

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144564.8 - phase: 0
(655 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
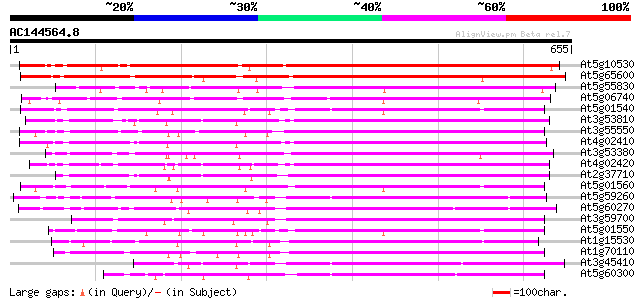
Score E
Sequences producing significant alignments: (bits) Value
At5g10530 lectin-like protein kinase - like 499 e-141
At5g65600 receptor protein kinase-like protein 483 e-136
At5g55830 serine/threonine-specific kinase like protein 356 2e-98
At5g06740 lectin-like protein kinase 330 2e-90
At5g01540 receptor like protein kinase 327 2e-89
At3g53810 serine/threonine-specific kinase like protein 319 4e-87
At3g55550 probable serine/threonine-specific protein kinase 318 5e-87
At4g02410 unknown protein 317 1e-86
At3g53380 receptor lectin kinase -like protein 317 1e-86
At4g02420 315 4e-86
At2g37710 putative receptor-like protein kinase 311 6e-85
At5g01560 receptor like protein kinase 311 1e-84
At5g59260 receptor-like protein kinase 306 2e-83
At5g60270 receptor like protein kinase 302 4e-82
At3g59700 serine/threonine-specific kinase lecRK1 precursor,lect... 302 4e-82
At5g01550 receptor like protein kinase 297 1e-80
At1g15530 hypothetical protein 296 2e-80
At1g70110 hypothetical protein 295 5e-80
At3g45410 receptor-like protein kinase 286 3e-77
At5g60300 receptor like protein kinase 285 6e-77
>At5g10530 lectin-like protein kinase - like
Length = 651
Score = 499 bits (1286), Expect = e-141
Identities = 291/648 (44%), Positives = 405/648 (61%), Gaps = 35/648 (5%)
Query: 12 TIMLF-LIIPLAHAHLVSFDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTDDPDKAS 70
+I+LF ++ L V F+ F + E+ GDA ++ ++LT D
Sbjct: 4 SILLFSFVLVLPFVCSVQFNISRFGSDVSEIA-YQGDAR---ANGAVELTNI----DYTC 55
Query: 71 SVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFSN---HSSYGDGLAFFLASSNLTKANR 127
G T K + L+N T++ DF T+FSF I + + +YG G AFFLA + +
Sbjct: 56 RAGWATYGKQVPLWNPGTSKPSDFSTRFSFRIDTRNVGYGNYGHGFAFFLAPARIQLPPN 115
Query: 128 IGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKNI-WDP--GFPHVGVNINSVVSDTNIE 184
GG GL N S+ + +V VEFDT N WDP HVG+N NS+VS
Sbjct: 116 -SAGGFLGLF--NGTNNQSSAFPLVYVEFDTFTNPEWDPLDVKSHVGINNNSLVSSNYTS 172
Query: 185 WFSNVSERMVYNCSIEYISRNNVLNVSFTGYRLNAWQEPQNFSHIINLREHLPEYVRVGI 244
W + + + I Y S L+VS+T + E + S+II+L + LP V +G
Sbjct: 173 WNATSHNQDIGRVLIFYDSARRNLSVSWTYDLTSDPLENSSLSYIIDLSKVLPSEVTIGF 232
Query: 245 SASTGKVDEEHMLLSWSFSTSQPSYFVVDPRKTK-----LWEGLAVGGVCLSWSLVAILI 299
SA++G V E + LLSW FS+S ++D +K++ + G++V G L + LI
Sbjct: 233 SATSGGVTEGNRLLSWEFSSSLE---LIDIKKSQNDKKGMIIGISVSGFVLLTFFITSLI 289
Query: 300 IFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGF 359
+FL +K + K+ E T TS ++++ + GAGP+K +Y +L +A NNF + +KLG+GGF
Sbjct: 290 VFLKRKQQKKKAEETENLTS---INEDLERGAGPRKFTYKDLASAANNFADDRKLGEGGF 346
Query: 360 GGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELI 419
G VY+GY + + AIK+ + S+QG +++ EVKIIS LRHRNLV+L GWCH+K+E +
Sbjct: 347 GAVYRGYLNSLDMMVAIKKFAGGSKQGKREFVTEVKIISSLRHRNLVQLIGWCHEKDEFL 406
Query: 420 LIYEYMPNGSLDFHLFRGGSILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIM 479
+IYE+MPNGSLD HLF L W++R I LGLASALLYL EEWE+CV+HRDIK+SN+M
Sbjct: 407 MIYEFMPNGSLDAHLFGKKPHLAWHVRCKITLGLASALLYLHEEWEQCVVHRDIKASNVM 466
Query: 480 LDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLL 539
LDS+FN KLGDFGLARLMDHE G +TT +AGT GY+APEYI T +A KESD++SFGVV L
Sbjct: 467 LDSNFNAKLGDFGLARLMDHELGPQTTGLAGTFGYMAPEYISTGRASKESDVYSFGVVTL 526
Query: 540 EIACGKKAIHHQE--LEGEVSLVEWVWELYGLRNLIVAADPKL-CGIFDVKQLECLLVVG 596
EI G+K++ ++ +E +LVE +W+LYG +I A D KL G FD KQ ECL++VG
Sbjct: 527 EIVTGRKSVDRRQGRVEPVTNLVEKMWDLYGKGEVITAIDEKLRIGGFDEKQAECLMIVG 586
Query: 597 LWCANPDITSRPSIKKVIKVLNFEAPLPILPLNMP---FLASLSPTTN 641
LWCA+PD+ +RPSIK+ I+VLN EAP+P LP MP + S S TT+
Sbjct: 587 LWCAHPDVNTRPSIKQAIQVLNLEAPVPHLPTKMPVATYHVSSSNTTS 634
>At5g65600 receptor protein kinase-like protein
Length = 675
Score = 483 bits (1244), Expect = e-136
Identities = 294/660 (44%), Positives = 406/660 (60%), Gaps = 44/660 (6%)
Query: 13 IMLFLIIPLAHAHLVSFDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTDDPDKASSV 72
+ LFL +P L F++ F GDAT D D + ++ ++ S V
Sbjct: 23 LSLFLFLPFVVDSLY-FNFTSFRQGDPGDIFYHGDAT-PDEDGTVNF----NNAEQTSQV 76
Query: 73 GRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFS-NHSSYGDGLAFFLASSNLT-KANRIGG 130
G +T K + +++ T + DF T FSF I + N S+ G G+ FFLA A +GG
Sbjct: 77 GWITYSKKVPIWSHKTGKASDFSTSFSFKIDARNLSADGHGICFFLAPMGAQLPAYSVGG 136
Query: 131 GGGFGLVPANEVALNSTEYSIVLVEFDTHKNI-WDPGF--PHVGVNINSVVSDTNIEWFS 187
N S+ + +V VEFDT N WDP HVG+N NS+VS W +
Sbjct: 137 FLNLFTRKNNY----SSSFPLVHVEFDTFNNPGWDPNDVGSHVGINNNSLVSSNYTSWNA 192
Query: 188 NVSERMVYNCSIEYISRNNVLNVSFTGYRLNAWQEPQ---NFSHIINLREHLPEYVRVGI 244
+ + + + I Y S L+V++ Y L A +P+ + S+II+L + LP V G
Sbjct: 193 SSHSQDICHAKISYDSVTKNLSVTWA-YELTATSDPKESSSLSYIIDLAKVLPSDVMFGF 251
Query: 245 SASTGKVDEEHMLLSWSFSTSQPSYFVVDPRKTKLWEGLAVG-----GVCLSWSLVAILI 299
A+ G EEH LLSW S+S +D K GL +G V L++ ++ ++
Sbjct: 252 IAAAGTNTEEHRLLSWELSSS------LDSDKADSRIGLVIGISASGFVFLTFMVITTVV 305
Query: 300 IFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGF 359
++ K+ K KE + + + ++D++ E AGP+K SY +L++ATN F +KLG+GGF
Sbjct: 306 VWSRKQRKKKERDIENMISINKDLERE----AGPRKFSYKDLVSATNRFSSHRKLGEGGF 361
Query: 360 GGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELI 419
G VY+G K+ N++ A+K++S DSRQG ++ EVKIIS+LRHRNLV+L GWC++KNE +
Sbjct: 362 GAVYEGNLKEINTMVAVKKLSGDSRQGKNEFLNEVKIISKLRHRNLVQLIGWCNEKNEFL 421
Query: 420 LIYEYMPNGSLDFHLF-RGGSILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNI 478
LIYE +PNGSL+ HLF + ++L W++RY I LGLASALLYL EEW++CV+HRDIK+SNI
Sbjct: 422 LIYELVPNGSLNSHLFGKRPNLLSWDIRYKIGLGLASALLYLHEEWDQCVLHRDIKASNI 481
Query: 479 MLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVL 538
MLDS+FN KLGDFGLARLM+HE GS TT +AGT GY+APEY+ A KESDI+SFG+VL
Sbjct: 482 MLDSEFNVKLGDFGLARLMNHELGSHTTGLAGTFGYMAPEYVMKGSASKESDIYSFGIVL 541
Query: 539 LEIACGKKAIHH-------QELEGEVSLVEWVWELYGLRNLIVA-ADPKLCGIFDVKQLE 590
LEI G+K++ E + E SLVE VWELYG + LI + D KL FD K+ E
Sbjct: 542 LEIVTGRKSLERTQEDNSDTESDDEKSLVEKVWELYGKQELITSCVDDKLGEDFDKKEAE 601
Query: 591 CLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLPILPLNMPF-LASLSPTTNEQFFSVPS 649
CLLV+GLWCA+PD SRPSIK+ I+V+NFE+PLP LPL P + +S TT+ SV S
Sbjct: 602 CLLVLGLWCAHPDKNSRPSIKQGIQVMNFESPLPDLPLKRPVAMYYISTTTSSSSPSVNS 661
>At5g55830 serine/threonine-specific kinase like protein
Length = 681
Score = 356 bits (914), Expect = 2e-98
Identities = 233/636 (36%), Positives = 353/636 (54%), Gaps = 78/636 (12%)
Query: 54 DNIIQLTGYTDDPDKASSVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFS---NHSSYG 110
+ ++ LT PD +S +P I+ Y+ +N F T FSFT+ + + +S G
Sbjct: 53 NGVVGLTRELGVPDTSSGTVIYNNP--IRFYDPDSNTTASFSTHFSFTVQNLNPDPTSAG 110
Query: 111 DGLAFFLASSNLTKANRIGGGGGF-GLVPANEVALNSTEYSIVLVEFDT----HKNIWDP 165
DGLAFFL+ N T +G GG+ GLV +++ N V +EFDT H N DP
Sbjct: 111 DGLAFFLSHDNDT----LGSPGGYLGLVNSSQPMKNR----FVAIEFDTKLDPHFN--DP 160
Query: 166 GFPHVGVNINSV----VSDTNIEWFSNVSERMVYNCSIEYISRNNVLNVSFTGYR--LNA 219
H+G++++S+ SD + ++ I+Y + +LNV F Y +
Sbjct: 161 NGNHIGLDVDSLNSISTSDPLLSSQIDLKSGKSITSWIDYKNDLRLLNV-FLSYTDPVTT 219
Query: 220 WQEPQN--FSHIINLREHLPEYVRVGISASTGKVDEEHMLLSWSFSTS------------ 265
++P+ S I+L L + VG S ST E H++ +WSF TS
Sbjct: 220 TKKPEKPLLSVNIDLSPFLNGEMYVGFSGSTEGSTEIHLIENWSFKTSGFLPVRSKSNHL 279
Query: 266 ---------QPSYFVVDPRKTKLWEGLAVGG-------VCLSWSLVAILIIFLWKKNKGK 309
V+ +K + LA+G +CL+ + + WK K +
Sbjct: 280 HNVSDSSVVNDDPVVIPSKKRRHRHNLAIGLGISCPVLICLALFVFGYFTLKKWKSVKAE 339
Query: 310 EDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKD 369
++ T T G ++ SY EL AT F ++ +G+G FG VY+ F
Sbjct: 340 KELKTELIT-------------GLREFSYKELYTATKGFHSSRVIGRGAFGNVYRAMFVS 386
Query: 370 SNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGS 429
S +++A+KR +S +G ++ AE+ II+ LRH+NLV+L GWC++K EL+L+YE+MPNGS
Sbjct: 387 SGTISAVKRSRHNSTEGKTEFLAELSIIACLRHKNLVQLQGWCNEKGELLLVYEFMPNGS 446
Query: 430 LDFHLFR----GGSILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFN 485
LD L++ G L W+ R NIA+GLASAL YL E E+ V+HRDIK+SNIMLD +FN
Sbjct: 447 LDKILYQESQTGAVALDWSHRLNIAIGLASALSYLHHECEQQVVHRDIKTSNIMLDINFN 506
Query: 486 TKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGK 545
+LGDFGLARL +H+K +T+ AGT GYLAPEY+ A +++D FS+GVV+LE+ACG+
Sbjct: 507 ARLGDFGLARLTEHDKSPVSTLTAGTMGYLAPEYLQYGTATEKTDAFSYGVVILEVACGR 566
Query: 546 KAIHHQ-ELEGEVSLVEWVWELYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDI 604
+ I + E + V+LV+WVW L+ ++ A D +L G FD + ++ LL+VGL CA+PD
Sbjct: 567 RPIDKEPESQKTVNLVDWVWRLHSEGRVLEAVDERLKGEFDEEMMKKLLLVGLKCAHPDS 626
Query: 605 TSRPSIKKVIKVLNFE---APLPILPLNMPFLASLS 637
RPS+++V+++LN E +P+P + + F LS
Sbjct: 627 NERPSMRRVLQILNNEIEPSPVPKMKPTLSFSCGLS 662
>At5g06740 lectin-like protein kinase
Length = 652
Score = 330 bits (846), Expect = 2e-90
Identities = 232/642 (36%), Positives = 333/642 (51%), Gaps = 54/642 (8%)
Query: 14 MLFLIIPL---AHAHLVSFDYPMFNHNCKELPELDGDATIEDSDNI----IQLT-GYTDD 65
+LFLI+ + FD+P FN + EL+ I D+ I IQ+T T
Sbjct: 9 LLFLILTCKIETQVKCLKFDFPGFNVS----NELE---LIRDNSYIVFGAIQVTPDVTGG 61
Query: 66 PDK--ASSVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFSNHSSYGDGLAFFLASSNLT 123
P A+ GR K +L+++ + F T F I + G+GLAF L
Sbjct: 62 PGGTIANQAGRALYKKPFRLWSKHKSAT--FNTTFVINISNKTDPGGEGLAFVLTPEETA 119
Query: 124 KANRIGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKNIWDP-GFPHVGVN---INSVVS 179
N G L NE + E IV VEFDT K+ D HV +N INSVV
Sbjct: 120 PQN----SSGMWLGMVNERTNRNNESRIVSVEFDTRKSHSDDLDGNHVALNVNNINSVVQ 175
Query: 180 DTNIEWFSNVSERMVYNCSIEYISRNNVLNVSFTGYRLNAWQEPQN-FSHIINLREHLPE 238
++ + + + Y +N + VS L+ +++ FS I+L +LPE
Sbjct: 176 ESLSGRGIKIDSGLDLTAHVRYDGKNLSVYVS---RNLDVFEQRNLVFSRAIDLSAYLPE 232
Query: 239 YVRVGISASTGKVDEEHMLLSWSFSTSQPSYFVVDPRKTKLWEGLAVGGVCLSWSLVAIL 298
V VG +AST E + + SWSF + +D LW + + V + +
Sbjct: 233 TVYVGFTASTSNFTELNCVRSWSFEGLK-----IDGDGNMLWLWITIPIVFIVGIGAFLG 287
Query: 299 IIFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGG 358
++L ++K E P E D A P+K EL AT NF KLGQGG
Sbjct: 288 ALYLRSRSKAGETNPDIEAELDN-------CAANPQKFKLRELKRATGNFGAENKLGQGG 340
Query: 359 FGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNEL 418
FG V+KG ++ + A+KR+S S QG +++ AE+ I L HRNLVKL GWC+++ E
Sbjct: 341 FGMVFKGKWQGRD--IAVKRVSEKSHQGKQEFIAEITTIGNLNHRNLVKLLGWCYERKEY 398
Query: 419 ILIYEYMPNGSLDFHLF---RGGSILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKS 475
+L+YEYMPNGSLD +LF + S L W R NI GL+ AL YL EK ++HRDIK+
Sbjct: 399 LLVYEYMPNGSLDKYLFLEDKSRSNLTWETRKNIITGLSQALEYLHNGCEKRILHRDIKA 458
Query: 476 SNIMLDSDFNTKLGDFGLARLMDHEKGS--ETTVVAGTRGYLAPEYIDTSKARKESDIFS 533
SN+MLDSDFN KLGDFGLAR++ + + T +AGT GY+APE +A E+D+++
Sbjct: 459 SNVMLDSDFNAKLGDFGLARMIQQSEMTHHSTKEIAGTPGYMAPETFLNGRATVETDVYA 518
Query: 534 FGVVLLEIACGKK----AIHHQELEGEVSLVEWVWELYGLRNLIVAADPKLCGIFDVKQL 589
FGV++LE+ GKK + + S+V W+WELY + AADP + +FD +++
Sbjct: 519 FGVLMLEVVSGKKPSYVLVKDNQNNYNNSIVNWLWELYRNGTITDAADPGMGNLFDKEEM 578
Query: 590 ECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLPILPLNMP 631
+ +L++GL C +P+ RPS+K V+KVL E P +P P
Sbjct: 579 KSVLLLGLACCHPNPNQRPSMKTVLKVLTGETSPPDVPTERP 620
>At5g01540 receptor like protein kinase
Length = 682
Score = 327 bits (837), Expect = 2e-89
Identities = 234/657 (35%), Positives = 348/657 (52%), Gaps = 75/657 (11%)
Query: 13 IMLFLIIPL-AHAHLVSFDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTDDPDKASS 71
+ LFL + + A +F + FN N ++ ++G A I+ D +++LT D+ S+
Sbjct: 16 LFLFLSVHVRAQRTTTNFAFRGFNGNQSKI-RIEGAAMIKP-DGLLRLT------DRKSN 67
Query: 72 V-GRVTSPKLIKLYNRSTNEVY--DFRTKFSFTIF-SNHSSYGDGLAFFLASSNLTKANR 127
V G K ++L NR++ V F T F F I S+ S+ G G F L+ + R
Sbjct: 68 VTGTAFYHKPVRLLNRNSTNVTIRSFSTSFVFVIIPSSSSNKGFGFTFTLSPTPY----R 123
Query: 128 IGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKNIWDPGFPHVG----VNINSVVSDTN- 182
+ G L N+ + VEFDT + D +G +N NS SD
Sbjct: 124 LNAGSAQYLGVFNKENNGDPRNHVFAVEFDTVQGSRDDNTDRIGNDIGLNYNSRTSDLQE 183
Query: 183 -IEWFSN----------VSERMVYNCSIEYISRNNVLNVSFTGYRLNAWQ-EPQNFSHII 230
+ +++N + +EY +LNV+ RL +P H+
Sbjct: 184 PVVYYNNDDHNKKEDFQLESGNPIQALLEYDGATQMLNVTVYPARLGFKPTKPLISQHVP 243
Query: 231 NLREHLPEYVRVGISASTGKVDEE-HMLLSWSFSTS--QPSYFVV--------DPRKTKL 279
L E + E + VG +ASTGK H ++ WSFS+ +P V+ P K K
Sbjct: 244 KLLEIVQEEMYVGFTASTGKGQSSAHYVMGWSFSSGGERPIADVLILSELPPPPPNKAKK 303
Query: 280 WEGLAVGGVCLSWSLVAILIIFL--------WKKNKGKEDEPTSETTSDQDMDDEFQMGA 331
EGL + + +L A++++ L +KK G+E ET D ++D
Sbjct: 304 -EGLNSQVIVMIVALSAVMLVMLVLLFFFVMYKKRLGQE-----ETLEDWEIDH------ 351
Query: 332 GPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYS 391
P+++ Y +L AT+ F++T +G GGFG V+KG +S+ +A +K+I SRQGV+++
Sbjct: 352 -PRRLRYRDLYVATDGFKKTGIIGTGGFGTVFKGKLPNSDPIA-VKKIIPSSRQGVREFV 409
Query: 392 AEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF----RGGSILPWNLRY 447
AE++ + +LRH+NLV L GWC KN+L+LIY+Y+PNGSLD L+ R G++L WN R+
Sbjct: 410 AEIESLGKLRHKNLVNLQGWCKHKNDLLLIYDYIPNGSLDSLLYTVPRRSGAVLSWNARF 469
Query: 448 NIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTV 507
IA G+AS LLYL EEWEK VIHRD+K SN+++DS N +LGDFGLARL + SETT
Sbjct: 470 QIAKGIASGLLYLHEEWEKIVIHRDVKPSNVLIDSKMNPRLGDFGLARLYERGTLSETTA 529
Query: 508 VAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELY 567
+ GT GY+APE SD+F+FGV+LLEI CG+K G LV+WV EL+
Sbjct: 530 LVGTIGYMAPELSRNGNPSSASDVFAFGVLLLEIVCGRKPTD----SGTFFLVDWVMELH 585
Query: 568 GLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLP 624
++ A DP+L +D + L VGL C + SRPS++ V++ LN E +P
Sbjct: 586 ANGEILSAIDPRLGSGYDGGEARLALAVGLLCCHQKPASRPSMRIVLRYLNGEENVP 642
>At3g53810 serine/threonine-specific kinase like protein
Length = 677
Score = 319 bits (817), Expect = 4e-87
Identities = 219/646 (33%), Positives = 347/646 (52%), Gaps = 63/646 (9%)
Query: 19 IPLAHAHLVSFDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTDDPDKASSVGRVTSP 78
I ++ + ++F Y F+ ++ L G AT+ + +++LT + G
Sbjct: 17 IMISSSQNLNFTYNGFHPPLTDI-SLQGLATVTPN-GLLKLTNTS-----VQKTGHAFCT 69
Query: 79 KLIKLYNRSTNEVYDFRTKFSFTIFSNHSSY-GDGLAFFLASSNLTKANRIGGGGGFGLV 137
+ I+ + V F T F F I S + G G+AF +A + G F L
Sbjct: 70 ERIRFKDSQNGNVSSFSTTFVFAIHSQIPTLSGHGIAFVVAPTL---------GLPFAL- 119
Query: 138 PANEVAL-------NSTEYSIVLVEFDTHKN--IWDPGFPHVGVNINSVVSDT------- 181
P+ + L N T + I VEFDT ++ DP HVG+++N + S
Sbjct: 120 PSQYIGLFNISNNGNDTNH-IFAVEFDTIQSSEFGDPNDNHVGIDLNGLRSANYSTAGYR 178
Query: 182 -NIEWFSNVS--ERMVYNCSIEYISRNNVLNVSFTGYRLNAWQEPQNFSHIINLREHLPE 238
+ + F N+S R I+Y +R++ ++V+ + + ++P S++ +L L E
Sbjct: 179 DDHDKFQNLSLISRKRIQVWIDYDNRSHRIDVTVAPFDSDKPRKPL-VSYVRDLSSILLE 237
Query: 239 YVRVGISASTGKVDEEHMLLSWSFS----------TSQPSYFVVDPRKTKLWEGLAVGGV 288
+ VG S++TG V EH L+ WSF + P +PR+ + + + +
Sbjct: 238 DMYVGFSSATGSVLSEHFLVGWSFRLNGEAPMLSLSKLPKLPRFEPRRISEFYKIGMPLI 297
Query: 289 CLSWSLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNF 348
LS I + F + K K +E D + EF G + + EL +AT F
Sbjct: 298 SLSLIFSIIFLAFYIVRRKKKYEEELD------DWETEF----GKNRFRFKELYHATKGF 347
Query: 349 EETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKL 408
+E LG GGFG VY+G + A+KR+S DS+QG+K++ AE+ I ++ HRNLV L
Sbjct: 348 KEKDLLGSGGFGRVYRGILPTTKLEVAVKRVSHDSKQGMKEFVAEIVSIGRMSHRNLVPL 407
Query: 409 TGWCHKKNELILIYEYMPNGSLDFHLFRGG-SILPWNLRYNIALGLASALLYLQEEWEKC 467
G+C ++ EL+L+Y+YMPNGSLD +L+ + L W R I G+AS L YL EEWE+
Sbjct: 408 LGYCRRRGELLLVYDYMPNGSLDKYLYNNPETTLDWKQRSTIIKGVASGLFYLHEEWEQV 467
Query: 468 VIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARK 527
VIHRD+K+SN++LD+DFN +LGDFGLARL DH +TT V GT GYLAPE+ T +A
Sbjct: 468 VIHRDVKASNVLLDADFNGRLGDFGLARLYDHGSDPQTTHVVGTLGYLAPEHSRTGRATT 527
Query: 528 ESDIFSFGVVLLEIACGKKAIH-HQELEGEVSLVEWVWELYGLRNLIVAADPKL-CGIFD 585
+D+++FG LLE+ G++ I H + LVEWV+ L+ N++ A DPKL +D
Sbjct: 528 TTDVYAFGAFLLEVVSGRRPIEFHSASDDTFLLVEWVFSLWLRGNIMEAKDPKLGSSGYD 587
Query: 586 VKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLP-ILPLNM 630
++++E +L +GL C++ D +RPS+++V++ L + LP + PL++
Sbjct: 588 LEEVEMVLKLGLLCSHSDPRARPSMRQVLQYLRGDMALPELTPLDL 633
>At3g55550 probable serine/threonine-specific protein kinase
Length = 684
Score = 318 bits (816), Expect = 5e-87
Identities = 218/649 (33%), Positives = 347/649 (52%), Gaps = 64/649 (9%)
Query: 12 TIMLFLIIPLAHAHLVS-----FDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTDDP 66
T + L++ + HLVS F + F L L+G A I + I+LT T
Sbjct: 4 TFAVILLLLIFLTHLVSSLIQDFSFIGFKKASPNLT-LNGVAEIAPT-GAIRLTTETQ-- 59
Query: 67 DKASSVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFSNHSSYGD-GLAFFLASSNLTKA 125
+G I+ N F T F+ + + G GLAF + + +
Sbjct: 60 ---RVIGHAFYSLPIRFKPIGVNRALSFSTSFAIAMVPEFVTLGGHGLAFAITPTPDLRG 116
Query: 126 NRIGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKNIW--DPGFPHVGVNINSVVSDTNI 183
+ GL+ ++ V +S ++ VEFDT +++ D HVG++INS+ S +
Sbjct: 117 SL--PSQYLGLLNSSRVNFSSHFFA---VEFDTVRDLEFEDINDNHVGIDINSMESSIST 171
Query: 184 E---WFSNVSERMVY-------NCSIEYISRNNVLNVSFTGYRLNAWQEPQN--FSHIIN 231
+ +N +++ ++ I+Y S L+V + + ++P+ S+ ++
Sbjct: 172 PAGYFLANSTKKELFLDGGRVIQAWIDYDSNKKRLDVKLSPFS----EKPKLSLLSYDVD 227
Query: 232 LREHLPEYVRVGISASTGKVDEEHMLLSWSFSTSQPSYFVVDP---------RKTKLWEG 282
L L + + VG SASTG + H +L W+F+ S ++ + P +K K
Sbjct: 228 LSSVLGDEMYVGFSASTGLLASSHYILGWNFNMSGEAFSLSLPSLPRIPSSIKKRKKKRQ 287
Query: 283 LAVGGVCLSWSLV--AILI---IFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKIS 337
+ GV L SL+ A+L+ +F+ +K K D+D +E+++ GP + S
Sbjct: 288 SLILGVSLLCSLLIFAVLVAASLFVVRKVK------------DEDRVEEWELDFGPHRFS 335
Query: 338 YYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKII 397
Y EL ATN F + + LG GGFG VYKG S+ A+KRIS +SRQGV+++ +EV I
Sbjct: 336 YRELKKATNGFGDKELLGSGGFGKVYKGKLPGSDEFVAVKRISHESRQGVREFMSEVSSI 395
Query: 398 SQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGS--ILPWNLRYNIALGLAS 455
LRHRNLV+L GWC ++++L+L+Y++MPNGSLD +LF IL W R+ I G+AS
Sbjct: 396 GHLRHRNLVQLLGWCRRRDDLLLVYDFMPNGSLDMYLFDENPEVILTWKQRFKIIKGVAS 455
Query: 456 ALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYL 515
LLYL E WE+ VIHRDIK++N++LDS+ N ++GDFGLA+L +H T V GT GYL
Sbjct: 456 GLLYLHEGWEQTVIHRDIKAANVLLDSEMNGRVGDFGLAKLYEHGSDPGATRVVGTFGYL 515
Query: 516 APEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVA 575
APE + K +D+++FG VLLE+ACG++ I L E+ +V+WVW + ++
Sbjct: 516 APELTKSGKLTTSTDVYAFGAVLLEVACGRRPIETSALPEELVMVDWVWSRWQSGDIRDV 575
Query: 576 ADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLP 624
D +L G FD +++ ++ +GL C+N RP++++V+ L + P P
Sbjct: 576 VDRRLNGEFDEEEVVMVIKLGLLCSNNSPEVRPTMRQVVMYLEKQFPSP 624
>At4g02410 unknown protein
Length = 674
Score = 317 bits (813), Expect = 1e-86
Identities = 219/647 (33%), Positives = 346/647 (52%), Gaps = 50/647 (7%)
Query: 12 TIMLFLIIPLAHAHLVSFDYPMFNHNCKELP----ELDGDATIEDSDNIIQLTGYTDDPD 67
TI F II L+ S F +N P + G AT+ S+ I++LT T
Sbjct: 7 TIFFFFIILLSKPLNSSSQSLNFTYNSFHRPPTNISIQGIATVT-SNGILKLTDKT---- 61
Query: 68 KASSVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFSNHSSY-GDGLAFFLASSNLTKAN 126
S G + I+ + + V F T F I+S + G G+AFF+A + + +
Sbjct: 62 -VISTGHAFYTEPIRFKDSPNDTVSSFSTTFVIGIYSGIPTISGHGMAFFIAPNPVLSSA 120
Query: 127 RIGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKN--IWDPGFPHVGVNINSVVSDTN-- 182
G N N T + I+ VEFDT N D HVG+NINS+ S +
Sbjct: 121 MASQYLGLFSSTNNG---NDTNH-ILAVEFDTIMNPEFDDTNDNHVGININSLTSVKSSL 176
Query: 183 ------IEWFSNVS--ERMVYNCSIEYISRNNVLNVSFTGYRLNAWQEPQNFSHIINLRE 234
I F+N++ R ++Y R N ++V+ + S + +L
Sbjct: 177 VGYWDEINQFNNLTLISRKRMQVWVDYDDRTNQIDVTMAPFG-EVKPRKALVSVVRDLSS 235
Query: 235 HLPEYVRVGISASTGKVDEEHMLLSWSFS-----------TSQPSYFVVDPRKTKLW--E 281
+ + +G SA+TG V EH + WSF + P + V P + +
Sbjct: 236 VFLQDMYLGFSAATGYVLSEHFVFGWSFMVKGKTAPPLTLSKVPKFPRVGPTSLQRFYKN 295
Query: 282 GLAVGGVCLSWSLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYEL 341
+ + + L L + +IFL + + + E +D + EF G ++ + +L
Sbjct: 296 RMPLFSLLLIPVLFVVSLIFLVRFIVRRRRKFAEEF---EDWETEF----GKNRLRFKDL 348
Query: 342 LNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLR 401
AT F++ LG GGFG VY+G + A+KR+S +SRQG+K++ AE+ I ++
Sbjct: 349 YYATKGFKDKDLLGSGGFGRVYRGVMPTTKKEIAVKRVSNESRQGLKEFVAEIVSIGRMS 408
Query: 402 HRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI-LPWNLRYNIALGLASALLYL 460
HRNLV L G+C +++EL+L+Y+YMPNGSLD +L+ + L W R+N+ +G+AS L YL
Sbjct: 409 HRNLVPLLGYCRRRDELLLVYDYMPNGSLDKYLYDCPEVTLDWKQRFNVIIGVASGLFYL 468
Query: 461 QEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYI 520
EEWE+ VIHRDIK+SN++LD+++N +LGDFGLARL DH +TT V GT GYLAP+++
Sbjct: 469 HEEWEQVVIHRDIKASNVLLDAEYNGRLGDFGLARLCDHGSDPQTTRVVGTWGYLAPDHV 528
Query: 521 DTSKARKESDIFSFGVVLLEIACGKKAIHHQ-ELEGEVSLVEWVWELYGLRNLIVAADPK 579
T +A +D+F+FGV+LLE+ACG++ I + E + V LV+ V+ + N++ A DP
Sbjct: 529 RTGRATTATDVFAFGVLLLEVACGRRPIEIEIESDESVLLVDSVFGFWIEGNILDATDPN 588
Query: 580 LCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLPIL 626
L ++D +++E +L +GL C++ D RP++++V++ L +A LP L
Sbjct: 589 LGSVYDQREVETVLKLGLLCSHSDPQVRPTMRQVLQYLRGDATLPDL 635
>At3g53380 receptor lectin kinase -like protein
Length = 715
Score = 317 bits (813), Expect = 1e-86
Identities = 228/655 (34%), Positives = 325/655 (48%), Gaps = 84/655 (12%)
Query: 43 ELDGDATIEDSDNIIQLTGYTDDPDKASSVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTI 102
+L GDA + S+ I+ LT P+ S G+V I+ T+ F + FSF+I
Sbjct: 35 KLLGDARL--SNGIVGLTRDLSVPN--SGAGKVLYSNPIRFRQPGTHFPTSFSSFFSFSI 90
Query: 103 FS-NHSSYGDGLAFFLASSNLTKANRIG-GGGGFGLVPANEVALNSTEYSIVLVEFDTHK 160
+ N SS G GLAF ++ AN IG GG GL N + V VEFDT
Sbjct: 91 TNVNPSSIGGGLAFVISPD----ANSIGIAGGSLGLTGPN-----GSGSKFVAVEFDTLM 141
Query: 161 NI--WDPGFPHVGVNINSVVSDT-------NIE---------WFSNVSERMVYNCSIEYI 202
++ D HVG ++N VVS NI+ W V+N S+ Y
Sbjct: 142 DVDFKDINSNHVGFDVNGVVSSVSGDLGTVNIDLKSGNTINSWIEYDGLTRVFNVSVSYS 201
Query: 203 SR-----------------NNVLNVSFTG--------YRLNAWQEPQNFSHIINLREHLP 237
+ N+ + V F+G + + W +F + P
Sbjct: 202 NLKPKVPILSFPLDLDRYVNDFMFVGFSGSTQGSTEIHSIEWWSFSSSFGSSLGSGSGSP 261
Query: 238 EYVRVGISASTGKVDEEHMLLSWSFSTSQP----------SYFVVDPRKTKLWEGLAVGG 287
++ V L S S++ P S R K G G
Sbjct: 262 PPRANLMNPKANSVKSPPPLASQPSSSAIPISSNTQLKTSSSSSCHSRFCKENPGTIAGV 321
Query: 288 VCLSWSLVAILI--IFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNAT 345
V +A+ +F K K E + S+ + PK+ SY EL T
Sbjct: 322 VTAGAFFLALFAGALFWVYSKKFKRVERSDSFASE--------IIKAPKEFSYKELKAGT 373
Query: 346 NNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNL 405
NF E++ +G G FG VY+G ++ + A+KR S S+ ++ +E+ II LRHRNL
Sbjct: 374 KNFNESRIIGHGAFGVVYRGILPETGDIVAVKRCSHSSQDKKNEFLSELSIIGSLRHRNL 433
Query: 406 VKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSILPWNLRYNIALGLASALLYLQEEWE 465
V+L GWCH+K E++L+Y+ MPNGSLD LF LPW+ R I LG+ASAL YL E E
Sbjct: 434 VRLQGWCHEKGEILLVYDLMPNGSLDKALFESRFTLPWDHRKKILLGVASALAYLHRECE 493
Query: 466 KCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKA 525
VIHRD+KSSNIMLD FN KLGDFGLAR ++H+K E TV AGT GYLAPEY+ T +A
Sbjct: 494 NQVIHRDVKSSNIMLDESFNAKLGDFGLARQIEHDKSPEATVAAGTMGYLAPEYLLTGRA 553
Query: 526 RKESDIFSFGVVLLEIACGKKAI------HHQELEGEVSLVEWVWELYGLRNLIVAADPK 579
+++D+FS+G V+LE+ G++ I + +LVEWVW LY + AAD +
Sbjct: 554 SEKTDVFSYGAVVLEVVSGRRPIEKDLNVQRHNVGVNPNLVEWVWGLYKEGKVSAAADSR 613
Query: 580 LCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLPILPLNMPFLA 634
L G FD ++ +LVVGL C++PD RP+++ V+++L EA +P++P + P ++
Sbjct: 614 LEGKFDEGEMWRVLVVGLACSHPDPAFRPTMRSVVQMLIGEADVPVVPKSRPTMS 668
>At4g02420
Length = 669
Score = 315 bits (808), Expect = 4e-86
Identities = 219/642 (34%), Positives = 348/642 (54%), Gaps = 64/642 (9%)
Query: 24 AHLVSFDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTDDPDKASSVGRVTSPKLIKL 83
+ ++ F Y F ++ L G ATI + +++LT T S G K I+
Sbjct: 23 SQIIDFTYNGFRPPPTDISIL-GIATITPN-GLLKLTNTT-----MQSTGHAFYTKPIRF 75
Query: 84 YNRSTNEVYDFRTKFSFTIFSNHSSYGDGLAFFLASSNLTKANRIGGGGGFGLVPANEVA 143
+ V F T F F I S G+AF +A + R+ G + V
Sbjct: 76 KDSPNGTVSSFSTTFVFAIHSQ-IPIAHGMAFVIAPNP-----RLPFGSPLQYLGLFNVT 129
Query: 144 LN-STEYSIVLVEFDTHKNIW--DPGFPHVGVNINSVVS---------DTNIEWFSNV-- 189
N + + VE DT NI D HVG++INS+ S D N + F N+
Sbjct: 130 NNGNVRNHVFAVELDTIMNIEFNDTNNNHVGIDINSLNSVKSSPAGYWDEN-DQFHNLTL 188
Query: 190 --SERMVYNCSIEYISRNNVLNVSFTGYRLNAWQEPQNFSHIINLREHLPEYVRVGISAS 247
S+RM +++ ++++V+ + ++P S + +L L + + VG S++
Sbjct: 189 ISSKRM--QVWVDFDGPTHLIDVTMAPFGEVKPRKPL-VSIVRDLSSVLLQDMFVGFSSA 245
Query: 248 TGKVDEEHMLLSWSFSTSQ----------PSYFVVDPRKTKL------WEGLAVGGVCLS 291
TG + E +L WSF + P V D + T++ W L + + +
Sbjct: 246 TGNIVSEIFVLGWSFGVNGEAQPLALSKLPRLPVWDLKPTRVYRFYKNWVPL-ISLLLIP 304
Query: 292 WSLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEET 351
+ L+ L+ F+ K+ + +E +D + EF G ++ + +L AT F++
Sbjct: 305 FLLIIFLVRFIMKRRRKFAEEV-------EDWETEF----GKNRLRFKDLYYATKGFKDK 353
Query: 352 QKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGW 411
LG GGFG VYKG + A+KR+S +SRQG+K++ AE+ I Q+ HRNLV L G+
Sbjct: 354 NILGSGGFGSVYKGIMPKTKKEIAVKRVSNESRQGLKEFVAEIVSIGQMSHRNLVPLVGY 413
Query: 412 CHKKNELILIYEYMPNGSLDFHLFRGGSI-LPWNLRYNIALGLASALLYLQEEWEKCVIH 470
C +++EL+L+Y+YMPNGSLD +L+ + L W R+ + G+ASAL YL EEWE+ VIH
Sbjct: 414 CRRRDELLLVYDYMPNGSLDKYLYNSPEVTLDWKQRFKVINGVASALFYLHEEWEQVVIH 473
Query: 471 RDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESD 530
RD+K+SN++LD++ N +LGDFGLA+L DH +TT V GT GYLAP++I T +A +D
Sbjct: 474 RDVKASNVLLDAELNGRLGDFGLAQLCDHGSDPQTTRVVGTWGYLAPDHIRTGRATTTTD 533
Query: 531 IFSFGVVLLEIACGKKAIHHQELEGE-VSLVEWVWELYGLRNLIVAADPKLCGIFDVKQL 589
+F+FGV+LLE+ACG++ I GE V LV+WV+ + N++ A DP L +D K++
Sbjct: 534 VFAFGVLLLEVACGRRPIEINNQSGERVVLVDWVFRFWMEANILDAKDPNLGSEYDQKEV 593
Query: 590 ECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLPIL-PLNM 630
E +L +GL C++ D +RP++++V++ L +A LP L PL++
Sbjct: 594 EMVLKLGLLCSHSDPLARPTMRQVLQYLRGDAMLPDLSPLDL 635
>At2g37710 putative receptor-like protein kinase
Length = 675
Score = 311 bits (798), Expect = 6e-85
Identities = 207/606 (34%), Positives = 323/606 (53%), Gaps = 52/606 (8%)
Query: 54 DNIIQLTGYTDDPDKASSVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFSNHSSY-GDG 112
+ +++LT T G K I+ + V F T F F I S + G G
Sbjct: 50 NGLLKLTNTT-----VQKTGHAFYTKPIRFKDSPNGTVSSFSTSFVFAIHSQIAILSGHG 104
Query: 113 LAFFLA-SSNLTKANRIGGGGGFGLVPANEVALNSTEYSIVLVEFDT--HKNIWDPGFPH 169
+AF +A +++L N G F L N T + + VE DT D H
Sbjct: 105 IAFVVAPNASLPYGNPSQYIGLFNLANNG----NETNH-VFAVELDTILSTEFNDTNDNH 159
Query: 170 VGVNINSVVSDTNIE---W-----FSNVS--ERMVYNCSIEYISRNNVLNVSFTGYRLNA 219
VG++INS+ S + W F N++ R ++Y R N ++V+ + +
Sbjct: 160 VGIDINSLKSVQSSPAGYWDEKGQFKNLTLISRKPMQVWVDYDGRTNKIDVTMAPFNEDK 219
Query: 220 WQEPQNFSHIINLREHLPEYVRVGISASTGKVDEEHMLLSWSFSTSQPSYFVVDPRKTKL 279
P + + +L L + + VG S++TG V EH +L WSF ++ + + R KL
Sbjct: 220 PTRPL-VTAVRDLSSVLLQDMYVGFSSATGSVLSEHYILGWSFGLNEKAPPLALSRLPKL 278
Query: 280 WE------------GLAVGGVCLSWSLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDEF 327
G+ + + L +S + ++ + ++ K E+ +E+
Sbjct: 279 PRFEPKRISEFYKIGMPLISLFLIFSFIFLVCYIVRRRRKFAEEL------------EEW 326
Query: 328 QMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGV 387
+ G + + +L AT F+E LG GGFG VYKG + A+KR+S +SRQG+
Sbjct: 327 EKEFGKNRFRFKDLYYATKGFKEKGLLGTGGFGSVYKGVMPGTKLEIAVKRVSHESRQGM 386
Query: 388 KQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI-LPWNLR 446
K++ AE+ I ++ HRNLV L G+C ++ EL+L+Y+YMPNGSLD +L+ + L W R
Sbjct: 387 KEFVAEIVSIGRMSHRNLVPLLGYCRRRGELLLVYDYMPNGSLDKYLYNTPEVTLNWKQR 446
Query: 447 YNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETT 506
+ LG+AS L YL EEWE+ VIHRD+K+SN++LD + N +LGDFGLARL DH +TT
Sbjct: 447 IKVILGVASGLFYLHEEWEQVVIHRDVKASNVLLDGELNGRLGDFGLARLYDHGSDPQTT 506
Query: 507 VVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIH-HQELEGEVSLVEWVWE 565
V GT GYLAPE+ T +A +D+F+FG LLE+ACG++ I QE + LV+WV+
Sbjct: 507 HVVGTLGYLAPEHTRTGRATMATDVFAFGAFLLEVACGRRPIEFQQETDETFLLVDWVFG 566
Query: 566 LYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLPI 625
L+ +++ A DP + D K++E +L +GL C++ D +RPS+++V+ L +A LP
Sbjct: 567 LWNKGDILAAKDPNMGSECDEKEVEMVLKLGLLCSHSDPRARPSMRQVLHYLRGDAKLPE 626
Query: 626 L-PLNM 630
L PL++
Sbjct: 627 LSPLDL 632
>At5g01560 receptor like protein kinase
Length = 685
Score = 311 bits (796), Expect = 1e-84
Identities = 215/653 (32%), Positives = 335/653 (50%), Gaps = 62/653 (9%)
Query: 13 IMLFLIIPLAHAHLVS------FDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTDDP 66
+ L L++ L AH+ + F + F N E+ + GD+TI S+ +++LT D
Sbjct: 2 VSLLLVLFLVRAHVATTETTTEFIFHGFKGNQSEI-HMQGDSTIT-SNGLLRLTDRNSDV 59
Query: 67 DKASSVGRVTSPKLIKLY--NRSTNEVYDFRTKFSFTIFSNHSSYGDGLAFFLASSNLTK 124
VG K ++L N + V F T F F I S+ +S G G F S
Sbjct: 60 -----VGTAFYHKPVRLLDSNSTNTTVRSFSTSFIFIIPSSSTSNG-GFGFTFTLSPTPN 113
Query: 125 ANRIGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKNIWDPGFP---HVGVNINSVVSDT 181
GL+ NE ++ + VEFDT + D H+G+N NS+ SD
Sbjct: 114 RTDADPEQYMGLL--NERNDGNSSNHVFAVEFDTVQGFKDGTNRIGNHIGLNFNSLSSDV 171
Query: 182 N--IEWFSNVSERMV---------YNCSIEYISRNNVLNVSFTGYRLNAWQE-PQNFSHI 229
+ +F+N + ++Y LN++ RL P +
Sbjct: 172 QEPVAYFNNNDSQKEEFQLVSGEPIQVFLDYHGPTKTLNLTVYPTRLGYKPRIPLISREV 231
Query: 230 INLREHLPEYVRVGISASTGKVDEE--HMLLSWSFSTS--QPSYFVVD--------PRKT 277
L + + + + VG +A+TG+ + H ++ WSF++ P ++D P K
Sbjct: 232 PKLSDIVVDEMFVGFTAATGRHGQSSAHYVMGWSFASGGEHPLAAMLDISQLPPPPPNKA 291
Query: 278 KL--WEGLAVGGVCLSWSLVAILIIFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKK 335
K + G + + ++++I+++ L+ K+ E D ++D P +
Sbjct: 292 KKRGYNGKVIALIVALSTVISIMLVLLFLFMMYKKRMQQEEILEDWEIDH-------PHR 344
Query: 336 ISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVK 395
Y +L AT F+E + +G GGFG VY+G + S+ A+K+I+ +S QGV+++ AE++
Sbjct: 345 FRYRDLYKATEGFKENRVVGTGGFGIVYRGNIRSSSDQIAVKKITPNSMQGVREFVAEIE 404
Query: 396 IISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLF----RGGSILPWNLRYNIAL 451
+ +LRH+NLV L GWC +N+L+LIY+Y+PNGSLD L+ R G++L WN R+ IA
Sbjct: 405 SLGRLRHKNLVNLQGWCKHRNDLLLIYDYIPNGSLDSLLYSKPRRSGAVLSWNARFQIAK 464
Query: 452 GLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGT 511
G+AS LLYL EEWE+ VIHRD+K SN+++DSD N +LGDFGLARL + S TTVV GT
Sbjct: 465 GIASGLLYLHEEWEQIVIHRDVKPSNVLIDSDMNPRLGDFGLARLYERGSQSCTTVVVGT 524
Query: 512 RGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRN 571
GY+APE + SD+F+FGV+LLEI G+K G + +WV EL
Sbjct: 525 IGYMAPELARNGNSSSASDVFAFGVLLLEIVSGRKPTD----SGTFFIADWVMELQASGE 580
Query: 572 LIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLP 624
++ A DP+L +D + L VGL C + SRP ++ V++ LN + +P
Sbjct: 581 ILSAIDPRLGSGYDEGEARLALAVGLLCCHHKPESRPLMRMVLRYLNRDEDVP 633
>At5g59260 receptor-like protein kinase
Length = 674
Score = 306 bits (785), Expect = 2e-83
Identities = 227/661 (34%), Positives = 328/661 (49%), Gaps = 75/661 (11%)
Query: 5 IFCYARGTIMLFLIIPLAHAHLVSFDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTD 64
+F G + FL+ H V +D+ + ELDG A + LT T+
Sbjct: 9 VFWLIIGIHVTFLVFAQEGDHFVYYDFRNADL------ELDGMANTNHGP--LHLTNNTN 60
Query: 65 DPDKASSVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFS-NHSSYGDGLAFFLASSNLT 123
+ G IK + S+ + F T+F F IF S+YG G+AF ++ T
Sbjct: 61 -----TGTGHAFYNIPIK-FTASSLSSFSFSTEFVFAIFPLQKSTYGHGMAFVVSP---T 111
Query: 124 KANRIGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKNI--WDPGFPHVGVNINSVVS-- 179
K R G L N N T I VE DT++N +D G VG++INS+VS
Sbjct: 112 KDLRSNGSANSNLGIFNRANDNKTATHIFAVELDTNQNSESFDKGGNDVGIDINSIVSVE 171
Query: 180 DTNIEWFS-----NVSERMVYNCSI----EYISRNNVLNVSFTGYRLNAWQEPQNFSHI- 229
+ +F+ N+S + SI +Y VLNV+ + P S I
Sbjct: 172 SADASYFNARKGKNISLPLASGKSILVWIDYDGIEKVLNVTLAPVQTPKPDSPYFSSFIK 231
Query: 230 ---------INLREHLPEYVRVGISASTGKVDEEHMLLSWSFST---------SQPSYFV 271
INL E E + VG S STG + +L WSF S+ S
Sbjct: 232 PKVPLLSRSINLSEIFTETMYVGFSGSTGSIKSNQYILGWSFKQGGKAESLDISRLSNPP 291
Query: 272 VDPRKTKLWEGLAVGGVCLSWSLVAIL----IIFLWKKNKGKEDEPTSETTSDQDMDDEF 327
P++ L E L + S +A L I++L+KK K E + +++
Sbjct: 292 PSPKRFPLKEVLGA-----TISTIAFLTLGGIVYLYKKKKYAE------------VLEQW 334
Query: 328 QMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGV 387
+ P++ S+ L AT F E Q LG GGFG VYKG +A +KR+ D+ QG+
Sbjct: 335 EKEYSPQRYSFRILYKATKGFRENQLLGAGGFGKVYKGILPSGTQIA-VKRVYHDAEQGM 393
Query: 388 KQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI--LPWNL 445
KQY AE+ + +LRH+NLV L G+C +K EL+L+Y+YMPNGSLD +LF + L W+
Sbjct: 394 KQYVAEIASMGRLRHKNLVHLLGYCRRKGELLLVYDYMPNGSLDDYLFHKNKLKDLTWSQ 453
Query: 446 RYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSET 505
R NI G+ASALLYL EEWE+ V+HRDIK+SNI+LD+D N KLGDFGLAR D E
Sbjct: 454 RVNIIKGVASALLYLHEEWEQVVLHRDIKASNILLDADLNGKLGDFGLARFHDRGVNLEA 513
Query: 506 TVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWE 565
T V GT GY+APE +D+++FG +LE+ CG++ + +V LV+WV
Sbjct: 514 TRVVGTIGYMAPELTAMGVTTTCTDVYAFGAFILEVVCGRRPVDPDAPREQVILVKWVAS 573
Query: 566 LYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLPI 625
L D KL F V++ + LL +G+ C+ + +RPS++++++ L +P
Sbjct: 574 CGKRDALTDTVDSKLID-FKVEEAKLLLKLGMLCSQINPENRPSMRQILQYLEGNVSVPA 632
Query: 626 L 626
+
Sbjct: 633 I 633
>At5g60270 receptor like protein kinase
Length = 668
Score = 302 bits (773), Expect = 4e-82
Identities = 228/658 (34%), Positives = 335/658 (50%), Gaps = 63/658 (9%)
Query: 11 GTIMLFLIIPLAHAHLVSFDYPMFNHNCKELPELDGDATIEDSDNIIQLTGYTDDPDKAS 70
G I + + + F Y F H + LDG A I S I+QLT T+ S
Sbjct: 8 GIIWMIFCVCSSFQQETPFVYNNFGH--VDHLHLDGSARIIPSGGILQLTNATN-----S 60
Query: 71 SVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFSNHSSYGDGLAFFLASSNLTKANRIGG 130
+G V K I+ ++E F T F + G G+ FF++ S K
Sbjct: 61 QIGHVFYEKPIEF---KSSESVSFSTYFVCALLPAGDPSGHGMTFFVSHSTDFKGAE--A 115
Query: 131 GGGFGLVPANEVALNSTEYSIVLVEFDTH--KNIWDPGFPHVGVNINSVVSDT--NIEWF 186
FG+ N ST ++ VE DT ++ D HVG+++NS S T N +F
Sbjct: 116 TRYFGIFNRN----GSTSTRVLAVELDTSLASDVKDISDNHVGIDVNSAESITSANASYF 171
Query: 187 SNVSERMVYNCSIEYISRNNV----------LNVSFTGYRLNAWQEPQNFSHIINLREHL 236
S+ + + I+ +S + + LNVS R P S INL + L
Sbjct: 172 SDKEGKKI---DIKLLSGDPIQVWVDYEGTTLNVSLAPLRNKKPSRPLLSSTSINLTDIL 228
Query: 237 P-EYVRVGISASTGKVDEEHMLLSWSFSTSQPSYFVVDPRK--------TKLWEGLAVGG 287
+ VG S STG +L WSFS S S +D K TK V
Sbjct: 229 QGRRMFVGFSGSTGSSMSYQYILGWSFSKSMASLPNIDISKLPKVPHSSTKKKSTSPVLS 288
Query: 288 VCL---SWSLVAILII-FLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLN 343
V L ++ ++ IL++ +L+++N E + +E++ GP + SY L
Sbjct: 289 VLLGLIAFIVLGILVVAYLYRRNLYSE------------VREEWEKEYGPIRYSYKSLYK 336
Query: 344 ATNNFEETQKLGQGGFGGVYKGYFKDSNSV--AAIKRISADSRQGVKQYSAEVKIISQLR 401
AT F ++ LG+GGFG VYKG S + A+KR+S D G+KQ+ AE+ + L+
Sbjct: 337 ATKGFNRSEFLGRGGFGEVYKGTLPRSRELREVAVKRVSHDGEHGMKQFVAEIVSMRSLK 396
Query: 402 HRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI-LPWNLRYNIALGLASALLYL 460
HR+LV L G+C +K+EL+L+ EYMPNGSLD +LF + LPW R I +ASAL YL
Sbjct: 397 HRSLVPLLGYCRRKHELLLVSEYMPNGSLDHYLFNHDRLSLPWWRRLAILRDIASALSYL 456
Query: 461 QEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYI 520
E ++ VIHRDIK++N+MLD++FN +LGDFG++RL D TT GT GY+APE +
Sbjct: 457 HTEADQVVIHRDIKAANVMLDAEFNGRLGDFGMSRLYDRGADPSTTAAVGTVGYMAPE-L 515
Query: 521 DTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVAADPKL 580
T A +D+++FGV LLE+ CG++ + E + L++WV E + +LI A DP+L
Sbjct: 516 TTMGASTGTDVYAFGVFLLEVTCGRRPVEPGLPEAKRFLIKWVSECWKRSSLIDARDPRL 575
Query: 581 CGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLPILPLNMPFLASLSP 638
F +++E +L +GL CAN SRP++++V++ LN LP N P + LSP
Sbjct: 576 TE-FSSQEVEKVLKLGLLCANLAPDSRPAMEQVVQYLNGNLALPEFWPNSPGIGVLSP 632
>At3g59700 serine/threonine-specific kinase lecRK1 precursor,lectin
receptor-like
Length = 661
Score = 302 bits (773), Expect = 4e-82
Identities = 195/582 (33%), Positives = 307/582 (52%), Gaps = 47/582 (8%)
Query: 73 GRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFSNHSSYGD-GLAFFLASSNLTKANRIGGG 131
G+ + +++ N ST + F F F I H+ G G+ F ++ + G
Sbjct: 52 GQAFENEHVEIKNSSTGVISSFSVNFFFAIVPEHNQQGSHGMTFVISPTR----GLPGAS 107
Query: 132 GGFGLVPANEVALNSTEYSIVLVEFDTHKN--IWDPGFPHVGVNINSVVS---------- 179
L N+ +++ +E D HK+ D HVG+NIN + S
Sbjct: 108 SDQYLGIFNKTNNGKASNNVIAIELDIHKDEEFGDIDDNHVGININGLRSVASASAGYYD 167
Query: 180 --DTNIEWFSNVSERMVYNCSIEYISRNNVLNVSFTGYRLNAWQEPQNFSHIINLREHLP 237
D + + S +S R V SI Y + LNV+ + S +L +L
Sbjct: 168 DKDGSFKKLSLIS-REVMRLSIVYSQPDQQLNVTLFPAEIPVPPLKPLLSLNRDLSPYLL 226
Query: 238 EYVRVGISASTGKVDEEHMLLSW------SFSTSQPSYFVVDPRKTKLWEGL-AVGGVCL 290
E + +G +ASTG V H L+ W + + S V+ P K V VCL
Sbjct: 227 EKMYLGFTASTGSVGAIHYLMGWLVNGVIEYPRLELSIPVLPPYPKKTSNRTKTVLAVCL 286
Query: 291 SWSLVAILI------IFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNA 344
+ S+ A + +F + K KE + +E+++ GP + +Y EL NA
Sbjct: 287 TVSVFAAFVASWIGFVFYLRHKKVKE------------VLEEWEIQYGPHRFAYKELFNA 334
Query: 345 TNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRN 404
T F+E Q LG+GGFG VYKG S++ A+KR S DSRQG+ ++ AE+ I +LRH N
Sbjct: 335 TKGFKEKQLLGKGGFGQVYKGTLPGSDAEIAVKRTSHDSRQGMSEFLAEISTIGRLRHPN 394
Query: 405 LVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGSI--LPWNLRYNIALGLASALLYLQE 462
LV+L G+C K L L+Y+YMPNGSLD +L R + L W R+ I +A+ALL+L +
Sbjct: 395 LVRLLGYCRHKENLYLVYDYMPNGSLDKYLNRSENQERLTWEQRFRIIKDVATALLHLHQ 454
Query: 463 EWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDT 522
EW + +IHRDIK +N+++D++ N +LGDFGLA+L D ET+ VAGT GY+APE++ T
Sbjct: 455 EWVQVIIHRDIKPANVLIDNEMNARLGDFGLAKLYDQGFDPETSKVAGTFGYIAPEFLRT 514
Query: 523 SKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVAADPKLCG 582
+A +D+++FG+V+LE+ CG++ I + E E LV+W+ EL+ + AA+ +
Sbjct: 515 GRATTSTDVYAFGLVMLEVVCGRRIIERRAAENEEYLVDWILELWENGKIFDAAEESIRQ 574
Query: 583 IFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEAPLP 624
+ Q+E +L +G+ C++ + RP++ V+++LN + LP
Sbjct: 575 EQNRGQVELVLKLGVLCSHQAASIRPAMSVVMRILNGVSQLP 616
>At5g01550 receptor like protein kinase
Length = 688
Score = 297 bits (760), Expect = 1e-80
Identities = 211/623 (33%), Positives = 326/623 (51%), Gaps = 70/623 (11%)
Query: 46 GDATIEDSDNIIQLTGYTDDPDKASSVGRVTSPKLIKLYNRSTNE-VYDFRTKFSFTIFS 104
G ATI+ D +++LT + S + +L++ STN + F T F F I
Sbjct: 42 GAATIK-LDGLLRLTDRNSNVTGTSFYHKPV--RLLETNTSSTNSTIRSFSTSFVFVIIP 98
Query: 105 NHSSYGD-GLAFFLASSNLTKANRIGGGGGFGLVPANEVALNSTEYSIVLVEFDT---HK 160
SS G G F L+ + +R G L N+ ++ + VEFDT K
Sbjct: 99 TSSSNGGFGFTFTLSPT----PDRTGAESAQYLGLLNKANDGNSTNHVFAVEFDTVQGFK 154
Query: 161 NIWDPGFPHVGVNINSVVSDTN--IEWFSNVSERMVYN----------CSIEYISRNNVL 208
+ D H+G+N NS+ SD + ++ N + ++Y L
Sbjct: 155 DGADRTGNHIGLNFNSLTSDVQEPVVYYDNEDPNRKEDFPLQSGDPIRAILDYDGPTQTL 214
Query: 209 NVSFTGYRLNAWQEPQN---FSHIINLREHLPEYVRVGISASTGKVDEE-HMLLSWSFST 264
N+ T Y N P + L + + E + VG +A+TG+ H ++ WSFS+
Sbjct: 215 NL--TVYPANLKSRPVRPLISRPVPKLSQIVQEEMYVGFTAATGRDQSSAHYVMGWSFSS 272
Query: 265 -----SQPSYFVVD----PRKTKLWEG---------LAVGGVCLSWSLVAILIIFLWKKN 306
++ + +++ P T G +A+ GV + L+A+L F+ K
Sbjct: 273 GGDLLTEDTLDLLELPRPPPNTAKKRGYNSQVLALIVALSGVTVI--LLALLFFFVMYKK 330
Query: 307 KGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGY 366
+ ++ E D +++ P ++ Y +L AT+ F+E + +G GGFG V++G
Sbjct: 331 RLQQ----GEVLEDWEINH-------PHRLRYKDLYAATDGFKENRIVGTGGFGTVFRGN 379
Query: 367 FKD-SNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYM 425
S+ A+K+I+ +S QGV+++ AE++ + +LRH+NLV L GWC +KN+L+LIY+Y+
Sbjct: 380 LSSPSSDQIAVKKITPNSMQGVREFIAEIESLGRLRHKNLVNLQGWCKQKNDLLLIYDYI 439
Query: 426 PNGSLDFHLF----RGGSILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLD 481
PNGSLD L+ + G +L WN R+ IA G+AS LLYL EEWEK VIHRDIK SN++++
Sbjct: 440 PNGSLDSLLYSRPRQSGVVLSWNARFKIAKGIASGLLYLHEEWEKVVIHRDIKPSNVLIE 499
Query: 482 SDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEI 541
D N +LGDFGLARL + S TTVV GT GY+APE K+ SD+F+FGV+LLEI
Sbjct: 500 DDMNPRLGDFGLARLYERGSQSNTTVVVGTIGYMAPELARNGKSSSASDVFAFGVLLLEI 559
Query: 542 ACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCAN 601
G++ G L +WV EL+ ++ A DP+L +D + LVVGL C +
Sbjct: 560 VSGRRPTD----SGTFFLADWVMELHARGEILHAVDPRLGFGYDGVEARLALVVGLLCCH 615
Query: 602 PDITSRPSIKKVIKVLNFEAPLP 624
TSRPS++ V++ LN + +P
Sbjct: 616 QRPTSRPSMRTVLRYLNGDDDVP 638
>At1g15530 hypothetical protein
Length = 656
Score = 296 bits (758), Expect = 2e-80
Identities = 204/600 (34%), Positives = 320/600 (53%), Gaps = 50/600 (8%)
Query: 50 IEDSDNIIQLTGYTDDPDKASSVGRVTSPKLIKLY---NRSTNEVYDFRTKFSFTIFSNH 106
IEDS + +D D S GRV P+ + + R+ + F T F F+I +
Sbjct: 49 IEDSRVESTVISLINDSDPLS-FGRVFYPQKLTIIPDPTRNPTRLSSFSTSFVFSILPDI 107
Query: 107 S-SYGDGLAFFLASSNLTKANRIGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKN--IW 163
S S G GL F L SN T FGL V N+ ++ VEFDT +N +
Sbjct: 108 STSPGFGLCFVL--SNSTSPPNAISSQYFGLFTNATVRFNAP---LLAVEFDTGRNSEVN 162
Query: 164 DPGFPHVGVNINSVVSDTNIE--WFSNVSERMV---------YNCSIEYISRNNVLNVSF 212
D HVG+++N++ S T++ ++ +V+ V I++ N +NVS
Sbjct: 163 DIDDNHVGIDLNNIESTTSVTAGYYDSVNGSFVRFNMRNGNNVRAWIDFDGPNFQINVSV 222
Query: 213 TGYRLNAWQEPQNFSHIINLREHLPEYVRVGISASTGKVDEEHMLLSWSFS--------- 263
+ + P + ++ + G SAS +E +L+WS S
Sbjct: 223 APVGVLRPRRPTLTFRDPVIANYVSADMYAGFSASKTNWNEARRILAWSLSDTGALREIN 282
Query: 264 TSQPSYFVVDPRKTKLWEGLAVGGVCLSWSLVAILIIF----LWKKNKGKEDEPTSETTS 319
T+ F ++ + L G A+ G+ + + LI F +WKK +E+E E
Sbjct: 283 TTNLPVFFLENSSSSLSTG-AIAGIVIGCVVFVALIGFGGYLIWKKLMREEEEEEIE--- 338
Query: 320 DQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRI 379
E+++ P + SY EL AT F + LG GGFG VY+G + NS A+K +
Sbjct: 339 ------EWELEFWPHRFSYEELAAATEVFSNDRLLGSGGFGKVYRGILSN-NSEIAVKCV 391
Query: 380 SADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGS 439
+ DS+QG++++ AE+ + +L+H+NLV++ GWC +KNEL+L+Y+YMPNGSL+ +F
Sbjct: 392 NHDSKQGLREFMAEISSMGRLQHKNLVQMRGWCRRKNELMLVYDYMPNGSLNQWIFDNPK 451
Query: 440 I-LPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMD 498
+PW R + +A L YL W++ VIHRDIKSSNI+LDS+ +LGDFGLA+L +
Sbjct: 452 EPMPWRRRRQVINDVAEGLNYLHHGWDQVVIHRDIKSSNILLDSEMRGRLGDFGLAKLYE 511
Query: 499 HEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVS 558
H TT V GT GYLAPE S + SD++SFGVV+LE+ G++ I + E E ++
Sbjct: 512 HGGAPNTTRVVGTLGYLAPELASASAPTEASDVYSFGVVVLEVVSGRRPIEYAE-EEDMV 570
Query: 559 LVEWVWELYGLRNLIVAADPKLCGIFD-VKQLECLLVVGLWCANPDITSRPSIKKVIKVL 617
LV+WV +LYG ++ AAD ++ + ++++E LL +GL C +PD RP++++++ +L
Sbjct: 571 LVDWVRDLYGGGRVVDAADERVRSECETMEEVELLLKLGLACCHPDPAKRPNMREIVSLL 630
>At1g70110 hypothetical protein
Length = 666
Score = 295 bits (755), Expect = 5e-80
Identities = 202/603 (33%), Positives = 323/603 (53%), Gaps = 54/603 (8%)
Query: 52 DSDNIIQLTGYTDDPDKASSVGRVTSPKLIKLYNRSTNEVYDFRTKFSFTIFSNHSSYGD 111
+++ +I+LT T + G+V ++ N V F T F F+I ++ YG
Sbjct: 43 NNNGLIRLTNSTPQ-----TTGQVFYNDQLRFKNSVNGTVSSFSTTFVFSIEFHNGIYGG 97
Query: 112 -GLAFFLASS-NLTKANRIGGGGGFGLVPANEVALNSTEYSIVLVEFDTH--KNIWDPGF 167
G+AF + + +L+ G F N + + IV VE DT + D
Sbjct: 98 YGIAFVICPTRDLSPTFPTTYLGLF-----NRSNMGDPKNHIVAVELDTKVDQQFEDKDA 152
Query: 168 PHVGVNINSVVSDTNI---EWFSNVSERMVYNCS-------IEYISRNNVLNVSFTGYRL 217
HVG++IN++VSDT + N + R + S IEY S+ +NV+ L
Sbjct: 153 NHVGIDINTLVSDTVALAGYYMDNGTFRSLLLNSGQPMQIWIEYDSKQKQINVT-----L 207
Query: 218 NAWQEPQNFSHIINLREHLPEYVR----VGISASTGKVDEEHMLLSWSFSTS------QP 267
+ P+ +++L + L Y+ VG +++TG + H +L W+F + P
Sbjct: 208 HPLYVPKPKIPLLSLEKDLSPYLLELMYVGFTSTTGDLTASHYILGWTFKMNGTTPDIDP 267
Query: 268 SYFVVDPRKTKLWEGLAVGGVCLSWSLVAILIIF-----LWKKNKGKEDEPTSETTSDQD 322
S PR + W G + +S ++ ++I+ LW K K+ E
Sbjct: 268 SRLPKIPRYNQPWIQSPNGILTISLTVSGVIILIILSLSLWLFLKRKKLLEVLE------ 321
Query: 323 MDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRISAD 382
++++ GP + ++ +L AT F++T+ LG+GGFG VYKG SN A+K +S D
Sbjct: 322 ---DWEVQFGPHRFAFKDLHIATKGFKDTEVLGKGGFGKVYKGTLPVSNVEIAVKMVSHD 378
Query: 383 SRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGS-IL 441
SRQG++++ AE+ I +LRH NLV+L G+C K EL L+Y+ M GSLD L+ + L
Sbjct: 379 SRQGMREFIAEIATIGRLRHPNLVRLQGYCRHKGELYLVYDCMAKGSLDKFLYHQQTGNL 438
Query: 442 PWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDHEK 501
W+ R+ I +AS L YL ++W + +IHRDIK +NI+LD++ N KLGDFGLA+L DH
Sbjct: 439 DWSQRFKIIKDVASGLYYLHQQWVQVIIHRDIKPANILLDANMNAKLGDFGLAKLCDHGT 498
Query: 502 GSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSLVE 561
+T+ VAGT GY++PE T KA SD+F+FG+V+LEIACG+K I + + E+ L +
Sbjct: 499 DPQTSHVAGTLGYISPELSRTGKASTRSDVFAFGIVMLEIACGRKPILPRASQREMVLTD 558
Query: 562 WVWELYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNFEA 621
WV E + +++ D K+ + +Q +L +GL+C++P RP++ VI++L+ A
Sbjct: 559 WVLECWENEDIMQVLDHKIGQEYVEEQAALVLKLGLFCSHPVAAIRPNMSSVIQLLDSVA 618
Query: 622 PLP 624
LP
Sbjct: 619 QLP 621
>At3g45410 receptor-like protein kinase
Length = 664
Score = 286 bits (732), Expect = 3e-77
Identities = 189/532 (35%), Positives = 289/532 (53%), Gaps = 48/532 (9%)
Query: 145 NSTEYSIVL-VEFDTHKNIW--DPGFPHVGVNINSVVSDTNI--EWFSNVSERMVYNCSI 199
N T S +L +E DT K + + PHVG+++NS +S + +FSN + N SI
Sbjct: 128 NGTSSSHLLAIELDTVKTVEFNELEKPHVGIDLNSPISVESALPSYFSNALGK---NISI 184
Query: 200 EYISRNNV----------LNVSFTGYRLNAWQEPQNFSHIINLREHLPEYVRVGISASTG 249
+S + LNV+ + +P S INL E E + VG S+STG
Sbjct: 185 NLLSGEPIQVWVDYDGSFLNVTLAPIEIKKPNQPL-ISRAINLSEIFQEKMYVGFSSSTG 243
Query: 250 KVDEEHMLLSWSFS-----------TSQPSYFVVDPRKTKLWEGLAVGGVCLSWSLVAIL 298
+ H +L WSFS ++ P + K KL L +G V L V ++
Sbjct: 244 NLLSNHYILGWSFSRRKEQLQSLNLSTLPRVPLPKEEKKKL-SPLLIGLVILLVIPVVMV 302
Query: 299 I--IFLWKKNKGKEDEPTSETTSDQDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQ 356
+ ++ +++ K E + + ++ GP + SY L ATN F + ++G+
Sbjct: 303 LGGVYWYRRKKYAE------------VKEWWEKEYGPHRFSYKSLYKATNGFRKDCRVGK 350
Query: 357 GGFGGVYKGYFKDSNSVAAIKRISADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKN 416
GGFG VYKG +A +KR+S D+ QG+KQ+ AEV + L+HRNLV L G+C +K
Sbjct: 351 GGFGEVYKGTLPGGRHIA-VKRLSHDAEQGMKQFVAEVVTMGNLQHRNLVPLLGYCRRKC 409
Query: 417 ELILIYEYMPNGSLDFHLFRGGSILP-WNLRYNIALGLASALLYLQEEWEKCVIHRDIKS 475
EL+L+ EYMPNGSLD +LF G+ P W R +I +ASAL YL ++ V+HRDIK+
Sbjct: 410 ELLLVSEYMPNGSLDQYLFHEGNPSPSWYQRISILKDIASALSYLHTGTKQVVLHRDIKA 469
Query: 476 SNIMLDSDFNTKLGDFGLARLMDHEKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFG 535
SN+MLDS+FN +LGDFG+A+ D T GT GY+APE I + K +D+++FG
Sbjct: 470 SNVMLDSEFNGRLGDFGMAKFHDRGTNLSATAAVGTIGYMAPELITMGTSMK-TDVYAFG 528
Query: 536 VVLLEIACGKKAIHHQELEGEVSLVEWVWELYGLRNLIVAADPKLCGIFDVKQLECLLVV 595
LLE+ CG++ + + G+ LV+WV+E + L DP+L F +++E +L +
Sbjct: 529 AFLLEVICGRRPVEPELPVGKQYLVKWVYECWKEACLFKTRDPRLGVEFLPEEVEMVLKL 588
Query: 596 GLWCANPDITSRPSIKKVIKVLNFEAPLPILPLNMPFLASLSPTTNEQFFSV 647
GL C N SRP++++V++ LN + PLPI + P + + P + E ++
Sbjct: 589 GLLCTNAMPESRPAMEQVVQYLNQDLPLPIFSPSTPGIGAFMPVSMEALSAI 640
>At5g60300 receptor like protein kinase
Length = 718
Score = 285 bits (729), Expect = 6e-77
Identities = 196/545 (35%), Positives = 292/545 (52%), Gaps = 53/545 (9%)
Query: 110 GDGLAFFLASS-NLTKANRIGGGGGFGLVPANEVALNSTEYSIVLVEFDTHKNIWDPGFP 168
G G+ F L+ S + T A G F N S+ Y ++ VE DT IW+P F
Sbjct: 98 GHGIVFVLSPSMDFTHAESTRYLGIF-----NASTNGSSSYHVLAVELDT---IWNPDFK 149
Query: 169 -----HVGVNINSVVSDTNIEWFSNVSERMVYNCSIEYISRNNV-LNVSFTGYRLNAWQE 222
HVG+++NS +S I S S+ N SI +S N + + V + G LN
Sbjct: 150 DIDHNHVGIDVNSPIS-VAIASASYYSDMKGSNESINLLSGNPIQVWVDYEGTLLNVSVA 208
Query: 223 PQN--------FSHIINLREHLPEYVRV--GISASTGKVDEEHMLLSWSFSTSQPSYFVV 272
P SH INL E P + G SA+TG + +L WSFS + S +
Sbjct: 209 PLEVQKPTRPLLSHPINLTELFPNRSSLFAGFSAATGTAISDQYILWWSFSIDRGSLQRL 268
Query: 273 DPRKT-----------KLWEGLAVGGVCLSWSLVAILI-IFLWKKNKGKEDEPTSETTSD 320
D K K+ + + VCL+ ++A+L ++ ++ K E T E D
Sbjct: 269 DISKLPEVPHPRAPHKKVSTLIILLPVCLAILVLAVLAGLYFRRRRKYSEVSETWEKEFD 328
Query: 321 QDMDDEFQMGAGPKKISYYELLNATNNFEETQKLGQGGFGGVYKGYFKDSNSVAAIKRIS 380
+ SY L AT F + + LG+GGFG VY+G +A +KR+S
Sbjct: 329 AH------------RFSYRSLFKATKGFSKDEFLGKGGFGEVYRGNLPQGREIA-VKRVS 375
Query: 381 ADSRQGVKQYSAEVKIISQLRHRNLVKLTGWCHKKNELILIYEYMPNGSLDFHLFRGGS- 439
+ +GVKQ+ AEV + L+HRNLV L G+C +K EL+L+ EYMPNGSLD HLF
Sbjct: 376 HNGDEGVKQFVAEVVSMRCLKHRNLVPLFGYCRRKRELLLVSEYMPNGSLDEHLFDDQKP 435
Query: 440 ILPWNLRYNIALGLASALLYLQEEWEKCVIHRDIKSSNIMLDSDFNTKLGDFGLARLMDH 499
+L W+ R + G+ASAL YL ++ V+HRD+K+SNIMLD++F+ +LGDFG+AR +H
Sbjct: 436 VLSWSQRLVVVKGIASALWYLHTGADQVVLHRDVKASNIMLDAEFHGRLGDFGMARFHEH 495
Query: 500 EKGSETTVVAGTRGYLAPEYIDTSKARKESDIFSFGVVLLEIACGKKAIHHQELEGEVSL 559
+ TT GT GY+APE I T A +D+++FGV +LE+ CG++ + Q + +
Sbjct: 496 GGNAATTAAVGTVGYMAPELI-TMGASTGTDVYAFGVFMLEVTCGRRPVEPQLQVEKRHM 554
Query: 560 VEWVWELYGLRNLIVAADPKLCGIFDVKQLECLLVVGLWCANPDITSRPSIKKVIKVLNF 619
++WV E + +L+ A DP+L G F +++E ++ +GL C+N SRP++++V+ LN
Sbjct: 555 IKWVCECWKKDSLLDATDPRLGGKFVAEEVEMVMKLGLLCSNIVPESRPTMEQVVLYLNK 614
Query: 620 EAPLP 624
PLP
Sbjct: 615 NLPLP 619
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.319 0.137 0.416
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 15,668,011
Number of Sequences: 26719
Number of extensions: 714437
Number of successful extensions: 5363
Number of sequences better than 10.0: 1011
Number of HSP's better than 10.0 without gapping: 858
Number of HSP's successfully gapped in prelim test: 153
Number of HSP's that attempted gapping in prelim test: 1830
Number of HSP's gapped (non-prelim): 1193
length of query: 655
length of database: 11,318,596
effective HSP length: 106
effective length of query: 549
effective length of database: 8,486,382
effective search space: 4659023718
effective search space used: 4659023718
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144564.8


