

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144564.12 + phase: 0
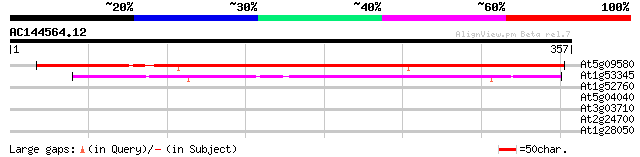
(357 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g09580 putative protein 366 e-102
At1g53345 unknown protein 151 6e-37
At1g52760 lipase like protein 29 4.5
At5g04040 unknown protein 28 7.7
At3g03710 putative polynucleotide phosphorylase 28 7.7
At2g24700 hypothetical protein 28 7.7
At1g28050 unknown protein 28 7.7
>At5g09580 putative protein
Length = 393
Score = 366 bits (940), Expect = e-102
Identities = 200/346 (57%), Positives = 249/346 (71%), Gaps = 17/346 (4%)
Query: 18 RSLRSDAALEAIAKASEDKVPNIVLYNYPSFSGAFSSLFAHLFHTRHNLPSLSLPFSSVP 77
RS RSDAALEAIA A E+KVPN+VLYNYPSFSGAFS+LFAHL+H R LP L LPFSS+
Sbjct: 53 RSFRSDAALEAIANALEEKVPNLVLYNYPSFSGAFSALFAHLYHYRLRLPCLILPFSSL- 111
Query: 78 SLAFSCYNIGLKICALKAFRHAICLTFFL-------QTNSLIIGFDHRKSVLSQIPSTNE 130
+ F ++ L+ F L F + +T I+ FDHR S L +I E
Sbjct: 112 -IPFRVEDL-----CLEGFERCYLLDFVVPKDFACQKTACEIVCFDHRNSALKRIGLIKE 165
Query: 131 -CPENIMINLNHEKSSSRAVYEYFTDKHEDIKTSNGVVPSLVDSKDKGRVELILKYIEDA 189
+ + I ++ E SSS+AVY+YF+ K D ++S SL+ +DK RVE +L YIED
Sbjct: 166 EHKKRLKIYVDTETSSSKAVYKYFSSKLTDQRSSEVEALSLLSVEDKTRVESVLDYIEDI 225
Query: 190 DLRHWSLPDIKPFNIGLSEWRSRFSCISNPYMFKQLLELSVEELIAKGNSSLLARRNAAS 249
DLR W LPDIK F+ GL +WRSR +CI NPYM++QLL++S +LIA GNS +R A
Sbjct: 226 DLRRWMLPDIKAFSFGLKDWRSRINCIINPYMYEQLLKISSADLIAYGNSYFSSRLIDAK 285
Query: 250 KLL--DKVFRVRLGRGFYGECLGVRADGNSNLSDEIGMLLSVKSASIGLRPIGAVIFMQR 307
KLL +K F++RLGRG YGECLG+RADGN LSDE+G LLS++S++ GLRPIGAV F+QR
Sbjct: 286 KLLKLNKAFKIRLGRGLYGECLGMRADGNHQLSDELGKLLSLQSSAAGLRPIGAVTFVQR 345
Query: 308 NNLKMCLRSSDNATDTSEVAKAYGGGGSASSSSFIIRMDEYNQWLS 353
NNLKMCLRS+D T+TSEVAKAYGGGG++SSSSFIIRMDEYNQW+S
Sbjct: 346 NNLKMCLRSTDAITNTSEVAKAYGGGGTSSSSSFIIRMDEYNQWIS 391
>At1g53345 unknown protein
Length = 325
Score = 151 bits (381), Expect = 6e-37
Identities = 99/320 (30%), Positives = 165/320 (50%), Gaps = 16/320 (5%)
Query: 41 VLYNYPSFSGAFSSLFAHLFHTRHNLPSLSLPFSSVPSLAFSCYNIGLKICALKAFRHAI 100
VLY+YP G F++L AHL+ + ++PSL P + + + + I L F
Sbjct: 10 VLYHYPCHDGVFAALAAHLYFSAKSIPSLFFPNTVYSPITINQLPLQ-DISHLYLFDFTG 68
Query: 101 CLTFFLQTNSLI---IGFDHRKSVLSQIPSTNECPENIMINLNHEKSSSRAVYEYFTDKH 157
F Q + + + DH K+ + + + +N+ L+ E+S + ++YFT K
Sbjct: 69 PPGFVHQVSPKVDNVVILDHHKTAIDSLGDVSLTCKNVTSVLDIERSGATIAFDYFTQKL 128
Query: 158 EDIKTSNGVVPSLVDSKDKGRVELILKYIEDADLRHWSLPDIKPFNIGLSEWRSRFSCIS 217
++ S G + D K R+ + +YIEDAD+ W LP+ K FN G+ + + +
Sbjct: 129 --VEESRGSCKEMNDFK---RMRRVFEYIEDADIWKWELPESKAFNSGILDLKIEYDFNK 183
Query: 218 NPYMFKQLLELSVEELIAKGNSSLLARRNAASKLLDKVFRVRLGRG-FYGECLGVRADGN 276
N +F QLL L E +I +G SL + ++L++ + + LG +G CL V AD
Sbjct: 184 NQSLFDQLLSLDHESVINRGKQSLSKKHKLIHEVLEQSYEIVLGGDEEFGRCLAVNADEI 243
Query: 277 SNLSDEIGMLLSVKSASIGLRPIGAVIFM-----QRNNLKMCLRSSDNATDTSEVAKAYG 331
+ L E+G L+ KS ++ LR +GAV++ N LK+ LRS DT+ V++ +G
Sbjct: 244 TELRSELGNQLAEKSKNLSLRGVGAVVYRVPELGDENKLKISLRSVAE-EDTTPVSQRFG 302
Query: 332 GGGSASSSSFIIRMDEYNQW 351
GGG ++SSF++ E+ QW
Sbjct: 303 GGGHKNASSFLLNSMEFEQW 322
>At1g52760 lipase like protein
Length = 332
Score = 28.9 bits (63), Expect = 4.5
Identities = 15/59 (25%), Positives = 29/59 (48%), Gaps = 5/59 (8%)
Query: 144 SSSRAVYEYFTDKHEDIKTSNGVVPSLVDSKDKGRVELILKYIEDADLRHWSLPDIKPF 202
+SS+ +YE + + +K G+ SL+ + E++LK D+R W +K +
Sbjct: 274 TSSKLLYEKASSADKTLKIYEGMYHSLIQGEPDENAEIVLK-----DMREWIDEKVKKY 327
>At5g04040 unknown protein
Length = 825
Score = 28.1 bits (61), Expect = 7.7
Identities = 15/60 (25%), Positives = 31/60 (51%)
Query: 130 ECPENIMINLNHEKSSSRAVYEYFTDKHEDIKTSNGVVPSLVDSKDKGRVELILKYIEDA 189
E PE++ +++ ++ +V E+ D +++ + NG V S+D G E + + DA
Sbjct: 766 ELPESVQLDIPEKEMDCSSVSEHEEDDNDNEEEHNGSSLVTVSSEDSGLQEPVSGSVIDA 825
>At3g03710 putative polynucleotide phosphorylase
Length = 948
Score = 28.1 bits (61), Expect = 7.7
Identities = 17/47 (36%), Positives = 27/47 (57%), Gaps = 1/47 (2%)
Query: 219 PYMFKQLLELSVEELI-AKGNSSLLARRNAASKLLDKVFRVRLGRGF 264
P ++K + EL+ EEL A S ++RR A S L +KV + +G+
Sbjct: 349 PELYKHVKELAGEELTKALQIKSKISRRKAISSLEEKVLTILTEKGY 395
>At2g24700 hypothetical protein
Length = 899
Score = 28.1 bits (61), Expect = 7.7
Identities = 28/98 (28%), Positives = 42/98 (42%), Gaps = 17/98 (17%)
Query: 249 SKLLDKVFRVRLGRGFYGECLGVRADGNSNLSDEIGMLLSVKSASIGLRPIGAVIFMQRN 308
S L+DK+ + LGRG G C ++ N S ML +V + I +P M N
Sbjct: 531 SLLMDKIGTMSLGRGSKGFCENIKI----NSSPLTTMLFNVSACDIRDKP------MPSN 580
Query: 309 NLKMCLRSSDNATDTSEVAK----AYGGGGSASSSSFI 342
N + D +V K + G S+ +SSF+
Sbjct: 581 N---DINKIDQLERVKKVRKNSSQSEAGSSSSGNSSFV 615
>At1g28050 unknown protein
Length = 433
Score = 28.1 bits (61), Expect = 7.7
Identities = 17/42 (40%), Positives = 21/42 (49%)
Query: 314 LRSSDNATDTSEVAKAYGGGGSASSSSFIIRMDEYNQWLSAN 355
L S DTS V AY G G+ASS + +D N+ S N
Sbjct: 268 LGQSRGPEDTSRVEAAYVGKGAASSFTINNFVDHMNETCSTN 309
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.320 0.135 0.393
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 8,044,035
Number of Sequences: 26719
Number of extensions: 341881
Number of successful extensions: 877
Number of sequences better than 10.0: 7
Number of HSP's better than 10.0 without gapping: 4
Number of HSP's successfully gapped in prelim test: 3
Number of HSP's that attempted gapping in prelim test: 864
Number of HSP's gapped (non-prelim): 9
length of query: 357
length of database: 11,318,596
effective HSP length: 100
effective length of query: 257
effective length of database: 8,646,696
effective search space: 2222200872
effective search space used: 2222200872
T: 11
A: 40
X1: 16 ( 7.4 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.8 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144564.12


