

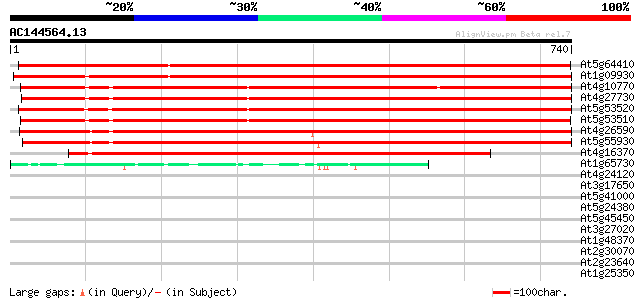
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144564.13 - phase: 0
(740 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g64410 Isp4-like protein 1241 0.0
At1g09930 unknown protein 1131 0.0
At4g10770 oligopeptide transporter like protein 940 0.0
At4g27730 unknown protein 889 0.0
At5g53520 isp4 protein 884 0.0
At5g53510 isp4 protein 868 0.0
At4g26590 isp4 like protein 858 0.0
At5g55930 sexual differentiation process protein ISP4-like 842 0.0
At4g16370 isp4 like protein 620 e-177
At1g65730 unknown protein 46 7e-05
At4g24120 unknown protein 42 0.001
At3g17650 unknown protein 35 0.15
At5g41000 unknown protein 34 0.26
At5g24380 putative protein 33 0.44
At5g45450 putative protein 33 0.75
At3g27020 unknown protein 32 0.98
At1g48370 unknown protein 32 0.98
At2g30070 high affinity K+ transporter (AtKUP1/AtKT1p) 32 1.3
At2g23640 putative seed maturation protein 30 3.7
At1g25350 unknown protein (At1g25350) 30 3.7
>At5g64410 Isp4-like protein
Length = 729
Score = 1241 bits (3212), Expect = 0.0
Identities = 575/728 (78%), Positives = 647/728 (87%), Gaps = 2/728 (0%)
Query: 12 TMKEEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPL 71
T E +E+ SPIEEVRLTV TDDPTLPVWTFRMWFLGL+SC+LLSFLNQFF+YRTEPL
Sbjct: 3 TADEFSDEDTSPIEEVRLTVTNTDDPTLPVWTFRMWFLGLISCSLLSFLNQFFSYRTEPL 62
Query: 72 IITQITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGA 131
+ITQITVQVATLP+GH +A VLP F +PG GS RFS NPGPFNMKEHVLI+IFANAG+
Sbjct: 63 VITQITVQVATLPIGHFLAKVLPKTRFGLPGCGSARFSLNPGPFNMKEHVLISIFANAGS 122
Query: 132 AFGSGSSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWP 191
AFGSGS+YAVGI+ IIKAFYGR+ISF+A WLLIITTQVLGYGWAGLLRKYVVEPAHMWWP
Sbjct: 123 AFGSGSAYAVGIITIIKAFYGRSISFIAGWLLIITTQVLGYGWAGLLRKYVVEPAHMWWP 182
Query: 192 GTLVQVSLFRTLHEKDDNPHQFSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVCWVFS 251
TLVQVSLFR LHEKDD + +RAKFF IALVCSF WYIVPGYLFTTLTSISWVCW F
Sbjct: 183 STLVQVSLFRALHEKDDQ--RMTRAKFFVIALVCSFGWYIVPGYLFTTLTSISWVCWAFP 240
Query: 252 KSVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIA 311
+SVTAQQIGSGM GLGLGA TLDW+AVASFLFSPLISPFFAI NVF+GY LL+Y V+P+A
Sbjct: 241 RSVTAQQIGSGMRGLGLGAFTLDWTAVASFLFSPLISPFFAIANVFIGYVLLIYFVLPLA 300
Query: 312 YWGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKYHEQGRIHLSTFFALSY 371
YWGF+ Y A RF IFSS L+T+ G Y+I IVND FE+DLAKY +QGRI+LS FFAL+Y
Sbjct: 301 YWGFDSYNATRFPIFSSHLFTSVGNTYDIPAIVNDNFELDLAKYEQQGRINLSMFFALTY 360
Query: 372 GFGFATIASTVTHVACFYGREIMERYRASKNGKEDIHTKLMKNYKDIPSWWFYLLLGVTF 431
G GFATIAST+THVA FYG+EI ER+R S GKEDIHT+LMK YKDIPSWWFY +L T
Sbjct: 361 GLGFATIASTLTHVALFYGKEISERFRVSYKGKEDIHTRLMKRYKDIPSWWFYSMLAATL 420
Query: 432 VVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEYIFGIIY 491
++SL +C+FLND++QMPWWGL+ ASA+AF+FTLPISIITATTNQTPGLNIITEY G+IY
Sbjct: 421 LISLALCVFLNDEVQMPWWGLVFASAMAFVFTLPISIITATTNQTPGLNIITEYAMGLIY 480
Query: 492 PGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVA 551
PGRPIANVCFK YGY+SMAQAVSFL+DFKLGHYMKIPPRSMFLVQFIGT+LAGTINI VA
Sbjct: 481 PGRPIANVCFKVYGYMSMAQAVSFLNDFKLGHYMKIPPRSMFLVQFIGTILAGTINITVA 540
Query: 552 WWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFL 611
WW L+S+KNIC ++LLP SPWTCP DRVFFDASV+WGLVGPKRIFGS G Y+ +NWFFL
Sbjct: 541 WWQLNSIKNICQEELLPPNSPWTCPGDRVFFDASVIWGLVGPKRIFGSQGNYAAMNWFFL 600
Query: 612 GGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIF 671
GGA+GP++VW LHKAFPK+SWIPL+NLPVLLGAT MMPPATA+NYNSWI+VGTIFN F+F
Sbjct: 601 GGALGPVIVWSLHKAFPKRSWIPLVNLPVLLGATAMMPPATAVNYNSWILVGTIFNLFVF 660
Query: 672 RYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLNWWGTAGEHCPLAACPTAKG 731
RYRK WWQRYNYVLSAA+DAGVAFMAVLLY ++G+E SL+WWGT GEHC LA CPTA+G
Sbjct: 661 RYRKSWWQRYNYVLSAAMDAGVAFMAVLLYFSVGMEEKSLDWWGTRGEHCDLAKCPTARG 720
Query: 732 IVVDGCPV 739
++VDGCPV
Sbjct: 721 VIVDGCPV 728
>At1g09930 unknown protein
Length = 734
Score = 1131 bits (2925), Expect = 0.0
Identities = 522/736 (70%), Positives = 623/736 (83%), Gaps = 6/736 (0%)
Query: 6 VDTEKGTMKEEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFA 65
++ K + +++++ESP+E+VRLTV DDP+LPVWTFRMWFLGLLSC LLSFLN FF
Sbjct: 4 IELHKPEINADDDDDESPVEQVRLTVSNHDDPSLPVWTFRMWFLGLLSCILLSFLNTFFG 63
Query: 66 YRTEPLIITQITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITI 125
YRT+PL+IT I+VQV TLPLG LMA VLP ++I GS FSFNPGPFN+KEHVLI++
Sbjct: 64 YRTQPLMITMISVQVVTLPLGKLMARVLPETKYKI---GSWEFSFNPGPFNVKEHVLISM 120
Query: 126 FANAGAAFGSGSSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEP 185
FANAGA FGSG++YAVGIV+II AFY R ISFLA+W+L+ITTQ+LGYGWAG++RK VV+P
Sbjct: 121 FANAGAGFGSGTAYAVGIVDIIMAFYKRKISFLASWILVITTQILGYGWAGIMRKLVVDP 180
Query: 186 AHMWWPGTLVQVSLFRTLHEKDDNPHQFSRAKFFFIALVCSFTWYIVPGYLFTTLTSISW 245
A MWWP +++QVSLFR LHEKD+ + SR KFF IA VCSF WYI P YLF TL+SISW
Sbjct: 181 AQMWWPTSVLQVSLFRALHEKDNA--RMSRGKFFVIAFVCSFAWYIFPAYLFLTLSSISW 238
Query: 246 VCWVFSKSVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVY 305
VCW F KS+TAQQ+GSGM+GLG+GA LDWS +AS+L SPL++PFFAIVNV VGY L++Y
Sbjct: 239 VCWAFPKSITAQQLGSGMSGLGIGAFALDWSVIASYLGSPLVTPFFAIVNVLVGYVLVMY 298
Query: 306 AVIPIAYWGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKYHEQGRIHLST 365
VIPI+YWG NVY AN+F IFSSDL+ QGQ YNIS IVN+KFE+D+ Y +QGR++LST
Sbjct: 299 MVIPISYWGMNVYEANKFPIFSSDLFDKQGQLYNISTIVNNKFELDMENYQQQGRVYLST 358
Query: 366 FFALSYGFGFATIASTVTHVACFYGREIMERYRASKNGKEDIHTKLMKNYKDIPSWWFYL 425
FFA+SYG GFA I ST+THVA F G+ I ++ RAS K DIHT+LMK YKDIP WWFY
Sbjct: 359 FFAISYGIGFAAIVSTLTHVALFNGKGIWQQVRASTKAKMDIHTRLMKKYKDIPGWWFYS 418
Query: 426 LLGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEY 485
LL ++ V+SL++CIF+ D+IQMPWWGLL+AS +A FT+P+SIITATTNQTPGLNIITEY
Sbjct: 419 LLAISLVLSLVLCIFMKDEIQMPWWGLLLASFMALTFTVPVSIITATTNQTPGLNIITEY 478
Query: 486 IFGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGT 545
+ G++ PGRPIANVCFKTYGYISM+QA+SFL+DFKLGHYMKIPPRSMFLVQFIGTV+AGT
Sbjct: 479 LMGVLLPGRPIANVCFKTYGYISMSQAISFLNDFKLGHYMKIPPRSMFLVQFIGTVIAGT 538
Query: 546 INIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYST 605
+NI VAW+LL SV+NIC K+LLP SPWTCPSDRVFFDASV+WGLVGPKRIFG LG Y
Sbjct: 539 VNISVAWYLLTSVENICQKELLPPNSPWTCPSDRVFFDASVIWGLVGPKRIFGRLGNYPA 598
Query: 606 LNWFFLGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTI 665
LNWFFLGG IGP+LVWLL KAFP ++WI INLPVLLGAT MPPAT++N+N WIIVG I
Sbjct: 599 LNWFFLGGLIGPVLVWLLQKAFPTKTWISQINLPVLLGATAAMPPATSVNFNCWIIVGVI 658
Query: 666 FNFFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLN-WWGTAGEHCPLA 724
FN+F+F+Y KKWWQRYNYVLSAALDAG+AFM VLLY +L + +S+N WWG GE+CPLA
Sbjct: 659 FNYFVFKYCKKWWQRYNYVLSAALDAGLAFMGVLLYFSLTMNGISINHWWGAKGENCPLA 718
Query: 725 ACPTAKGIVVDGCPVF 740
+CPTA G++VDGCPVF
Sbjct: 719 SCPTAPGVLVDGCPVF 734
>At4g10770 oligopeptide transporter like protein
Length = 766
Score = 940 bits (2429), Expect = 0.0
Identities = 436/729 (59%), Positives = 559/729 (75%), Gaps = 14/729 (1%)
Query: 15 EEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLIIT 74
EEEEEE SPI +V LTV TDDP+LPV TFRMW LG LSC LLSFLNQFF YRTEPL I+
Sbjct: 48 EEEEEENSPIRQVALTVPTTDDPSLPVLTFRMWVLGTLSCILLSFLNQFFWYRTEPLTIS 107
Query: 75 QITVQVATLPLGHLMASVLPSKTFRIPGFGSK-RFSFNPGPFNMKEHVLITIFANAGAAF 133
I+ Q+A +PLG LMA+ + + F GSK +F+ NPGPFN+KEHVLITIFANAGA
Sbjct: 108 AISAQIAVVPLGRLMAAKITDRVFFQ---GSKWQFTLNPGPFNVKEHVLITIFANAGA-- 162
Query: 134 GSGSSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGT 193
GS YA+ +V ++KAFY +NI+F ++++I+TTQVLG+GWAG+ RKY+VEPA MWWP
Sbjct: 163 --GSVYAIHVVTVVKAFYMKNITFFVSFIVIVTTQVLGFGWAGIFRKYLVEPAAMWWPAN 220
Query: 194 LVQVSLFRTLHEKDDNPHQ-FSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVCWVFSK 252
LVQVSLFR LHEK++ +R +FF IA VCSF +Y+ PGYLF +TS+SWVCW F
Sbjct: 221 LVQVSLFRALHEKEERTKGGLTRTQFFVIAFVCSFAYYVFPGYLFQIMTSLSWVCWFFPS 280
Query: 253 SVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIAY 312
SV AQQIGSG++GLG+GA+ LDWS ++S+L SPL SP+FA NV VG+ L++Y ++PI Y
Sbjct: 281 SVMAQQIGSGLHGLGVGAIGLDWSTISSYLGSPLASPWFATANVGVGFVLVIYVLVPICY 340
Query: 313 WGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKYHEQGRIHLSTFFALSYG 372
W +VY A F IFSS L+++QG YNI++I++ F +DL Y QG ++L TFFA+SYG
Sbjct: 341 W-LDVYKAKTFPIFSSSLFSSQGSKYNITSIIDSNFHLDLPAYERQGPLYLCTFFAISYG 399
Query: 373 FGFATIASTVTHVACFYGREIMERYRAS-KNGKEDIHTKLMKNYKDIPSWWFYLLLGVTF 431
GFA +++T+ HVA F+GREI E+ + S K K D+H +LM+ YK +P WWF+ +L
Sbjct: 400 VGFAALSATIMHVALFHGREIWEQSKESFKEKKLDVHARLMQRYKQVPEWWFWCILVTNV 459
Query: 432 VVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEYIFGIIY 491
++ C + NDQ+Q+PWWG+L+A +A IFTLPI IITA TNQ PGLNIITEYI G IY
Sbjct: 460 GATIFACEYYNDQLQLPWWGVLLACTVAIIFTLPIGIITAITNQAPGLNIITEYIIGYIY 519
Query: 492 PGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVA 551
PG P+AN+CFK YGYISM QA++FL DFKLGHYMKIPPR+MF+ Q +GT+++ + + A
Sbjct: 520 PGYPVANMCFKVYGYISMQQAITFLQDFKLGHYMKIPPRTMFMAQIVGTLISCFVYLTTA 579
Query: 552 WWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFL 611
WWL++++ NIC+ S WTCPSD+VF+DASV+WGL+GP+RIFG LG Y ++NWFFL
Sbjct: 580 WWLMETIPNICDS---VTNSVWTCPSDKVFYDASVIWGLIGPRRIFGDLGLYKSVNWFFL 636
Query: 612 GGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIF 671
GAI PILVWL + FP+Q WI LIN+PVL+ AT MPPATA+NY +W++ G + F +F
Sbjct: 637 VGAIAPILVWLASRMFPRQEWIKLINMPVLISATSSMPPATAVNYTTWVLAGFLSGFVVF 696
Query: 672 RYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLNWWGTAGEHCPLAACPTAKG 731
RYR WQRYNYVLS ALDAG+AFM VLLY+ LGLENVSL+WWG + CPLA+CPTA G
Sbjct: 697 RYRPNLWQRYNYVLSGALDAGLAFMGVLLYMCLGLENVSLDWWGNELDGCPLASCPTAPG 756
Query: 732 IVVDGCPVF 740
I+V+GCP++
Sbjct: 757 IIVEGCPLY 765
>At4g27730 unknown protein
Length = 736
Score = 889 bits (2296), Expect = 0.0
Identities = 415/732 (56%), Positives = 544/732 (73%), Gaps = 15/732 (2%)
Query: 16 EEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLIITQ 75
+++++ + EV LTV KTDD TLPV TFRMW LG+ +C +LSF+NQFF YRT PL IT
Sbjct: 13 DDDDDRCVVPEVELTVPKTDDSTLPVLTFRMWVLGIGACIVLSFINQFFWYRTMPLSITG 72
Query: 76 ITVQVATLPLGHLMASVLPSKTFRIPGFGSK-RFSFNPGPFNMKEHVLITIFANAGAAFG 134
I+ Q+A +PLGHLMA VLP+K F G++ +F+ NPG FN+KEHVLITIFAN+GA
Sbjct: 73 ISAQIAVVPLGHLMARVLPTKRFLE---GTRFQFTLNPGAFNVKEHVLITIFANSGA--- 126
Query: 135 SGSSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGTL 194
GS YA I++ IK +Y R++ FL A+L++ITTQ+LG+GWAGL RK++VEP MWWP L
Sbjct: 127 -GSVYATHILSAIKLYYKRSLPFLPAFLVMITTQILGFGWAGLFRKHLVEPGEMWWPSNL 185
Query: 195 VQVSLFRTLHEKDDNPHQ-FSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVCWVFSKS 253
VQVSLF LHEK+ SR +FF I LV SF +YI PGYLFT LTSISWVCW+ KS
Sbjct: 186 VQVSLFGALHEKEKKSRGGMSRTQFFLIVLVASFAYYIFPGYLFTMLTSISWVCWLNPKS 245
Query: 254 VTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIAYW 313
+ Q+GSG +GLG+G++ DW ++++L SPL SP FA VNV +G+ L++Y V P+ YW
Sbjct: 246 ILVNQLGSGEHGLGIGSIGFDWVTISAYLGSPLASPLFASVNVAIGFVLVMYIVTPVCYW 305
Query: 314 GFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKYHEQGRIHLSTFFALSYGF 373
N+Y A F IFSS L+ G Y++ +I++ KF +D Y G I++STFFA++YG
Sbjct: 306 -LNIYDAKTFPIFSSQLFMGNGSRYDVLSIIDSKFHLDRVVYSRTGSINMSTFFAVTYGL 364
Query: 374 GFATIASTVTHVACFYGREIMERYRAS--KNGKEDIHTKLMK-NYKDIPSWWFYLLLGVT 430
GFAT+++T+ HV F G ++ ++ R + KN K DIHT++MK NY+++P WWF ++L +
Sbjct: 365 GFATLSATIVHVLVFNGSDLWKQTRGAFQKNKKMDIHTRIMKKNYREVPLWWFLVILLLN 424
Query: 431 FVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEYIFGII 490
+ + I + N +Q+PWWG+L+A A+A FT I +I ATTNQ PGLNIITEY+ G I
Sbjct: 425 IALIMFISVHYNATVQLPWWGVLLACAIAISFTPLIGVIAATTNQAPGLNIITEYVIGYI 484
Query: 491 YPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGV 550
YP RP+AN+CFK YGYISM QA++F+SDFKLGHYMKIPPRSMF+ Q GT++A + G
Sbjct: 485 YPERPVANMCFKVYGYISMTQALTFISDFKLGHYMKIPPRSMFMAQVAGTLVAVVVYTGT 544
Query: 551 AWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFF 610
AWWL++ + ++C+ LLP S WTCP DRVFFDASV+WGLVGP+R+FG LGEYS +NWFF
Sbjct: 545 AWWLMEEIPHLCDTSLLPSDSQWTCPMDRVFFDASVIWGLVGPRRVFGDLGEYSNVNWFF 604
Query: 611 LGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFI 670
L GAI P+LVWL K FP Q+WI I++PVL+GAT MMPPATA+N+ SW+IV IF FI
Sbjct: 605 LVGAIAPLLVWLATKMFPAQTWIAKIHIPVLVGATAMMPPATAVNFTSWLIVAFIFGHFI 664
Query: 671 FRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLNWWGTAGEH--CPLAACPT 728
F+YR+ WW +YNYVLS LDAG AFM +LL+LALG + + + WWG +G+ CPLA+CPT
Sbjct: 665 FKYRRVWWTKYNYVLSGGLDAGSAFMTILLFLALGRKGIEVQWWGNSGDRDTCPLASCPT 724
Query: 729 AKGIVVDGCPVF 740
AKG+VV GCPVF
Sbjct: 725 AKGVVVKGCPVF 736
>At5g53520 isp4 protein
Length = 733
Score = 884 bits (2283), Expect = 0.0
Identities = 405/736 (55%), Positives = 545/736 (74%), Gaps = 16/736 (2%)
Query: 12 TMKEEE-EEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEP 70
T+ E E ++E S + +V LTV KTDDPT P TFRMW LG+ +C LLSFLNQFF YRT P
Sbjct: 7 TISESECDDEISIVPQVELTVPKTDDPTSPTVTFRMWVLGITACVLLSFLNQFFWYRTNP 66
Query: 71 LIITQITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSF--NPGPFNMKEHVLITIFAN 128
L I+ ++ Q+A +P+GHLMA VLP++ F F R+SF NPGPF+ KEHVLIT+FAN
Sbjct: 67 LTISSVSAQIAVVPIGHLMAKVLPTRRF----FEGTRWSFTMNPGPFSTKEHVLITVFAN 122
Query: 129 AGAAFGSGSSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHM 188
+G SG+ YA I++ +K +Y R + FL A L++ITTQVLG+GWAGL RK++VEP M
Sbjct: 123 SG----SGAVYATHILSAVKLYYKRRLDFLPALLVMITTQVLGFGWAGLYRKHLVEPGEM 178
Query: 189 WWPGTLVQVSLFRTLHEKDDNPHQ-FSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVC 247
WWP LVQVSLFR LHEK++ SR +FF I L+ SF++Y++PGYLFT LT++SW+C
Sbjct: 179 WWPSNLVQVSLFRALHEKENKSKWGISRNQFFVITLITSFSYYLLPGYLFTVLTTVSWLC 238
Query: 248 WVFSKSVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAV 307
W+ KS+ Q+GSG GLG+G+ LDWS +AS+L SPL SPFFA N+ G+ L++Y +
Sbjct: 239 WISPKSILVNQLGSGSAGLGIGSFGLDWSTIASYLGSPLASPFFASANIAAGFFLVMYVI 298
Query: 308 IPIAYWGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKYHEQGRIHLSTFF 367
P+ Y+ ++Y A F I+S L+ A G+ Y +++I++ F +D Y E G +H+STFF
Sbjct: 299 TPLCYY-LDLYNAKTFPIYSGKLFVASGKEYKVTSIIDANFRLDRQAYAETGPVHMSTFF 357
Query: 368 ALSYGFGFATIASTVTHVACFYGREIMERYRAS--KNGKEDIHTKLMK-NYKDIPSWWFY 424
A++YG GFAT+++++ HV F G+++ + R + KN K DIHTK+MK NYK++P WWF
Sbjct: 358 AVTYGLGFATLSASIFHVLIFNGKDLWTQTRGAFGKNKKMDIHTKIMKRNYKEVPLWWFL 417
Query: 425 LLLGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITE 484
+ V V + ICI+ QIQ+PWWG +A +A FT + +I ATTNQ PGLNIITE
Sbjct: 418 SIFAVNLAVIVFICIYYKTQIQLPWWGAFLACLIAIFFTPLVGVIMATTNQAPGLNIITE 477
Query: 485 YIFGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAG 544
YI G YP RP+AN+CFKTYGYISM+Q+++FLSD KLG YMKIPPR+MF+ Q +GT++A
Sbjct: 478 YIIGYAYPERPVANICFKTYGYISMSQSLTFLSDLKLGTYMKIPPRTMFMAQVVGTLVAV 537
Query: 545 TINIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYS 604
G AWWL+ + N+C+ +LLP GS WTCPSDRVFFDASV+WGLVGP+R+FG LGEYS
Sbjct: 538 IAYAGTAWWLMAEIPNLCDTNLLPPGSQWTCPSDRVFFDASVIWGLVGPRRMFGDLGEYS 597
Query: 605 TLNWFFLGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGT 664
+NWFF+GGAI P LV+L + FP + WI I++PVL+GAT +MPPA+A+N+ SW+++
Sbjct: 598 NINWFFVGGAIAPALVYLASRLFPNKKWISDIHIPVLIGATAIMPPASAVNFTSWLVMAF 657
Query: 665 IFNFFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLNWWGTAGEHCPLA 724
+F F+F+YR++WWQRYNYVLS +DAG FM+VLL+LAL ++++WWG +GE CP+A
Sbjct: 658 VFGHFVFKYRREWWQRYNYVLSGGMDAGTGFMSVLLFLALQRSEIAIDWWGNSGEGCPVA 717
Query: 725 ACPTAKGIVVDGCPVF 740
CPTAKG+VV GCPVF
Sbjct: 718 KCPTAKGVVVHGCPVF 733
>At5g53510 isp4 protein
Length = 741
Score = 868 bits (2242), Expect = 0.0
Identities = 399/732 (54%), Positives = 540/732 (73%), Gaps = 15/732 (2%)
Query: 15 EEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLIIT 74
+ ++ + +EEV LTV KTDDPTLPV TFRMW LGL +C +LSF+NQFF YR PL I+
Sbjct: 17 QNDDLDRCVVEEVELTVPKTDDPTLPVLTFRMWTLGLGACIILSFINQFFWYRQMPLTIS 76
Query: 75 QITVQVATLPLGHLMASVLPSKTFRIPGFGSK-RFSFNPGPFNMKEHVLITIFANAGAAF 133
I+ Q+A +PLGHLMA VLP++ F GSK FS NPGPFN+KEHVLITIFAN+GA
Sbjct: 77 GISAQIAVVPLGHLMAKVLPTRMFLE---GSKWEFSMNPGPFNVKEHVLITIFANSGA-- 131
Query: 134 GSGSSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGT 193
G+ YA I++ IK +Y R++ FL A+LL+ITTQ LG+GWAGL RK++VEP MWWP
Sbjct: 132 --GTVYATHILSAIKLYYKRSLPFLPAFLLMITTQFLGFGWAGLFRKHLVEPGEMWWPSN 189
Query: 194 LVQVSLFRTLHEKDDNPHQ-FSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVCWVFSK 252
LVQVSLF LHEK+ +R +FF I LV SF +YI+PGYLFT +TSISW+CW+ K
Sbjct: 190 LVQVSLFSALHEKEKKKKGGMTRIQFFLIVLVTSFAYYILPGYLFTMITSISWICWLGPK 249
Query: 253 SVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIAY 312
SV Q+GSG GLG+GA+ +DW+ ++S+L SPL SP FA +NV +G+ +++Y PI Y
Sbjct: 250 SVLVHQLGSGEQGLGIGAIGIDWATISSYLGSPLASPLFATINVTIGFVVIMYVATPICY 309
Query: 313 WGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKYHEQGRIHLSTFFALSYG 372
W N+Y A + IFSS L+ G Y++ +I++ KF +D Y + G I++STFFA++YG
Sbjct: 310 W-LNIYKAKTYPIFSSGLFMGNGSSYDVLSIIDKKFHLDRDIYAKTGPINMSTFFAVTYG 368
Query: 373 FGFATIASTVTHVACFYGREIMERYRAS--KNGKEDIHTKLMK-NYKDIPSWWFYLLLGV 429
GFAT+++T+ HV F GR++ ++ R + +N K D HT++MK NY+++P WWFY++L +
Sbjct: 369 LGFATLSATIVHVLLFNGRDLWKQTRGAFQRNKKMDFHTRIMKKNYREVPMWWFYVILVL 428
Query: 430 TFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEYIFGI 489
+ + I + N +Q+PWWG+L+A A+A FT I +I ATTNQ PGLN+ITEY+ G
Sbjct: 429 NIALIMFISFYYNATVQLPWWGVLLACAIAVFFTPLIGVIAATTNQEPGLNVITEYVIGY 488
Query: 490 IYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIG 549
+YP RP+AN+CFK YGYISM QA++F+ DFKLG YMKIPPRSMF+ Q +GT+++ + G
Sbjct: 489 LYPERPVANMCFKVYGYISMTQALTFIQDFKLGLYMKIPPRSMFMAQVVGTLVSVVVYTG 548
Query: 550 VAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWF 609
AWWL+ + ++C+K LLP S WTCP DRVFFDASV+WGLVGP+R+FG+LGEY+ +NWF
Sbjct: 549 TAWWLMVDIPHLCDKSLLPPDSEWTCPMDRVFFDASVIWGLVGPRRMFGNLGEYAAINWF 608
Query: 610 FLGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFF 669
FL GAI P VWL KAFP WI I+ PV+LGAT MMPPA A+N+ SW IV +F F
Sbjct: 609 FLVGAIAPFFVWLATKAFPAHKWISKIHFPVILGATSMMPPAMAVNFTSWCIVAFVFGHF 668
Query: 670 IFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSLNWWGTAGE--HCPLAACP 727
+++Y+++WW++YNYVLS LDAG AFM +L++L++G + + L WWG A + +C LA+CP
Sbjct: 669 LYKYKRQWWKKYNYVLSGGLDAGTAFMTILIFLSVGRKGIGLLWWGNADDSTNCSLASCP 728
Query: 728 TAKGIVVDGCPV 739
TAKG+++ GCPV
Sbjct: 729 TAKGVIMHGCPV 740
>At4g26590 isp4 like protein
Length = 753
Score = 858 bits (2218), Expect = 0.0
Identities = 404/736 (54%), Positives = 533/736 (71%), Gaps = 15/736 (2%)
Query: 14 KEEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLII 73
+EEE+E +SPIEEVRLTV TDDP+LPV TFR WFLG++SC +L+F+N FF YR+ PL +
Sbjct: 24 EEEEDENDSPIEEVRLTVPITDDPSLPVLTFRTWFLGMVSCVVLAFVNNFFGYRSNPLTV 83
Query: 74 TQITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGAAF 133
+ + Q+ TLPLG LMA+ LP+ R+PG S NPGPFNMKEHVLITIFAN GA
Sbjct: 84 SSVVAQIITLPLGKLMATTLPTTKLRLPGTNWS-CSLNPGPFNMKEHVLITIFANTGA-- 140
Query: 134 GSGSSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGT 193
G +YA I+ I+KAFY RN++ AA LL+ TTQ+LGYGWAG+ RKY+V+ +MWWP
Sbjct: 141 --GGAYATSILTIVKAFYHRNLNPAAAMLLVQTTQLLGYGWAGMFRKYLVDSPYMWWPAN 198
Query: 194 LVQVSLFRTLHEKDDNPH-QFSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVCWVFSK 252
LVQVSLFR LHEK++ + ++ +FF I SFT+YIVPGYLF +++ +S+VCW++++
Sbjct: 199 LVQVSLFRALHEKEEKREGKQTKLRFFLIVFFLSFTYYIVPGYLFPSISYLSFVCWIWTR 258
Query: 253 SVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIAY 312
SVTAQQIGSG++GLG+G+ LDWS VA FL SPL PFFAI N F G+ + Y ++PI Y
Sbjct: 259 SVTAQQIGSGLHGLGIGSFGLDWSTVAGFLGSPLAVPFFAIANSFGGFIIFFYIILPIFY 318
Query: 313 WGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDK-FEIDLAKYHEQGRIHLSTFFALSY 371
W N Y A +F ++S + GQ YN + I+N K F IDL Y +++LS FAL Y
Sbjct: 319 WS-NAYEAKKFPFYTSHPFDHTGQRYNTTRILNQKTFNIDLPAYESYSKLYLSILFALIY 377
Query: 372 GFGFATIASTVTHVACFYGREIMERYR----ASKNGKEDIHTKLMK-NYKDIPSWWFYLL 426
G F T+ +T++HVA F G+ I E ++ +K+ D+HT+LMK NYK++P WWF +
Sbjct: 378 GLSFGTLTATISHVALFDGKFIWELWKKATLTTKDKFGDVHTRLMKKNYKEVPQWWFVAV 437
Query: 427 LGVTFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEYI 486
L +FV++L C Q+Q+PWWGLL+A A+AF FTLPI +I ATTNQ GLN+I+E I
Sbjct: 438 LAASFVLALYACEGFGKQLQLPWWGLLLACAIAFTFTLPIGVILATTNQRMGLNVISELI 497
Query: 487 FGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTI 546
G +YPG+P+ANV FKTYG +S+AQA+ F+ DFKLGHYMKIPPRSMF+VQ + T++A T+
Sbjct: 498 IGFLYPGKPLANVAFKTYGSVSIAQALYFVGDFKLGHYMKIPPRSMFIVQLVATIVASTV 557
Query: 547 NIGVAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTL 606
+ G WWLL SV+NICN D+LPK SPWTCP D VF++AS++WG++GP R+F S G Y +
Sbjct: 558 SFGTTWWLLSSVENICNTDMLPKSSPWTCPGDVVFYNASIIWGIIGPGRMFTSKGIYPGM 617
Query: 607 NWFFLGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIF 666
NWFFL G + P+ VW + FP++ WI I++P++ +MP A A++Y SW VG +F
Sbjct: 618 NWFFLIGFLAPVPVWFFARKFPEKKWIHQIHIPLIFSGANVMPMAKAVHYWSWFAVGIVF 677
Query: 667 NFFIFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSL-NWWGTAG-EHCPLA 724
N++IFR K WW R+NY+LSAALDAG A M VL+Y AL N+SL +WWG +HCPLA
Sbjct: 678 NYYIFRRYKGWWARHNYILSAALDAGTAVMGVLIYFALQNNNISLPDWWGNENTDHCPLA 737
Query: 725 ACPTAKGIVVDGCPVF 740
CPT KGIV GCPVF
Sbjct: 738 NCPTEKGIVAKGCPVF 753
>At5g55930 sexual differentiation process protein ISP4-like
Length = 755
Score = 842 bits (2174), Expect = 0.0
Identities = 397/733 (54%), Positives = 524/733 (71%), Gaps = 15/733 (2%)
Query: 17 EEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYRTEPLIITQI 76
EEE ++PIEEVRLTV TDDPTLPV TFR W LGL SC LL+F+NQFF +R+ L ++ +
Sbjct: 29 EEENDNPIEEVRLTVPITDDPTLPVLTFRTWTLGLFSCILLAFVNQFFGFRSNQLWVSSV 88
Query: 77 TVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGAAFGSG 136
Q+ TLPLG LMA LP+K F PG +SFNPGPFNMKEHVLITIFAN GA G
Sbjct: 89 AAQIVTLPLGKLMAKTLPTKKFGFPGTNWS-WSFNPGPFNMKEHVLITIFANTGA----G 143
Query: 137 SSYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGTLVQ 196
YA I+ I+KAFY R ++ AA LL TTQ+LGYGWAG+ RK++V+ +MWWP LVQ
Sbjct: 144 GVYATSIITIVKAFYNRQLNVAAAMLLTQTTQLLGYGWAGIFRKFLVDSPYMWWPSNLVQ 203
Query: 197 VSLFRTLHEKDD-NPHQFSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVCWVFSKSVT 255
VSLFR LHEK+D Q +R +FF I SF +YI+PGYLF ++++IS+VCW++ SVT
Sbjct: 204 VSLFRALHEKEDLQKGQQTRFRFFIIVFCVSFAYYIIPGYLFPSISAISFVCWIWKSSVT 263
Query: 256 AQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIAYWGF 315
AQ +GSG+ GLG+G+ LDWS VA FL SPL PFFAI N F G+ + +Y V+PI YW
Sbjct: 264 AQIVGSGLKGLGIGSFGLDWSTVAGFLGSPLAVPFFAIANFFGGFFIFLYIVLPIFYWT- 322
Query: 316 NVYGANRFSIFSSDLYTAQGQPYNISNIVNDK-FEIDLAKYHEQGRIHLSTFFALSYGFG 374
N Y A +F ++S + G YNI+ I+N+K F+I+L Y+ +++LS FAL YG
Sbjct: 323 NAYDAQKFPFYTSHTFDQTGHTYNITRILNEKNFDINLDAYNGYSKLYLSVMFALLYGLS 382
Query: 375 FATIASTVTHVACFYGREIMERYRASKNGKED----IHTKLMK-NYKDIPSWWFYLLLGV 429
F ++ +T++HVA + G+ I ++ +K +D +H++LMK NY+ +P WWF +L +
Sbjct: 383 FGSLCATISHVALYDGKFIWGMWKKAKTATKDKYGDVHSRLMKKNYQSVPQWWFIAVLVI 442
Query: 430 TFVVSLMICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEYIFGI 489
+F +L C + Q+Q+PWWGL++A A+A FTLPI +I ATTNQ GLN+ITE I G
Sbjct: 443 SFAFALYACEGFDKQLQLPWWGLILACAIALFFTLPIGVIQATTNQQMGLNVITELIIGY 502
Query: 490 IYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIG 549
+YPG+P+ANV FKTYGYISM+QA+ F+ DFKLGHYMKIPPRSMF+VQ + TV+A T+ G
Sbjct: 503 LYPGKPLANVAFKTYGYISMSQALYFVGDFKLGHYMKIPPRSMFIVQLVATVVASTVCFG 562
Query: 550 VAWWLLDSVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWF 609
WWL+ SV+NICN DLLP GSPWTCP D VF++AS++WG++GP R+F G Y +NWF
Sbjct: 563 TTWWLITSVENICNVDLLPVGSPWTCPGDEVFYNASIIWGVIGPGRMFTKEGIYPGMNWF 622
Query: 610 FLGGAIGPILVWLLHKAFPKQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFF 669
FL G + P+ W L K FP++ W+ I++P++ A MP A A++Y SW IVG +FN++
Sbjct: 623 FLIGLLAPVPFWYLSKKFPEKKWLKQIHVPLIFSAVSAMPQAKAVHYWSWAIVGVVFNYY 682
Query: 670 IFRYRKKWWQRYNYVLSAALDAGVAFMAVLLYLALGLENVSL-NWWGTA-GEHCPLAACP 727
IFR K WW R+NY+LSAALDAG A M VL++ A ++SL +WWG +HCPLA CP
Sbjct: 683 IFRRFKTWWARHNYILSAALDAGTAIMGVLIFFAFQNNDISLPDWWGLENSDHCPLAHCP 742
Query: 728 TAKGIVVDGCPVF 740
AKG+VV+GCPVF
Sbjct: 743 LAKGVVVEGCPVF 755
>At4g16370 isp4 like protein
Length = 637
Score = 620 bits (1598), Expect = e-177
Identities = 281/558 (50%), Positives = 397/558 (70%), Gaps = 4/558 (0%)
Query: 78 VQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITIFANAGAAFGSGS 137
+Q+A LP+G MA LP+ + + G+ FS NPGPFN+KEHV+ITIFAN G A+G G
Sbjct: 1 MQIAGLPIGKFMARTLPTTSHNLLGWS---FSLNPGPFNIKEHVIITIFANCGVAYGGGD 57
Query: 138 SYAVGIVNIIKAFYGRNISFLAAWLLIITTQVLGYGWAGLLRKYVVEPAHMWWPGTLVQV 197
+Y++G + ++KA+Y +++SF+ +++TTQ+LGYGWAG+LR+Y+V+P MWWP L QV
Sbjct: 58 AYSIGAITVMKAYYKQSLSFICGLFIVLTTQILGYGWAGILRRYLVDPVDMWWPSNLAQV 117
Query: 198 SLFRTLHEKDDNPHQFSRAKFFFIALVCSFTWYIVPGYLFTTLTSISWVCWVFSKSVTAQ 257
SLFR LHEK++ +R KFF +AL SF +Y +PGYLF LT SWVCW + S+TAQ
Sbjct: 118 SLFRALHEKENKSKGLTRMKFFLVALGASFIYYALPGYLFPILTFSSWVCWAWPNSITAQ 177
Query: 258 QIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVNVFVGYALLVYAVIPIAYWGFNV 317
Q+GSG +GLG+GA TLDW+ ++++ SPL++P+ +I+NV VG+ + +Y ++P+ YW FN
Sbjct: 178 QVGSGYHGLGVGAFTLDWAGISAYHGSPLVAPWSSILNVGVGFIMFIYIIVPVCYWKFNT 237
Query: 318 YGANRFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKYHEQGRIHLSTFFALSYGFGFAT 377
+ A +F I S+ L+T GQ Y+ + I+ +F++D+ Y+ G+++LS FALS G GFA
Sbjct: 238 FDARKFPISSNQLFTTSGQKYDTTKILTPQFDLDIGAYNNYGKLYLSPLFALSIGSGFAR 297
Query: 378 IASTVTHVACFYGREIMER-YRASKNGKEDIHTKLMKNYKDIPSWWFYLLLGVTFVVSLM 436
+T+THVA F GR+I ++ + A K DIH KLM++YK +P WWFY+LL + +SL+
Sbjct: 298 FTATLTHVALFNGRDIWKQTWSAVNTTKLDIHGKLMQSYKKVPEWWFYILLAGSVAMSLL 357
Query: 437 ICIFLNDQIQMPWWGLLIASALAFIFTLPISIITATTNQTPGLNIITEYIFGIIYPGRPI 496
+ + +Q+PWWG+L A ALAFI TLPI +I ATTNQ PG +II ++I G I PG+PI
Sbjct: 358 MSFVWKESVQLPWWGMLFAFALAFIVTLPIGVIQATTNQQPGYDIIGQFIIGYILPGKPI 417
Query: 497 ANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLD 556
AN+ FK YG IS A+SFL+D KLGHYMKIPP M+ Q +GTV+AG +N+GVAWW+L+
Sbjct: 418 ANLIFKIYGRISTVHALSFLADLKLGHYMKIPPPCMYTAQLVGTVVAGVVNLGVAWWMLE 477
Query: 557 SVKNICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIG 616
S+++IC+ + SPWTCP RV FDASV+WGL+GP+R+FG G Y L FFL GA+
Sbjct: 478 SIQDICDIEGDHPNSPWTCPKYRVTFDASVIWGLIGPRRLFGPGGMYRNLVGFFLIGAVL 537
Query: 617 PILVWLLHKAFPKQSWIP 634
P+ + PKQ P
Sbjct: 538 PVPRVGAEQDLPKQEVDP 555
>At1g65730 unknown protein
Length = 688
Score = 46.2 bits (108), Expect = 7e-05
Identities = 118/589 (20%), Positives = 216/589 (36%), Gaps = 103/589 (17%)
Query: 1 MENINVDTEKGTMKEEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFL 60
+E D + + EEEE +E R+ + + P P W ++ F L+ +L+ L
Sbjct: 3 VERSKKDDDLNNGSKSNEEEEISVE--RIFEESNEIP--PPWQKQLTFRALIVSFILAIL 58
Query: 61 NQFFAYRTEPLIITQITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEH 120
F ++ ++ + +P ++ A +L + + F PF +E+
Sbjct: 59 FTF--------VVMKLNLTTGIIPSLNISAGLLGFFFVKSWTKILNKAGFLKQPFTRQEN 110
Query: 121 VLIT--IFANAGAAFGSG-SSYAVGIVNIIKA----------FYGRNISFLAAWLLIITT 167
+I + A++G AF G SY G+ +++ ++ ++ +L +++
Sbjct: 111 TVIQTCVVASSGIAFSGGFGSYLFGMSDVVAKQSAEANTPLNIKNPHLGWMIGFLFVVS- 169
Query: 168 QVLGYGWAGLLRKYVVEPAHMWWPGTLVQVSLFRTLHEKDDNPHQFSRAKFFFIALVCSF 227
LG LRK ++ + +P L + H P AK AL F
Sbjct: 170 -FLGLFSVVPLRKIMIVDFKLTYPSGTATAHLINSFH----TPQGAKLAKKQVRALGKFF 224
Query: 228 TWYIVPGYLFTTLTSISWVCWVFSKSVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLI 287
++ + G+ W F+ GL + A+++ +I
Sbjct: 225 SFSFLWGFFQ----------WFFATGDGCGFANFPTFGLKAYENKFYFDFSATYVGVGMI 274
Query: 288 SPFFAIVNVFVGYALLVYAVIPIAYWGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDK 347
P+ V++ +G A+L + V+ + + GA + +++DL +
Sbjct: 275 CPYLINVSLLIG-AILSWGVM------WPLIGAQKGKWYAADLSSTS------------- 314
Query: 348 FEIDLAKYHEQGRIHLSTFFALSYGFGFATIASTVTHVACFYGREIMERYRASKNGKEDI 407
H + A+ G G + GR + Y+ KN +D+
Sbjct: 315 -------LHGLQGYRVFIAIAMILGDGLYNFIKVL-------GRTVFGLYKQFKN--KDV 358
Query: 408 -----HTKLMK---NYKD-----------IPSWWFYLLLGVTFVVSLMICIFLNDQIQMP 448
HT +Y D IPSW+ V +VS++ + Q++
Sbjct: 359 LPINDHTSTAPVTISYDDKRRTELFLKDRIPSWFAVTGYVVLAIVSIITVPHIFHQLK-- 416
Query: 449 WWGLLI----ASALAFIFTLPISIITATTNQTPG-LNIITEYIFGIIYPGRPIANVCFKT 503
W+ +LI A LAF + + T G L I T + G +A +
Sbjct: 417 WYHILIMYIIAPVLAFCNAYGCGLTDWSLASTYGKLAIFTIGAWAGASNGGVLAGLAACG 476
Query: 504 YGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAW 552
++ A + DFK G+ PRSMFL Q IGT + I+ V W
Sbjct: 477 VMMNIVSTASDLMQDFKTGYMTLASPRSMFLSQAIGTAMGCVISPCVFW 525
>At4g24120 unknown protein
Length = 665
Score = 42.0 bits (97), Expect = 0.001
Identities = 128/591 (21%), Positives = 225/591 (37%), Gaps = 105/591 (17%)
Query: 13 MKEEEEEEE-----SPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAYR 67
MK E EEEE S EE T ++ T+ WT ++ G+ ++ + A +
Sbjct: 1 MKREGEEEEDNNQLSLQEEEPDTEEEMSGRTIEPWTKQITVRGVFVSIVIGVVFSVIAQK 60
Query: 68 TEPL--IITQITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLITI 125
I+ + A L + +K + GF +K PF +E+ +I
Sbjct: 61 LNLTTGIVPNLNSSAALLAFVFVQTW---TKILKKSGFVAK-------PFTRQENTMIQT 110
Query: 126 FANA--GAAFGSG-SSYAVGIVNIIKAFYGRNIS-------------FLAAWLLIITTQV 169
A A G A G G +SY +G+ + G N+ ++ A+L ++
Sbjct: 111 SAVACYGIAVGGGFASYLLGLNHKTYVLSGVNLEGNSPKSVKEPGLGWMTAYLFVVC--F 168
Query: 170 LGYGWAGLLRKYVVEPAHMWWPGTLVQVSLFRTLHEKDDNPHQFSRAKFFFIALVCSFTW 229
+G LRK ++ + +P L L H + D + + + F SF W
Sbjct: 169 IGLFVLIPLRKVMIVDLKLTYPSGLATAVLINGFHTQGDAQAK-KQVRGFMKYFSFSFLW 227
Query: 230 YIVPGYLFTTLTSISWVCWVFSKSVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISP 289
+ W FS GL T + +F+ + +I
Sbjct: 228 --------------GFFQWFFSGIEDCGFAQFPTFGLKAWKQTFFFDFSMTFVGAGMICS 273
Query: 290 FFAIVNVFVGYALLVYAVIPIAYWGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFE 349
+++ +G A+L Y ++ + + + S F +L +N+ +I K
Sbjct: 274 HLVNLSLLLG-AILSYGLM------WPLLDKLKGSWFPDNL-----DEHNMKSIYGYKVF 321
Query: 350 IDLAKYHEQGRIHLSTFFALSYGFGFATIASTVTHVACFYGREIMERYRASKNGKEDI-H 408
+ +A G L TF + F TIA+ + R + N +D+ H
Sbjct: 322 LSVALILGDG---LYTFVKIL----FVTIAN------------VNARLKNKPNDLDDVGH 362
Query: 409 TKLMKNYKDIPSW-------WFYLLLGVTFV-VSLMICIFLNDQIQMPWWGLLIASALAF 460
K K+ K+ ++ WF + +TF VS ++ + Q++ W+ +++A +
Sbjct: 363 KKQRKDLKEDENFLRDKIPMWFAVSGYLTFAAVSTVVVPLIFPQLK--WYYVIVA----Y 416
Query: 461 IFTLPISIITATTNQTPGLNIITEY----IFGII-YPGRPIANVC-FKTYGYISMAQAVS 514
IF ++ A +N+ Y +F I GR V G I +VS
Sbjct: 417 IFAPSLAFCNAYGAGLTDINMAYNYGKIGLFVIAAVTGRENGVVAGLAGCGLIKSVVSVS 476
Query: 515 --FLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICN 563
+ DFK HY P++MF Q IGTV+ G I ++++L +I N
Sbjct: 477 CILMQDFKTAHYTMTSPKAMFASQMIGTVV-GCIVTPLSFFLFYKAFDIGN 526
>At3g17650 unknown protein
Length = 714
Score = 35.0 bits (79), Expect = 0.15
Identities = 108/576 (18%), Positives = 198/576 (33%), Gaps = 82/576 (14%)
Query: 7 DTEKGTMKEEEEEEESPIEEVRLTVKKTDDPTLPVWTFRMWFLGLLSCALLSFLNQFFAY 66
+TEK K EE+EE E V K + +P W ++ + +LS L F
Sbjct: 27 ETEKVKNKNFEEDEEEEDESVE---KIFESREVPSWKKQLTVRAFVVSFMLSILFSF--- 80
Query: 67 RTEPLIITQITVQVATLPLGHLMASVLPSKTFRIPGFGSKRFSFNPGPFNMKEHVLIT-- 124
I+ ++ + +P ++ A +L + R PF +E+ +I
Sbjct: 81 -----IVMKLNLTTGIIPSLNVSAGLLGFFFVKTWTKMLHRSGLLKQPFTRQENTVIQTC 135
Query: 125 IFANAGAAFGSG-SSYAVGIVNIIKAFYGR--------NISFLAAWLLIITTQVLGYGWA 175
+ A++G AF G +Y G+ I G ++ ++ +L +++ LG
Sbjct: 136 VVASSGIAFSGGFGTYLFGMSERIATQSGDVSRGVKDPSLGWIIGFLFVVS--FLGLFSV 193
Query: 176 GLLRKYVVEPAHMWWPGTLVQVSLFRTLHEKDDNPHQFSRAKFFFIALVCSFTWYIVPGY 235
LRK +V + +P L + H + + SF W
Sbjct: 194 VPLRKIMVIDFKLTYPSGTATAHLINSFHTPQGAKLAKKQVRVLGKFFSLSFFW------ 247
Query: 236 LFTTLTSISWVCWVFSKSVTAQQIGSGMNGLGLGALTLDWSAVASFLFSPLISPFFAIVN 295
S+ W F+ GL + A+++ +I P+ ++
Sbjct: 248 --------SFFQWFFTGGENCGFSNFPTFGLKAYQYKFYFDFSATYVGVGMICPYIINIS 299
Query: 296 VFVGYALLVYAVIPIAYWGFNVYGANRFSIFSSDLYTAQGQPYNISNIVNDKFEIDLAKY 355
V +G L + P+ D + ++ + K I +A
Sbjct: 300 VLLGGILSWGIMWPL------------IETKKGDWFPDNVPSSSMHGLQAYKVFIAVAII 347
Query: 356 HEQGRIHLSTFFALSYGFGFATIASTVTHVACFYGREIMERYRASKNGKEDIHTKLM--- 412
G + + + F + T ++ R S +ED H +
Sbjct: 348 LGDGLYNFCKVLSRTLSGLFVQLRGPTTSIS-----------RTSFTLEEDPHASPLSPK 396
Query: 413 KNYKD-----------IPSWWFYLLLGVTFVVSLMICIFLNDQIQMPWWGLLI----ASA 457
++Y D IP+W + + G + + I + Q+ W+ +L+ A
Sbjct: 397 QSYDDQRRTRFFLKDQIPTW--FAVGGYITIAATSTAILPHMFHQLRWYYILVIYICAPV 454
Query: 458 LAFIFTLPISIITATTNQTPG-LNIITEYIFGIIYPGRPIANVCFKTYGYISMAQAVSFL 516
LAF + + T G L I T + G +A + ++ A
Sbjct: 455 LAFCNAYGAGLTDWSLASTYGKLAIFTIGAWAGSEHGGMLAGLAACGVMMNIVSTASDLT 514
Query: 517 SDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAW 552
DFK G+ P+SMF+ Q IGT + ++ V W
Sbjct: 515 QDFKTGYLTLSSPKSMFVSQVIGTAMGCVVSPCVFW 550
>At5g41000 unknown protein
Length = 670
Score = 34.3 bits (77), Expect = 0.26
Identities = 44/184 (23%), Positives = 79/184 (42%), Gaps = 18/184 (9%)
Query: 509 MAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLP 568
++ A + DFK G+ +SMF+ Q +GT + G I + +WL + +I + D L
Sbjct: 478 VSTAADLMQDFKTGYLTLSSAKSMFVTQLLGTAM-GCIIAPLTFWLFWTAFDIGDPDGLY 536
Query: 569 KGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHKAFP 628
K +P+ V + + G+ G F L ++ G I ++V L+ P
Sbjct: 537 K-APYA-----VIYREMAILGVEG----FAKLPKHCLA--LCCGFFIAALIVNLIRDMTP 584
Query: 629 KQSWIPLINLPVLLGATGMMPPATALNYNSWIIVGTIFNFFIFRYRKKWWQRYNYVLSAA 688
+ LI LP+ + + A++ + VGT+ R KK Y+ +++
Sbjct: 585 PKI-SKLIPLPMAMAGPFYIGAYFAID----MFVGTVIMLVWERMNKKDADDYSGAVASG 639
Query: 689 LDAG 692
L G
Sbjct: 640 LICG 643
>At5g24380 putative protein
Length = 652
Score = 33.5 bits (75), Expect = 0.44
Identities = 34/137 (24%), Positives = 63/137 (45%), Gaps = 14/137 (10%)
Query: 512 AVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVKNICNKDLLPKGS 571
+ + DFK GH + PRSM + Q IGT + G + + ++L ++ N++
Sbjct: 462 SADLMHDFKTGHLTQTSPRSMLVAQAIGTAI-GCVVAPLTFFLFYKAFDVGNQN-----G 515
Query: 572 PWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPILVWLLHKAFPKQS 631
+ P ++ + +++ G+ GP + E L + F A+ L L P +
Sbjct: 516 EYKAPYAMIYRNMAII-GVQGPSALPKHCLE---LCYGFFAFAVAANLARDLLPDKPGK- 570
Query: 632 WIPL---INLPVLLGAT 645
WIPL + +P L+G +
Sbjct: 571 WIPLPMAMAVPFLVGGS 587
>At5g45450 putative protein
Length = 216
Score = 32.7 bits (73), Expect = 0.75
Identities = 15/44 (34%), Positives = 24/44 (54%)
Query: 509 MAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAW 552
+++A DFK G+ P+SMF+ Q IGT + ++ V W
Sbjct: 15 VSRASDLTQDFKTGYLTLSSPKSMFVSQVIGTAMGCVVSPCVFW 58
>At3g27020 unknown protein
Length = 676
Score = 32.3 bits (72), Expect = 0.98
Identities = 52/210 (24%), Positives = 92/210 (43%), Gaps = 28/210 (13%)
Query: 447 MPWWGLL----IASALAFIFTLPISIIT---ATTNQTPGLNIITEYIFGIIYPGRPIANV 499
+ W+ +L IA ALAF + + A+T GL II + G IA +
Sbjct: 415 LKWYFVLCSYFIAPALAFCNSYGTGLTDWSLASTYGKIGLFIIASVVGS---DGGVIAGL 471
Query: 500 CFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSMFLVQFIGTVLAGTINIGVAWWLLDSVK 559
++ A + DFK G+ +SMF+ Q +GT + G + + +WL +
Sbjct: 472 AACGVMMSIVSTAADLMQDFKTGYLTLSSAKSMFVSQLVGTAM-GCVIAPLTFWLFWTAF 530
Query: 560 NICNKDLLPKGSPWTCPSDRVFFDASVVWGLVGPKRIFGSLGEYSTLNWFFLGGAIGPIL 619
+I + P G P+ P +F + +++ G+ G F L ++ + G I ++
Sbjct: 531 DIGD----PNG-PYKAPYAVIFREMAIL-GIEG----FAELPKHCLALCY--GFFIAALI 578
Query: 620 VWLLHKAFPKQ--SWIPL---INLPVLLGA 644
V LL P + +IP+ + +P +GA
Sbjct: 579 VNLLRDITPPKISQFIPIPMAMAVPFYIGA 608
>At1g48370 unknown protein
Length = 724
Score = 32.3 bits (72), Expect = 0.98
Identities = 34/140 (24%), Positives = 58/140 (41%), Gaps = 7/140 (5%)
Query: 418 IPSWWFYLLLGVTFVVSLMICIFLNDQIQMPWWGLLI----ASALAFIFTLPISIITATT 473
IPSW + + G + ++ I + Q+ W+ +++ A LAF + +
Sbjct: 423 IPSW--FAVGGYVVISAVSTAILPHMFSQLRWYYIIVIYIFAPILAFCNAYGAGLTDWSL 480
Query: 474 NQTPG-LNIITEYIFGIIYPGRPIANVCFKTYGYISMAQAVSFLSDFKLGHYMKIPPRSM 532
T G L I T + G +A + ++ A DFK G+ PR+M
Sbjct: 481 ASTYGKLAIFTIGAWAGSDHGGLLAGLAACGVMMNIVSTASDLTQDFKTGYLTLSSPRAM 540
Query: 533 FLVQFIGTVLAGTINIGVAW 552
F+ Q IGT + ++ V W
Sbjct: 541 FVSQVIGTAMGCLVSPCVFW 560
>At2g30070 high affinity K+ transporter (AtKUP1/AtKT1p)
Length = 712
Score = 32.0 bits (71), Expect = 1.3
Identities = 22/93 (23%), Positives = 43/93 (45%), Gaps = 8/93 (8%)
Query: 580 VFFDASVVWGLVGPKRIFGSLGEYSTLNWF-FLGGAIGPILVWLLHKAFPKQSWIPLINL 638
+F S W L S+G Y+T+ W + A+ P+ ++ ++ + W+ L
Sbjct: 223 IFAPISTAWLLS-----ISSIGVYNTIKWNPRIVSALSPVYMYKFLRSTGVEGWVSLGG- 276
Query: 639 PVLLGATGMMPPATALNYNSWIIVGTIFNFFIF 671
V+L TG+ L + S + + F+FF++
Sbjct: 277 -VVLSITGVETMFADLGHFSSLSIKVAFSFFVY 308
>At2g23640 putative seed maturation protein
Length = 206
Score = 30.4 bits (67), Expect = 3.7
Identities = 12/28 (42%), Positives = 18/28 (63%)
Query: 219 FFIALVCSFTWYIVPGYLFTTLTSISWV 246
F LV + TW ++ Y FTT+T +SW+
Sbjct: 29 FSTLLVSTSTWILLSFYGFTTITIVSWI 56
>At1g25350 unknown protein (At1g25350)
Length = 795
Score = 30.4 bits (67), Expect = 3.7
Identities = 13/37 (35%), Positives = 23/37 (62%)
Query: 2 ENINVDTEKGTMKEEEEEEESPIEEVRLTVKKTDDPT 38
E D EK T K+E++E+ + +EE + V+ T +P+
Sbjct: 184 EKTAADNEKPTKKKEKKEKPAKVEEKKAVVETTAEPS 220
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.327 0.142 0.461
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 16,866,128
Number of Sequences: 26719
Number of extensions: 740739
Number of successful extensions: 3302
Number of sequences better than 10.0: 26
Number of HSP's better than 10.0 without gapping: 20
Number of HSP's successfully gapped in prelim test: 6
Number of HSP's that attempted gapping in prelim test: 3204
Number of HSP's gapped (non-prelim): 47
length of query: 740
length of database: 11,318,596
effective HSP length: 107
effective length of query: 633
effective length of database: 8,459,663
effective search space: 5354966679
effective search space used: 5354966679
T: 11
A: 40
X1: 15 ( 7.1 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 40 (21.7 bits)
S2: 64 (29.3 bits)
Medicago: description of AC144564.13


