

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144617.10 + phase: 0
(337 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
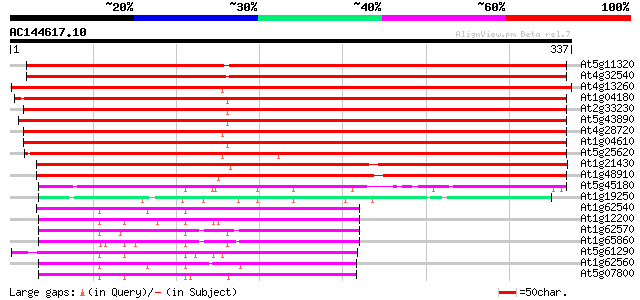
Score E
Sequences producing significant alignments: (bits) Value
At5g11320 putative protein 492 e-139
At4g32540 dimethylaniline monooxygenase - like protein 456 e-128
At4g13260 unknown protein 431 e-121
At1g04180 putative dimethylaniline monooxygenase 425 e-119
At2g33230 putative flavin-containing monooxygenase 416 e-117
At5g43890 dimethylaniline monooxygenase-like 414 e-116
At4g28720 unknown protein 412 e-115
At1g04610 putative dimethylaniline monooxygenase 410 e-115
At5g25620 unknown protein 407 e-114
At1g21430 flavin-containing monooxygenases, putative 288 4e-78
At1g48910 hypothetical protein 283 9e-77
At5g45180 dimethylaniline monooxygenase (N-oxide-forming)-like p... 78 8e-15
At1g19250 hypothetical protein 75 5e-14
At1g62540 unknown protein 71 1e-12
At1g12200 unknown protein 70 2e-12
At1g62570 similar to glutamate synthase 69 3e-12
At1g65860 flavin-containing monooxygenase FMO3, putative 68 8e-12
At5g61290 unknown protein 66 2e-11
At1g62560 similar to flavin-containing monooxygenase (sp|P36366)... 66 2e-11
At5g07800 dimethylaniline monooxygenase-like protein 66 3e-11
>At5g11320 putative protein
Length = 411
Score = 492 bits (1266), Expect = e-139
Identities = 236/324 (72%), Positives = 276/324 (84%), Gaps = 2/324 (0%)
Query: 11 VWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHFCE 70
+++ GPIIVGAGPSG+AVAACLS +GVPS+ILER+DC+ASLWQ RTYDRLKLHLPKHFCE
Sbjct: 12 IFVPGPIIVGAGPSGLAVAACLSNRGVPSVILERTDCLASLWQKRTYDRLKLHLPKHFCE 71
Query: 71 LPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMVRTKEGD 130
LP+M FP+ FPKYP+K FISY+ESYA F+I P FNQTV AEFD S +W V+T++G
Sbjct: 72 LPLMPFPKNFPKYPSKQLFISYVESYAARFNIKPVFNQTVEKAEFDDASGLWNVKTQDG- 130
Query: 131 FQYFSPWLIVATGENAEPVFPTIHGMEHFHGPVVHTSDYKSGSEYKNKKVLVIGCGNSGM 190
Y S WL+VATGENAEPVFP I G++ F GPVVHTS YKSGS + N+KVLV+GCGNSGM
Sbjct: 131 -VYTSTWLVVATGENAEPVFPNIPGLKKFTGPVVHTSAYKSGSAFANRKVLVVGCGNSGM 189
Query: 191 EVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVDKFLLLVSSFF 250
EVSLDLCR+NA+PH+V RNSVH+LPRD FG ST+GIAM L KW PLKLVDKFLLL+++
Sbjct: 190 EVSLDLCRYNALPHMVVRNSVHVLPRDFFGLSTFGIAMTLLKWFPLKLVDKFLLLLANST 249
Query: 251 LGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVKEITRNGAKFLD 310
LGNT+ G++RPKTGPIELK TGKTPVLDVG I+ I+SG IKV + VKEITRNGAKFL+
Sbjct: 250 LGNTDLLGLRRPKTGPIELKNVTGKTPVLDVGAISLIRSGQIKVTQAVKEITRNGAKFLN 309
Query: 311 GQEKEFDAIILATGYKSNVPSWLK 334
G+E EFD+IILATGYKSNVP WLK
Sbjct: 310 GKEIEFDSIILATGYKSNVPDWLK 333
>At4g32540 dimethylaniline monooxygenase - like protein
Length = 414
Score = 456 bits (1172), Expect = e-128
Identities = 209/325 (64%), Positives = 271/325 (83%), Gaps = 2/325 (0%)
Query: 11 VWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHFCE 70
+ +HGPII+GAGPSG+A +ACLS +GVPSLILERSD IASLW+++TYDRL+LHLPKHFC
Sbjct: 16 ILVHGPIIIGAGPSGLATSACLSSRGVPSLILERSDSIASLWKSKTYDRLRLHLPKHFCR 75
Query: 71 LPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMVRTKEGD 130
LP++ FP+ +PKYP+K++F++Y+ESYA HF I PRFN+ V +A +DS+S W V+T +
Sbjct: 76 LPLLDFPEYYPKYPSKNEFLAYLESYASHFRIAPRFNKNVQNAAYDSSSGFWRVKTHDNT 135
Query: 131 FQYFSPWLIVATGENAEPVFPTIHGMEHFHG-PVVHTSDYKSGSEYKNKKVLVIGCGNSG 189
+Y S WLIVATGENA+P FP I G + F G +VH S+YKSG E++ +KVLV+GCGNSG
Sbjct: 136 -EYLSKWLIVATGENADPYFPEIPGRKKFSGGKIVHASEYKSGEEFRRQKVLVVGCGNSG 194
Query: 190 MEVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVDKFLLLVSSF 249
ME+SLDL RHNA PHLV RN+VH+LPR++ G ST+G+ M L K LPL+LVDKFLLL+++
Sbjct: 195 MEISLDLVRHNASPHLVVRNTVHVLPREILGVSTFGVGMTLLKCLPLRLVDKFLLLMANL 254
Query: 250 FLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVKEITRNGAKFL 309
GNT+ G++RPKTGP+ELK TGK+PVLDVG ++ I+SG I++MEGVKEIT+ GAKF+
Sbjct: 255 SFGNTDRLGLRRPKTGPLELKNVTGKSPVLDVGAMSLIRSGMIQIMEGVKEITKKGAKFM 314
Query: 310 DGQEKEFDAIILATGYKSNVPSWLK 334
DGQEK+FD+II ATGYKSNVP+WL+
Sbjct: 315 DGQEKDFDSIIFATGYKSNVPTWLQ 339
>At4g13260 unknown protein
Length = 415
Score = 431 bits (1107), Expect = e-121
Identities = 202/340 (59%), Positives = 256/340 (74%), Gaps = 4/340 (1%)
Query: 2 DPFKEKVKCVWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLK 61
DP+ E+ +C+ I GPIIVG+GPSG+A AACL + +PSLILERS CIASLWQ++TYDRL+
Sbjct: 14 DPYVEETRCLMIPGPIIVGSGPSGLATAACLKSRDIPSLILERSTCIASLWQHKTYDRLR 73
Query: 62 LHLPKHFCELPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQI 121
LHLPK FCELP+M FP ++P YPTK QF+ Y+ESYA+HF + P FNQTV A+FD +
Sbjct: 74 LHLPKDFCELPLMPFPSSYPTYPTKQQFVQYLESYAEHFDLKPVFNQTVEEAKFDRRCGL 133
Query: 122 WMVRT----KEGDFQYFSPWLIVATGENAEPVFPTIHGMEHFHGPVVHTSDYKSGSEYKN 177
W VRT K+ +Y S WL+VATGENAE V P I G+ F GP++HTS YKSG +
Sbjct: 134 WRVRTTGGKKDETMEYVSRWLVVATGENAEEVMPEIDGIPDFGGPILHTSSYKSGEIFSE 193
Query: 178 KKVLVIGCGNSGMEVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLK 237
KK+LV+GCGNSGMEV LDLC NA+P LV R+SVH+LP++M G ST+GI+ L KW P+
Sbjct: 194 KKILVVGCGNSGMEVCLDLCNFNALPSLVVRDSVHVLPQEMLGISTFGISTSLLKWFPVH 253
Query: 238 LVDKFLLLVSSFFLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEG 297
+VD+FLL +S LG+T+ G+ RPK GP+E K+ GKTPVLDVG +A+I+SG+IKV
Sbjct: 254 VVDRFLLRMSRLVLGDTDRLGLVRPKLGPLERKIKCGKTPVLDVGTLAKIRSGHIKVYPE 313
Query: 298 VKEITRNGAKFLDGQEKEFDAIILATGYKSNVPSWLKVKN 337
+K + A+F+DG+ FDAIILATGYKSNVP WLK N
Sbjct: 314 LKRVMHYSAEFVDGRVDNFDAIILATGYKSNVPMWLKGVN 353
>At1g04180 putative dimethylaniline monooxygenase
Length = 421
Score = 425 bits (1092), Expect = e-119
Identities = 196/334 (58%), Positives = 262/334 (77%), Gaps = 4/334 (1%)
Query: 4 FKEKVKCVWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLH 63
F E+ +CVW++GP+IVGAGPSG+A AACL +QGVP +++ERSDCIASLWQ RTYDRLKLH
Sbjct: 14 FSER-RCVWVNGPVIVGAGPSGLATAACLHDQGVPFVVVERSDCIASLWQKRTYDRLKLH 72
Query: 64 LPKHFCELPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWM 123
LPK FC+LP M FP +P+YPTK QFI Y+ESYA+ F I P FN++V SA FD TS +W
Sbjct: 73 LPKKFCQLPKMPFPDHYPEYPTKRQFIDYLESYANRFDIKPEFNKSVESARFDETSGLWR 132
Query: 124 VRTKEG--DFQYFSPWLIVATGENAEPVFPTIHG-MEHFHGPVVHTSDYKSGSEYKNKKV 180
VRT + +Y WL+VATGENAE V P I+G M F G V+H +YKSG +++ K+V
Sbjct: 133 VRTTSDGEEMEYICRWLVVATGENAERVVPEINGLMTEFDGEVIHACEYKSGEKFRGKRV 192
Query: 181 LVIGCGNSGMEVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVD 240
LV+GCGNSGMEVSLDL HNA+ +V R+SVH+LPR++ G ST+GI++ + KWLPL LVD
Sbjct: 193 LVVGCGNSGMEVSLDLANHNAITSMVVRSSVHVLPREIMGKSTFGISVMMMKWLPLWLVD 252
Query: 241 KFLLLVSSFFLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVKE 300
K LL++S LG+ ++YG+KRP GP+ELK TGKTPVLD+G + +IKSG+++++ +K+
Sbjct: 253 KLLLILSWLVLGSLSNYGLKRPDIGPMELKSMTGKTPVLDIGALEKIKSGDVEIVPAIKQ 312
Query: 301 ITRNGAKFLDGQEKEFDAIILATGYKSNVPSWLK 334
+R+ + +DGQ+ + DA++LATGY+SNVPSWL+
Sbjct: 313 FSRHHVELVDGQKLDIDAVVLATGYRSNVPSWLQ 346
>At2g33230 putative flavin-containing monooxygenase
Length = 431
Score = 416 bits (1070), Expect = e-117
Identities = 190/333 (57%), Positives = 253/333 (75%), Gaps = 7/333 (2%)
Query: 9 KCVWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHF 68
+C+W++GP+IVGAGPSG+AVAA L Q VP +ILER++CIASLWQNRTYDRLKLHLPK F
Sbjct: 25 RCIWVNGPVIVGAGPSGLAVAADLKRQEVPFVILERANCIASLWQNRTYDRLKLHLPKQF 84
Query: 69 CELPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMVRTKE 128
C+LP + FP+ P+YPTK+QFI Y+ESYA HF + P+FN+TV SA++D +W V+T
Sbjct: 85 CQLPNLPFPEDIPEYPTKYQFIEYLESYATHFDLRPKFNETVQSAKYDKRFGLWRVQTVL 144
Query: 129 G-------DFQYFSPWLIVATGENAEPVFPTIHGMEHFHGPVVHTSDYKSGSEYKNKKVL 181
+F+Y WL+VATGENAE V P G+E F G V+H DYKSG Y+ K+VL
Sbjct: 145 RSELLGYCEFEYICRWLVVATGENAEKVVPEFEGLEDFGGDVLHAGDYKSGERYRGKRVL 204
Query: 182 VIGCGNSGMEVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVDK 241
V+GCGNSGMEVSLDLC H+A P +V R+SVH+LPR++ G ST+ +++ + KW+P+ LVDK
Sbjct: 205 VVGCGNSGMEVSLDLCNHDASPSMVVRSSVHVLPREVLGKSTFELSVTMMKWMPVWLVDK 264
Query: 242 FLLLVSSFFLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVKEI 301
LL+++ LGNT+ YG+KRP+ GP+ELK GKTPVLD+G I+ IKSG IK++ G+ +
Sbjct: 265 TLLVLTRLLLGNTDKYGLKRPEIGPLELKNTAGKTPVLDIGAISMIKSGKIKIVAGIAKF 324
Query: 302 TRNGAKFLDGQEKEFDAIILATGYKSNVPSWLK 334
+ +DG+ + D++ILATGY+SNVPSWLK
Sbjct: 325 GPGKVELVDGRVLQIDSVILATGYRSNVPSWLK 357
>At5g43890 dimethylaniline monooxygenase-like
Length = 424
Score = 414 bits (1065), Expect = e-116
Identities = 187/335 (55%), Positives = 254/335 (75%), Gaps = 6/335 (1%)
Query: 6 EKVKCVWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLP 65
++ +C+W++GP+IVGAGPSG+A AACL E+GVP ++LER+DCIASLWQ RTYDR+KLHLP
Sbjct: 15 DRRRCIWVNGPVIVGAGPSGLATAACLREEGVPFVVLERADCIASLWQKRTYDRIKLHLP 74
Query: 66 KHFCELPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMVR 125
K C+LP M FP+ +P+YPTK QFI Y+ESYA+ F I P+FN+ V SA +D TS +W ++
Sbjct: 75 KKVCQLPKMPFPEDYPEYPTKRQFIEYLESYANKFEITPQFNECVQSARYDETSGLWRIK 134
Query: 126 TKEG-----DFQYFSPWLIVATGENAEPVFPTIHGME-HFHGPVVHTSDYKSGSEYKNKK 179
T + +Y WL+VATGENAE V P I G+ F G V+H+ +YKSG +Y+ K
Sbjct: 135 TTSSSSSGSEMEYICRWLVVATGENAEKVVPEIDGLTTEFEGEVIHSCEYKSGEKYRGKS 194
Query: 180 VLVIGCGNSGMEVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLV 239
VLV+GCGNSGMEVSLDL HNA +V R+SVH+LPR++ G S++ I+M L KW PL LV
Sbjct: 195 VLVVGCGNSGMEVSLDLANHNANASMVVRSSVHVLPREILGKSSFEISMMLMKWFPLWLV 254
Query: 240 DKFLLLVSSFFLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVK 299
DK LL+++ LGN YG+KRP GP+ELK+ +GKTPVLD+G + +IKSG ++++ G+K
Sbjct: 255 DKILLILAWLILGNLTKYGLKRPTMGPMELKIVSGKTPVLDIGAMEKIKSGEVEIVPGIK 314
Query: 300 EITRNGAKFLDGQEKEFDAIILATGYKSNVPSWLK 334
+R+ + +DGQ + DA++LATGY+SNVPSWL+
Sbjct: 315 RFSRSHVELVDGQRLDLDAVVLATGYRSNVPSWLQ 349
>At4g28720 unknown protein
Length = 426
Score = 412 bits (1058), Expect = e-115
Identities = 187/332 (56%), Positives = 247/332 (74%), Gaps = 6/332 (1%)
Query: 9 KCVWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHF 68
+C+W++GP+IVGAGPSG+A AACL EQ VP ++LER+DCIASLWQ RTYDRLKLHLPK F
Sbjct: 18 RCIWVNGPVIVGAGPSGLATAACLHEQNVPFVVLERADCIASLWQKRTYDRLKLHLPKQF 77
Query: 69 CELPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMVRT-- 126
C+LP M FP+ FP+YPTK QFI Y+ESYA F I+P+FN+ V +A FD TS +W V+T
Sbjct: 78 CQLPKMPFPEDFPEYPTKRQFIDYLESYATRFEINPKFNECVQTARFDETSGLWRVKTVS 137
Query: 127 ----KEGDFQYFSPWLIVATGENAEPVFPTIHGMEHFHGPVVHTSDYKSGSEYKNKKVLV 182
+ + +Y WL+VATGENAE V P I G+ F G V+H DYKSG ++ KKVLV
Sbjct: 138 KSESTQTEVEYICRWLVVATGENAERVMPEIDGLSEFSGEVIHACDYKSGEKFAGKKVLV 197
Query: 183 IGCGNSGMEVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVDKF 242
+GCGNSGMEVSLDL H A P +V R+S+H++PR++ G ST+ +AM + +W PL LVDK
Sbjct: 198 VGCGNSGMEVSLDLANHFAKPSMVVRSSLHVMPREVMGKSTFELAMKMLRWFPLWLVDKI 257
Query: 243 LLLVSSFFLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVKEIT 302
LL++S LGN YG+KRP+ GP+ELK GKTPVLD+G I +I+ G I V+ G+K
Sbjct: 258 LLVLSWMVLGNIEKYGLKRPEMGPMELKSVKGKTPVLDIGAIEKIRLGKINVVPGIKRFN 317
Query: 303 RNGAKFLDGQEKEFDAIILATGYKSNVPSWLK 334
N + ++G++ + D+++LATGY+SNVP WL+
Sbjct: 318 GNKVELVNGEQLDVDSVVLATGYRSNVPYWLQ 349
>At1g04610 putative dimethylaniline monooxygenase
Length = 437
Score = 410 bits (1055), Expect = e-115
Identities = 188/333 (56%), Positives = 251/333 (74%), Gaps = 7/333 (2%)
Query: 9 KCVWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHF 68
+C+W++GP+IVGAGPSG+AVAA L +GVP +ILER++CIASLWQNRTYDRLKLHLPK F
Sbjct: 30 RCIWVNGPVIVGAGPSGLAVAAGLKREGVPFIILERANCIASLWQNRTYDRLKLHLPKQF 89
Query: 69 CELPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMVRTKE 128
C+LP FP FP+YPTK QFI Y+ESYA +F I+P+FN+TV SA++D T +W V+T
Sbjct: 90 CQLPNYPFPDEFPEYPTKFQFIQYLESYAANFDINPKFNETVQSAKYDETFGLWRVKTIS 149
Query: 129 G-------DFQYFSPWLIVATGENAEPVFPTIHGMEHFHGPVVHTSDYKSGSEYKNKKVL 181
+F+Y W++VATGENAE V P G+E F G V+H DYKSG Y+ KKVL
Sbjct: 150 NMGQLGSCEFEYICRWIVVATGENAEKVVPDFEGLEDFGGDVLHAGDYKSGGRYQGKKVL 209
Query: 182 VIGCGNSGMEVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVDK 241
V+GCGNSGMEVSLDL H A P +V R++VH+LPR++FG ST+ + + + K++P+ L DK
Sbjct: 210 VVGCGNSGMEVSLDLYNHGANPSMVVRSAVHVLPREIFGKSTFELGVTMMKYMPVWLADK 269
Query: 242 FLLLVSSFFLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVKEI 301
+L ++ LGNT+ YG+KRPK GP+ELK GKTPVLD+G + +I+SG IK++ G+ +
Sbjct: 270 TILFLARIILGNTDKYGLKRPKIGPLELKNKEGKTPVLDIGALPKIRSGKIKIVPGIIKF 329
Query: 302 TRNGAKFLDGQEKEFDAIILATGYKSNVPSWLK 334
+ + +DG+ E D++ILATGY+SNVPSWLK
Sbjct: 330 GKGKVELIDGRVLEIDSVILATGYRSNVPSWLK 362
>At5g25620 unknown protein
Length = 417
Score = 407 bits (1047), Expect = e-114
Identities = 197/329 (59%), Positives = 248/329 (74%), Gaps = 5/329 (1%)
Query: 10 CVWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHFC 69
CV + GP+IVGAGPSG+A AACL E+G+ S++LERS+CIASLWQ +TYDRL LHLPK FC
Sbjct: 27 CV-VTGPVIVGAGPSGLATAACLKERGITSVLLERSNCIASLWQLKTYDRLHLHLPKQFC 85
Query: 70 ELPMMSFPQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMVRT--K 127
ELP++ FP FP YPTK QFI Y+E YA F I P FNQTV SA FD +W V + +
Sbjct: 86 ELPIIPFPGDFPTYPTKQQFIEYLEDYARRFDIKPEFNQTVESAAFDENLGMWRVTSVGE 145
Query: 128 EGDFQYFSPWLIVATGENAEPVFPTIHGMEHFH--GPVVHTSDYKSGSEYKNKKVLVIGC 185
EG +Y WL+ ATGENAEPV P GM+ F G V HT YK+G ++ K+VLV+GC
Sbjct: 146 EGTTEYVCRWLVAATGENAEPVVPRFEGMDKFAAAGVVKHTCHYKTGGDFAGKRVLVVGC 205
Query: 186 GNSGMEVSLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVDKFLLL 245
GNSGMEV LDLC A P LV R++VH+LPR+M G ST+G++M L KWLP++LVD+FLL+
Sbjct: 206 GNSGMEVCLDLCNFGAQPSLVVRDAVHVLPREMLGTSTFGLSMFLLKWLPIRLVDRFLLV 265
Query: 246 VSSFFLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVKEITRNG 305
VS F LG+T G+ RP+ GP+ELK +GKTPVLDVG +A+IK+G+IKV G++ + R+
Sbjct: 266 VSRFILGDTTLLGLNRPRLGPLELKNISGKTPVLDVGTLAKIKTGDIKVCSGIRRLKRHE 325
Query: 306 AKFLDGQEKEFDAIILATGYKSNVPSWLK 334
+F +G+ + FDAIILATGYKSNVPSWLK
Sbjct: 326 VEFDNGKTERFDAIILATGYKSNVPSWLK 354
>At1g21430 flavin-containing monooxygenases, putative
Length = 391
Score = 288 bits (736), Expect = 4e-78
Identities = 143/322 (44%), Positives = 207/322 (63%), Gaps = 8/322 (2%)
Query: 17 IIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHFCELPMMSF 76
+I+GAGP+G+A +ACL+ +P++++ER C ASLW+ R+YDRLKLHL K FC+LP M F
Sbjct: 10 LIIGAGPAGLATSACLNRLNIPNIVVERDVCSASLWKRRSYDRLKLHLAKQFCQLPHMPF 69
Query: 77 PQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMVRTKEGDF--QYF 134
P P + +K FI+Y++ YA F+++PR+N+ V SA F I V K Y
Sbjct: 70 PSNTPTFVSKLGFINYLDEYATRFNVNPRYNRNVKSAYFKDGQWIVKVVNKTTALIEVYS 129
Query: 135 SPWLIVATGENAEPVFPTIHGM-EHFHGPVVHTSDYKSGSEYKNKKVLVIGCGNSGMEVS 193
+ +++ ATGEN E V P I G+ E F G +H+S+YK+G ++ K VLV+GCGNSGME++
Sbjct: 130 AKFMVAATGENGEGVIPEIPGLVESFQGKYLHSSEYKNGEKFAGKDVLVVGCGNSGMEIA 189
Query: 194 LDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVDKFLLLVSSFFLGN 253
DL + NA +V R+ VH+L R I M L ++ P+KLVD+ LL++ N
Sbjct: 190 YDLSKCNANVSIVVRSQVHVLTR-----CIVRIGMSLLRFFPVKLVDRLCLLLAELRFRN 244
Query: 254 TNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVMEGVKEITRNGAKFLDGQE 313
T+ YG+ RP GP KL TG++ +DVG + +IKSG I+V+ +K I +F+DG
Sbjct: 245 TSRYGLVRPNNGPFLNKLITGRSATIDVGCVGEIKSGKIQVVTSIKRIEGKTVEFIDGNT 304
Query: 314 KEFDAIILATGYKSNVPSWLKV 335
K D+I+ ATGYKS+V WL+V
Sbjct: 305 KNVDSIVFATGYKSSVSKWLEV 326
>At1g48910 hypothetical protein
Length = 383
Score = 283 bits (724), Expect = 9e-77
Identities = 136/323 (42%), Positives = 212/323 (65%), Gaps = 10/323 (3%)
Query: 17 IIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHFCELPMMSF 76
+IVGAGP+G+A + CL++ +P++ILE+ D ASLW+ R YDRLKLHL K FC+LP M
Sbjct: 6 VIVGAGPAGLATSVCLNQHSIPNVILEKEDIYASLWKKRAYDRLKLHLAKEFCQLPFMPH 65
Query: 77 PQTFPKYPTKHQFISYMESYADHFHIHPRFNQTVLSAEFDSTSQIWMV---RTKEGDFQ- 132
+ P + +K F++Y+++Y F I+PR+N+TV S+ FD ++ W V T G+ +
Sbjct: 66 GREVPTFMSKELFVNYLDAYVARFDINPRYNRTVKSSTFDESNNKWRVVAENTVTGETEV 125
Query: 133 YFSPWLIVATGENAEPVFPTIHGMEHFHGPVVHTSDYKSGSEYKNKKVLVIGCGNSGMEV 192
Y+S +L+VATGEN + P + G++ F G ++H+S+YKSG ++K+K VLV+G GNSGME+
Sbjct: 126 YWSEFLVVATGENGDGNIPMVEGIDTFGGEIMHSSEYKSGRDFKDKNVLVVGGGNSGMEI 185
Query: 193 SLDLCRHNAMPHLVARNSVHILPRDMFGFSTYGIAMGLYKWLPLKLVDKFLLLVSSFFLG 252
S DLC A ++ R H++ +++ + M L K+ P+ +VD + ++ G
Sbjct: 186 SFDLCNFGANTTILIRTPRHVVTKEVI-----HLGMTLLKYAPVAMVDTLVTTMAKILYG 240
Query: 253 NTNHYGIKRPKTGPIELKLATGKTPVLDVGQIAQIKSGNIKVME-GVKEITRNGAKFLDG 311
+ + YG+ RPK GP KL TGK PV+DVG + +I+ G I+V+ G+ I F +G
Sbjct: 241 DLSKYGLFRPKQGPFATKLFTGKAPVIDVGTVEKIRDGEIQVINGGIGSINGKTLTFENG 300
Query: 312 QEKEFDAIILATGYKSNVPSWLK 334
+++FDAI+ ATGYKS+V +WL+
Sbjct: 301 HKQDFDAIVFATGYKSSVCNWLE 323
>At5g45180 dimethylaniline monooxygenase (N-oxide-forming)-like
protein
Length = 453
Score = 77.8 bits (190), Expect = 8e-15
Identities = 96/369 (26%), Positives = 148/369 (40%), Gaps = 76/369 (20%)
Query: 18 IVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHFCELPMMSFP 77
I+GAG SG+A A L+ + E SD I +W+ TY+ KL + EL +P
Sbjct: 9 IIGAGVSGLAAAKHLARHHPQ--VFEASDSIGGVWRKCTYETTKLQSVRVSYELSDFLWP 66
Query: 78 QTFPK-YPTKHQFISYMESYADHFHIHP--RFNQTVLSAEFDSTSQ-------------- 120
+PT + Y+E+YA HF++ +FN V+ F +
Sbjct: 67 NRGESSFPTYVDVLDYLEAYAKHFNLVKFIKFNSKVVELRFIGDGKTLQMGDLGAYGNLL 126
Query: 121 ----IW--MVRTKEGDFQYFS-PWLIVATGENAE----PVFPTIHGMEHFHGPVVHTSDY 169
+W V T +GD Q+ + +++V G+ + P FP G E F G V+H+ DY
Sbjct: 127 PGKPVWEVAVNTGDGDIQWHAFEYVVVCAGKYGDVPRTPTFPVKKGPEIFKGKVLHSMDY 186
Query: 170 ------KSGSEYKNKKVLVIGCGNSGMEVSLDLCRHNAMPH-----LVARNSVHILPRDM 218
K+ KKV VIG S ++++L+ N +V R ++P
Sbjct: 187 SKLQKEKASQLLHGKKVAVIGFKKSAIDLALESALANQGKEGKTCTMVVRTPHWVIP--- 243
Query: 219 FGFSTYGIAMGLYKWLPLKLVDKFLLLVSSFFLGN--TNHYGIKRPKTGPIELKLATGKT 276
+ W V KF + S+ L YG+K E A+ +
Sbjct: 244 ------------HYW--RATVSKF---IESYVLWKLPLEKYGLKPDHA--FEEDYASCQM 284
Query: 277 PVLDVGQIAQIKSGNIKVMEGVK-EITRNGAKFLDGQEKEFDAIILATGY------KSNV 329
++ + G I+ G +F DG E D +ILATGY K+ V
Sbjct: 285 ALVPEKFFEEADKGMIRFKRTTNWWFYDEGIEFEDGTTLEADVVILATGYDGMKKLKAIV 344
Query: 330 P----SWLK 334
P SWL+
Sbjct: 345 PEPFRSWLE 353
>At1g19250 hypothetical protein
Length = 498
Score = 75.1 bits (183), Expect = 5e-14
Identities = 88/342 (25%), Positives = 139/342 (39%), Gaps = 42/342 (12%)
Query: 18 IVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQNRTYDRLKLHLPKHFCELPMMSFP 77
I+GAG SG+A A L + E SD + +W++ TY+ KL + E +P
Sbjct: 15 IIGAGVSGLAAAKNLVHHN--PTVFEASDSVGGVWRSCTYETTKLQSARVDYEFSDFPWP 72
Query: 78 Q-----TFPKYPTKHQFISYMESYADHFHI--HPRFNQTVLSAEF--DSTSQIWMVRTKE 128
TFP Y + + Y+ESYA HF + +F V+ F D + +
Sbjct: 73 NNRDDTTFPPYL---EILDYLESYAKHFDLLKFMKFGSKVIEVRFIGDGETPQMVDLGAY 129
Query: 129 GDFQYFSP---WLIVATGENAE----PVFPTIHGMEHFHGPVVHTSDY------KSGSEY 175
G+ P +++V TG+ + P FP G E F G V+H+ DY ++ +
Sbjct: 130 GNLLPGKPVWEFVVVCTGKYGDVPRIPAFPAKKGPEMFQGKVMHSMDYCKLEKEEASTLL 189
Query: 176 KNKKVLVIGCGNSGMEVSLDLCRHN-----AMPHLVARNSVHILPR------DMFGFSTY 224
KKV VIG S ++++L+ N +V R + +P F F +
Sbjct: 190 SGKKVAVIGFKKSAIDLALESALANQGEGGKACTMVVRTTHWGIPHYWVWGLPFFLFYSS 249
Query: 225 GIAMGLYKWLPLKLVDKFLLLVSSFFLGNTNHYGIKRPKTGPIELKLATGKTPVLDVGQI 284
+ L+ + L+ S L YG+K E A+ + ++
Sbjct: 250 RASQFLHDRPNQSFLRTLFCLLFSLLL-PLEKYGLK--PNHSFEEDYASCQMAIIPENFF 306
Query: 285 AQIKSGNIKVMEGVK-EITRNGAKFLDGQEKEFDAIILATGY 325
+ G I+ + K G F DG E D +ILATGY
Sbjct: 307 EEADKGMIRFKKSSKWWFYEEGIVFEDGTTLEADVVILATGY 348
>At1g62540 unknown protein
Length = 457
Score = 70.9 bits (172), Expect = 1e-12
Identities = 50/224 (22%), Positives = 99/224 (43%), Gaps = 30/224 (13%)
Query: 17 IIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLW--------------------QNRT 56
+++GAG +G+ A LS +G ++LER + LW +
Sbjct: 14 VVIGAGAAGLVAARELSREGHTVVVLEREKEVGGLWIYSPKAESDPLSLDPTRSIVHSSV 73
Query: 57 YDRLKLHLPKHFCELPMMSFPQTFP-------KYPTKHQFISYMESYADHFHIHP--RFN 107
Y+ L+ +LP+ F F +YP+ + ++Y++ +A F++ RF
Sbjct: 74 YESLRTNLPRECMGFTDFPFVPRFDDESRDSRRYPSHMEVLAYLQDFAREFNLEEMVRFE 133
Query: 108 QTVLSAE-FDSTSQIWMVRTKEGDFQYFSPWLIVATGENAEPVFPTIHGMEHFHGPVVHT 166
V+ E + ++W + ++V +G EP I G++ + G +H+
Sbjct: 134 IEVVRVEPVNGKWRVWSKTSGGVSHDEIFDAVVVCSGHYTEPNVAHIPGIKSWPGKQIHS 193
Query: 167 SDYKSGSEYKNKKVLVIGCGNSGMEVSLDLCRHNAMPHLVARNS 210
+Y+ ++N+ V+VIG SG ++S D+ + H+ +R S
Sbjct: 194 HNYRVPGPFENEVVVVIGNFASGADISRDIAKVAKEVHIASRAS 237
>At1g12200 unknown protein
Length = 465
Score = 69.7 bits (169), Expect = 2e-12
Identities = 52/228 (22%), Positives = 101/228 (43%), Gaps = 35/228 (15%)
Query: 18 IVGAGPSGIAVAACLSEQGVPSLILERSDCIASLW--------------------QNRTY 57
++GAG +G+ A L +G ++LER I +W + Y
Sbjct: 16 VIGAGAAGLVAARELRREGHSVVVLERGSQIGGVWAYTSQVEPDPLSLDPTRPVVHSSLY 75
Query: 58 DRLKLHLPKH---FCELPMMSFPQTFPKYPTKH----QFISYMESYADHFHIHP--RFNQ 108
L+ ++P+ F + P + P + P +H + ++Y+ +A F I RF
Sbjct: 76 RSLRTNIPRECMGFTDFPFATRPHDGSRDPRRHPAHTEVLAYLRDFAKEFDIEEMVRFET 135
Query: 109 TVLSAEFDSTSQI----WMV--RTKEGDFQYFSPWLIVATGENAEPVFPTIHGMEHFHGP 162
V+ AE + W V R+ +G ++V G EP I G++ + G
Sbjct: 136 EVVKAEQVAAEGEERGKWRVESRSSDGVVDEIYDAVVVCNGHYTEPRHALITGIDSWPGK 195
Query: 163 VVHTSDYKSGSEYKNKKVLVIGCGNSGMEVSLDLCRHNAMPHLVARNS 210
+H+ +Y+ ++K++ V+VIG SG+++ D+ + H+ +R++
Sbjct: 196 QIHSHNYRVPDQFKDQVVIVIGSSASGVDICRDIAQVAKEVHVSSRST 243
>At1g62570 similar to glutamate synthase
Length = 461
Score = 69.3 bits (168), Expect = 3e-12
Identities = 58/227 (25%), Positives = 102/227 (44%), Gaps = 38/227 (16%)
Query: 18 IVGAGPSGIAVAACLSEQGVPSLILERSDCIASLW--------------------QNRTY 57
++GAG +G+ A L +G ++L+R + LW + Y
Sbjct: 15 VIGAGAAGLVAARELRREGHTVVVLDREKQVGGLWVYTPETESDELGLDPTRPIVHSSVY 74
Query: 58 DRLKLHLP------KHFCELPMMSFP-QTFPKYPTKHQFISYMESYADHFHIHP--RFNQ 108
L+ +LP K F +P P + +YP+ + ++Y++ +A F+I RF
Sbjct: 75 KSLRTNLPRECMGYKDFPFVPRGDDPSRDSRRYPSHREVLAYLQDFATEFNIEEMIRFET 134
Query: 109 TVLSAEFDSTSQIWMVRTKEG-----DFQYFSPWLIVATGENAEPVFPTIHGMEHFHGPV 163
VL E + W V++K G D Y + +++ G AEP I G+E + G
Sbjct: 135 EVLRVE--PVNGKWRVQSKTGGGFSNDEIYDA--VVMCCGHFAEPNIAQIPGIESWPGRQ 190
Query: 164 VHTSDYKSGSEYKNKKVLVIGCGNSGMEVSLDLCRHNAMPHLVARNS 210
H+ Y+ +K++ V+VIG SG ++S D+ + H+ +R S
Sbjct: 191 THSHSYRVPDPFKDEVVVVIGNFASGADISRDISKVAKEVHIASRAS 237
>At1g65860 flavin-containing monooxygenase FMO3, putative
Length = 459
Score = 67.8 bits (164), Expect = 8e-12
Identities = 54/226 (23%), Positives = 101/226 (43%), Gaps = 36/226 (15%)
Query: 18 IVGAGPSGIAVAACLSEQGVPSLILERSDCIASLWQ--------------NRT------Y 57
++GAG +G+ A L +G ++ +R + LW RT Y
Sbjct: 15 VIGAGAAGLVTARELRREGHTVVVFDREKQVGGLWNYSSKADSDPLSLDTTRTIVHTSIY 74
Query: 58 DRLKLHLPKH---FCELPMM----SFPQTFPKYPTKHQFISYMESYADHFHIHP--RFNQ 108
+ L+ +LP+ F + P + + +YP+ + ++Y++ +A F I RF
Sbjct: 75 ESLRTNLPRECMGFTDFPFVPRIHDISRDSRRYPSHREVLAYLQDFAREFKIEEMVRFET 134
Query: 109 TVLSAEFDSTSQIWMVRTKEG----DFQYFSPWLIVATGENAEPVFPTIHGMEHFHGPVV 164
V+ E + W VR+K + F ++V +G EP I G++ + G +
Sbjct: 135 EVVCVE--PVNGKWSVRSKNSVGFAAHEIFDA-VVVCSGHFTEPNVAHIPGIKSWPGKQI 191
Query: 165 HTSDYKSGSEYKNKKVLVIGCGNSGMEVSLDLCRHNAMPHLVARNS 210
H+ +Y+ + N+ V+VIG SG ++S D+ + H+ +R S
Sbjct: 192 HSHNYRVPGPFNNEVVVVIGNYASGADISRDIAKVAKEVHIASRAS 237
>At5g61290 unknown protein
Length = 461
Score = 66.2 bits (160), Expect = 2e-11
Identities = 55/244 (22%), Positives = 111/244 (44%), Gaps = 41/244 (16%)
Query: 2 DPFKEKVKCVWIHGPIIVGAGPSGIAVAACLSEQGVPSLILERSDCIASLW--------- 52
+P K + K V ++GAGPSG+ A L ++G +++E++ + W
Sbjct: 7 EPIKSQSKTV-----CVIGAGPSGLVSARELKKEGHKVVVMEQNHDVGGQWLYQPNVDEE 61
Query: 53 -----------QNRTYDRLKLHLPKH---FCELPMMSFP-QTFPKYPTKHQFISYMESYA 97
+ Y L+L P+ F + P ++ + ++P + + Y++ +
Sbjct: 62 DTLGKTKTLKVHSSVYSSLRLASPREVMGFSDFPFIAKEGRDSRRFPGHEELLLYLKDFC 121
Query: 98 DHFHIHP--RFNQTV-----LSAEFDSTSQI--WMVRT--KEGD-FQYFSPWLIVATGEN 145
F + RFN V ++ + D + WMV++ K G+ + ++VA+G
Sbjct: 122 QVFGLREMIRFNVRVEFVGMVNEDDDDDDDVKKWMVKSVKKSGEVMEEVFDAVVVASGHY 181
Query: 146 AEPVFPTIHGMEHFHGPVVHTSDYKSGSEYKNKKVLVIGCGNSGMEVSLDLCRHNAMPHL 205
+ P PTI GM+ + +H+ Y+ + ++ V+V+GC SG ++S++L HL
Sbjct: 182 SYPRLPTIKGMDLWKRKQLHSHIYRVPEPFCDEVVVVVGCSMSGQDISIELVEVAKEVHL 241
Query: 206 VARN 209
++
Sbjct: 242 STKS 245
>At1g62560 similar to flavin-containing monooxygenase (sp|P36366);
similar to ESTs gb|R30018, gb|H36886, gb|N37822, and
gb|T88100
Length = 462
Score = 66.2 bits (160), Expect = 2e-11
Identities = 51/223 (22%), Positives = 97/223 (42%), Gaps = 33/223 (14%)
Query: 18 IVGAGPSGIAVAACLSEQGVPSLILERSDCIASLW--------------------QNRTY 57
++GAGP+G+ + L +G ++ ER + LW + Y
Sbjct: 15 VIGAGPAGLITSRELRREGHSVVVFEREKQVGGLWVYTPKSDSDPLSLDPTRSKVHSSIY 74
Query: 58 DRLKLHLPKHFCELPMMSFPQTFP-------KYPTKHQFISYMESYADHFHIHP--RFNQ 108
+ L+ ++P+ + F F +YP + ++Y++ +A F I RF
Sbjct: 75 ESLRTNVPRESMGVRDFPFLPRFDDESRDARRYPNHREVLAYIQDFAREFKIEEMIRFET 134
Query: 109 TVLSAEFDSTSQIWMVRTKE-GDFQYFSPW--LIVATGENAEPVFPTIHGMEHFHGPVVH 165
V+ E W V++K G F + ++V G EP I G++ + G +H
Sbjct: 135 EVVRVEPVDNGN-WRVQSKNSGGFLEDEIYDAVVVCNGHYTEPNIAHIPGIKSWPGKQIH 193
Query: 166 TSDYKSGSEYKNKKVLVIGCGNSGMEVSLDLCRHNAMPHLVAR 208
+ +Y+ ++N+ V+VIG SG ++S D+ + H+ +R
Sbjct: 194 SHNYRVPDPFENEVVVVIGNFASGADISRDIAKVAKEVHIASR 236
>At5g07800 dimethylaniline monooxygenase-like protein
Length = 460
Score = 65.9 bits (159), Expect = 3e-11
Identities = 50/229 (21%), Positives = 105/229 (45%), Gaps = 38/229 (16%)
Query: 18 IVGAGPSGIAVAACLSEQGVPSLILERSDCIASLW------------------------- 52
++GAGP+G+ A L ++G ++LE+++ + W
Sbjct: 18 VIGAGPAGLVSARELRKEGHKVVVLEQNEDVGGQWFYQPNVEEEDPLGRSSGSINGELKV 77
Query: 53 QNRTYDRLKLHLPKH---FCELPMMSFP-QTFPKYPTKHQFISYMESYADHFHIHP--RF 106
+ Y L+L P+ + + P ++ + ++P + Y++ +++ F + RF
Sbjct: 78 HSSIYSSLRLTSPREIMGYSDFPFLAKKGRDMRRFPGHKELWLYLKDFSEAFGLREMIRF 137
Query: 107 N---QTVLSAEFDSTSQIWMVRTKEGD----FQYFSPWLIVATGENAEPVFPTIHGMEHF 159
N + V E + + W+VR++E + ++VATG + P P+I GM+ +
Sbjct: 138 NVRVEFVGEKEEEDDVKKWIVRSREKFSGKVMEEIFDAVVVATGHYSHPRLPSIKGMDSW 197
Query: 160 HGPVVHTSDYKSGSEYKNKKVLVIGCGNSGMEVSLDLCRHNAMPHLVAR 208
+H+ Y+ ++N+ V+V+G SG ++S++L HL A+
Sbjct: 198 KRKQIHSHVYRVPDPFRNEVVVVVGNSMSGQDISMELVEVAKEVHLSAK 246
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.322 0.139 0.433
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,286,387
Number of Sequences: 26719
Number of extensions: 358816
Number of successful extensions: 894
Number of sequences better than 10.0: 53
Number of HSP's better than 10.0 without gapping: 46
Number of HSP's successfully gapped in prelim test: 7
Number of HSP's that attempted gapping in prelim test: 781
Number of HSP's gapped (non-prelim): 73
length of query: 337
length of database: 11,318,596
effective HSP length: 100
effective length of query: 237
effective length of database: 8,646,696
effective search space: 2049266952
effective search space used: 2049266952
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.9 bits)
S2: 60 (27.7 bits)
Medicago: description of AC144617.10


