +I+V+WDL SAK+ N PEP
Sbjct: 79 ISVASA-DQNPSTTLKLNTSSRFFRKKKGNKSVDSDLGKIEVFWDLSSAKYDSNLCGPEP 137
Query: 127 CSDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKS--FSTKAK 184
+ +YV+++ D ++ LLLGD ++ ++ + LV R+ F + +STK +
Sbjct: 138 INGFYVIVLVDGQMGLLLGDSSEETLRKKGFSGDIGFDFSLVSRQEHFTGNNTFYSTKVR 197
Query: 185 FDEKRKESDIVV------ESSTAGNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQ 238
F E +IV+ E N P + + ID +I +K LQW FRGNQT+ ++
Sbjct: 198 FVETGDSHEIVIRCNKETEGLKQSNHYPVLSVCIDKKTVIKVKRLQWNFRGNQTIFLDGL 257
Query: 239 PVHVFWDVYDWLFSVSGS-GPGLFIFK 264
V + WDV+DW FS G+ G +F+F+
Sbjct: 258 LVDLMWDVHDWFFSNQGACGRAVFMFR 284
>At2g25200 hypothetical protein
Length = 354
Score = 107 bits (267), Expect = 8e-24
Identities = 76/257 (29%), Positives = 122/257 (46%), Gaps = 34/257 (13%)
Query: 34 TAQSTVTCLYQTSIGGQWRNISVLWCKNLINHTLNLKVDSARGEQFHQNC---------- 83
+ +T TC Y T++G + + W ++ + +L+L S + +
Sbjct: 45 SGDATTTCQYHTNVGVFFLS----WSRSFLRRSLHLHFYSCNSTNCYLHSLDCYRHSIPF 100
Query: 84 --KIEVKPWCFWNRKGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVV 141
++E+KP FW + G + + I V WDL AKF G+ P+P S +YV + EV
Sbjct: 101 AFRLEIKPLTFWRKNGSKKLSRKPD-IRVVWDLTHAKF-GSGPDPESGFYVAVFVSGEVG 158
Query: 142 LLLGDYKKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTA 201
LL+G K R RQ LV K+EN+F + +STK K +E I V+
Sbjct: 159 LLVGGGNLKQRPR---RQILVS-----KKENLFGNRVYSTKIMIQGKLREISIDVK---V 207
Query: 202 GNKDPEMWISIDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSG---- 257
N D + S+D ++ I LQWKFRGN ++++ + + WDV++WLF
Sbjct: 208 VNDDASLRFSVDDKSVLKISQLQWKFRGNTKIVIDGVTIQISWDVFNWLFGGKDKVKPDK 267
Query: 258 -PGLFIFKPGPTEVESD 273
P +F+ + EVE +
Sbjct: 268 IPAVFLLRFENQEVEGN 284
>At2g36470 unknown protein
Length = 327
Score = 72.4 bits (176), Expect = 3e-13
Identities = 41/156 (26%), Positives = 78/156 (49%), Gaps = 20/156 (12%)
Query: 109 DVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAYKRMKMRQALVEATLLV 168
++ WDL A++ N PEP ++V++V + E+ L +GD + R ++ V
Sbjct: 100 EILWDLSEAEYENNGPEPIRRFFVVVVVNSEITLGVGDVDHE-------RDTSSSSSWRV 152
Query: 169 KRENVFAKKSF-STKAKFDEKRKESDIVVESSTAGN-----------KDPE-MWISIDGI 215
+ F+ + +TKA+F + ++ +I ++ G K PE M + +D
Sbjct: 153 SKTERFSGTCWLTTKAQFSDVGRKHEIQIQCGGGGGGGGEEGYLWKVKSPETMSVYVDKR 212
Query: 216 VLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLF 251
+ +K L+W FRGNQT+ + + + WD++DW +
Sbjct: 213 KVFSVKKLKWNFRGNQTMFFDGMLIDMMWDLHDWFY 248
>At4g03640 hypothetical protein
Length = 330
Score = 34.3 bits (77), Expect = 0.090
Identities = 23/66 (34%), Positives = 33/66 (49%), Gaps = 1/66 (1%)
Query: 157 MRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAGNKDPEMWISIDGIV 216
++ L + L ++REN+F T A F + DIV E S A DP +SID +
Sbjct: 239 LKGKLFQFLLCMQRENIFGGYDSFTVAIFYTGNIDDDIVFEDSDA-YVDPSSIVSIDQVC 297
Query: 217 LIHIKN 222
LI + N
Sbjct: 298 LITLNN 303
>At5g02440 putative protein
Length = 176
Score = 30.8 bits (68), Expect = 0.99
Identities = 13/29 (44%), Positives = 16/29 (54%)
Query: 64 NHTLNLKVDSARGEQFHQNCKIEVKPWCF 92
N T N + +AR Q H+N K KPW F
Sbjct: 91 NRTRNDQTGAARNHQHHKNLKTYYKPWIF 119
>At5g36760 hypothetical protein
Length = 226
Score = 29.3 bits (64), Expect = 2.9
Identities = 20/59 (33%), Positives = 29/59 (48%), Gaps = 5/59 (8%)
Query: 252 SVSGSGPGLFIFKPG---PTEVESDKEEKGLEGESDDGSATSGYYSTKSYTPFESCLVL 307
SV S GL + P P +VE +K E+ + + AT+G S + P +SCL L
Sbjct: 14 SVVSSPNGLVLLAPEMTLPVDVEENKPEESKDSAHERNCATAGVESPSN--PVDSCLKL 70
>At5g36670 putative protein
Length = 1030
Score = 29.3 bits (64), Expect = 2.9
Identities = 20/59 (33%), Positives = 29/59 (48%), Gaps = 5/59 (8%)
Query: 252 SVSGSGPGLFIFKPG---PTEVESDKEEKGLEGESDDGSATSGYYSTKSYTPFESCLVL 307
SV S GL + P P +VE +K E+ + + AT+G S + P +SCL L
Sbjct: 818 SVVSSPNGLVLLAPEMTLPVDVEENKPEESKDSAHERNCATAGVESPSN--PVDSCLKL 874
>At4g10710 putative transcriptional regulator
Length = 1074
Score = 28.9 bits (63), Expect = 3.8
Identities = 36/130 (27%), Positives = 51/130 (38%), Gaps = 20/130 (15%)
Query: 167 LVKRENV-FAKKSFSTKAKFDEKRKES---DIVVESSTAGNKDPEMWISIDGIVLIHIK- 221
+V E V F +K+F F + +K+ D V SS G K+ W+ I K
Sbjct: 869 IVNLERVGFGQKNFDMAIIFKDFKKDVLRVDSVPTSSLEGIKE---WLDTTDIKYYESKL 925
Query: 222 NLQWKFRGNQTVMVNKQPVHVFWDVYDWLF--------SVSGSGPGLFIFKPGPTEVESD 273
NL W+ Q + F D W F GS ++P EVES+
Sbjct: 926 NLNWR----QILKTITDDPQSFIDDGGWEFLNLDGSDSESGGSEESDKGYEPSDVEVESE 981
Query: 274 KEEKGLEGES 283
E++ E ES
Sbjct: 982 SEDEASESES 991
>At4g37950 hypothetical protein
Length = 296
Score = 28.5 bits (62), Expect = 4.9
Identities = 25/99 (25%), Positives = 41/99 (41%), Gaps = 11/99 (11%)
Query: 96 KGYRSFDVDGNQIDVYWDLRSAKFAGNCPEPCSDYYVMLVSDEEVVLLLGDYKKKAYKRM 155
+GY DVD +QI + + L + KF D+ M VSD ++ D + K
Sbjct: 106 RGYSWPDVDMDQIRIVFKLNTTKF---------DF--MAVSDNRQKIMPFDTDRDITKGR 154
Query: 156 KMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDI 194
A EA L+ +N K K + + K++ +
Sbjct: 155 ASPLAYKEAVHLINPQNHMLKGQVDDKYMYSVENKDNKV 193
>At3g27390 unknown protein
Length = 599
Score = 28.5 bits (62), Expect = 4.9
Identities = 20/52 (38%), Positives = 26/52 (49%), Gaps = 2/52 (3%)
Query: 212 IDGIVLIHIKNLQWKFRGNQTVMVNKQPVHVFWDVYDWLFSVSGSGPGLFIF 263
I GIVL + L GN V+++ PVH+ W Y + S GP L IF
Sbjct: 30 IKGIVLCPLVCLVVTI-GNSAVILSLLPVHIVWTFYS-IVSAKQVGPILKIF 79
>At4g04170 putative protein
Length = 319
Score = 28.1 bits (61), Expect = 6.4
Identities = 16/56 (28%), Positives = 32/56 (56%), Gaps = 2/56 (3%)
Query: 147 YKKKAYKRMKMRQALVEATLLVKRENVFAKKSFSTKAKFDEKRKESDIVVESSTAG 202
YK +A KRM+ + L+ +++ K E ++ S++ + +E + SD ES+ +G
Sbjct: 144 YKARAKKRMRHIENLMVVSMIKKSEEIY--NEVSSRIQEEEFQLCSDNTTESAASG 197
>At1g26750 unknown protein
Length = 195
Score = 28.1 bits (61), Expect = 6.4
Identities = 18/76 (23%), Positives = 34/76 (44%), Gaps = 14/76 (18%)
Query: 149 KKAYKRMKMRQALVEATL-----------LVKRENVFAKKSFST---KAKFDEKRKESDI 194
KKAYKR+K + + L+ + AK F T + ++ + E D+
Sbjct: 118 KKAYKRLKELAVMQDKDPPPKPYPSAKKGLITQSKTSAKDRFQTPSVRRLVNQLKNEKDV 177
Query: 195 VVESSTAGNKDPEMWI 210
+++ T G+ + + WI
Sbjct: 178 LLQDRTGGSANQDNWI 193
>At5g48310 putative protein
Length = 1156
Score = 27.7 bits (60), Expect = 8.4
Identities = 12/42 (28%), Positives = 23/42 (54%)
Query: 270 VESDKEEKGLEGESDDGSATSGYYSTKSYTPFESCLVLCAYK 311
VE K + E ++ + SG Y+ +S +++C+ LC Y+
Sbjct: 255 VEDSKISEICSDELEECHSISGQYAWQSLLAYDACIRLCLYE 296
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.407
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,319,086
Number of Sequences: 26719
Number of extensions: 324083
Number of successful extensions: 926
Number of sequences better than 10.0: 22
Number of HSP's better than 10.0 without gapping: 14
Number of HSP's successfully gapped in prelim test: 8
Number of HSP's that attempted gapping in prelim test: 872
Number of HSP's gapped (non-prelim): 24
length of query: 313
length of database: 11,318,596
effective HSP length: 99
effective length of query: 214
effective length of database: 8,673,415
effective search space: 1856110810
effective search space used: 1856110810
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144618.9


