

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144563.4 + phase: 0
(588 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
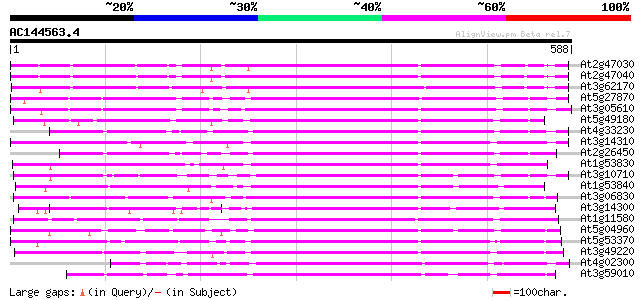
Score E
Sequences producing significant alignments: (bits) Value
At2g47030 putative pectinesterase 382 e-106
At2g47040 putative pectinesterase 362 e-100
At3g62170 PECTINESTERASE-like protein 350 2e-96
At5g27870 pectin methyl-esterase - like protein 333 2e-91
At3g05610 putative pectinesterase 327 1e-89
At5g49180 pectin methylesterase 325 4e-89
At4g33230 pectinesterase - like protein 325 6e-89
At3g14310 pectin methylesterase like protein 323 2e-88
At2g26450 putative pectinesterase 322 5e-88
At1g53830 unknown protein 309 3e-84
At3g10710 putative pectinesterase 305 6e-83
At1g53840 unknown protein 300 1e-81
At3g06830 pectin methylesterase like protein 298 4e-81
At3g14300 putative pectin methylesterase 296 3e-80
At1g11580 unknown protein 290 1e-78
At5g04960 pectinesterase 287 1e-77
At5g53370 pectinesterase 286 2e-77
At3g49220 pectinesterase - like protein 278 6e-75
At4g02300 putative pectinesterase 277 1e-74
At3g59010 pectinesterase precursor-like protein 275 4e-74
>At2g47030 putative pectinesterase
Length = 588
Score = 382 bits (982), Expect = e-106
Identities = 231/620 (37%), Positives = 339/620 (54%), Gaps = 68/620 (10%)
Query: 1 MKGKVIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLCHDTLS 60
M GKV++SV S++L+VGV IGVV I KNG D + Q + ++ +CQ+ D+ C TL
Sbjct: 1 MIGKVVVSVASILLIVGVAIGVVAFINKNG-DANLSPQMKAVQGICQSTSDKASCVKTLE 59
Query: 61 SVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGK--KNGAKMALDDCKDLMQF 118
V+ + DP I A + A D + K+ N + + G K LD CK + +
Sbjct: 60 PVK---SEDPNKLIKAFMLATKDELTKSSNFTGQTEVNMGSSISPNNKAVLDYCKRVFMY 116
Query: 119 ALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFH 178
AL+ L + + ++ + + ++ WL V +Y+ C++ ++ +D K I E
Sbjct: 117 ALEDLATIIEEMGE-DLSQIGSKIDQLKQWLIGVYNYQTDCLDDIEE-DDLRKAIGE--- 171
Query: 179 VQSLYSVQKVTAVALDIVTGLSDILQQFNLNF-------------------------DIK 213
+ + + +T A+DI + + + N D
Sbjct: 172 --GIANSKILTTNAIDIFHTVVSAMAKINNKVDDLKNMTGGIPTPGAPPVVDESPVADPD 229
Query: 214 PPSRRLLNSEVTVDDQGYPSWISSSGRKLLAKMQR-----KGWRANIRPNAVVANDGSGQ 268
P+RRLL +D+ G P+W+S + RKL+AK R +G A +R N VVA DGSGQ
Sbjct: 230 GPARRLLED---IDETGIPTWVSGADRKLMAKAGRGRRGGRGGGARVRTNFVVAKDGSGQ 286
Query: 269 FKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKN-- 326
FKT+Q A+ + P+ N+ R +IY+KAG+Y E + +PK+ NI M+GDG KT+++ ++
Sbjct: 287 FKTVQQAVDACPENNRGRCIIYIKAGLYREQVIIPKKKNNIFMFGDGARKTVISYNRSVA 346
Query: 327 QMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQD 386
GT T +AT + GF+ K M F+NTAGP G QA A R GD + + C GYQD
Sbjct: 347 LSRGTTTSLSATVQVESEGFMAKWMGFKNTAGPMGHQAAAIRVNGDRAVIFNCRFDGYQD 406
Query: 387 TLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPL-V 445
TLYV RQFYRNCV+SGT+DFIFG SAT+IQ++ I+VRKG+ +YN + ADG+ L +
Sbjct: 407 TLYVNNGRQFYRNCVVSGTVDFIFGKSATVIQNTLIVVRKGS--KGQYNTVTADGNELGL 464
Query: 446 NMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWP 505
M GIV+Q+C I+P+ L PE+ TV +YLGRPW+ S VIM + +GD I +GW W
Sbjct: 465 GMKIGIVLQNCRIVPDRKLTPERLTVATYLGRPWKKFSTTVIMSTEMGDLIRPEGWKIWD 524
Query: 506 EEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPK 565
E +C + EY N GPGA RRV W K S +E +TA+ WL GP
Sbjct: 525 GES------FHKSCRYVEYNNRGPGAFANRRVNWA--KVARSAAEVNGFTAANWL--GP- 573
Query: 566 TAPKSAVEWLHDLHVPHYLG 585
+ W+ + +VP +G
Sbjct: 574 ------INWIQEANVPVTIG 587
>At2g47040 putative pectinesterase
Length = 595
Score = 362 bits (929), Expect = e-100
Identities = 227/627 (36%), Positives = 331/627 (52%), Gaps = 75/627 (11%)
Query: 1 MKGKVIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLCHDTLS 60
M GKV++SV S++L+VGV IGVV I KNG D + Q + +R +C+ D+ C TL
Sbjct: 1 MIGKVVVSVASILLIVGVAIGVVAYINKNG-DANLSPQMKAVRGICEATSDKASCVKTLE 59
Query: 61 SVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGK--KNGAKMALDDCKDLMQF 118
V+ + DP I A + A D + ++ N + + G K LD CK + +
Sbjct: 60 PVK---SDDPNKLIKAFMLATRDAITQSSNFTGKTEGNLGSGISPNNKAVLDYCKKVFMY 116
Query: 119 ALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFH 178
AL+ L + ++ + + + ++ WL+ V +Y+ C++ ++ +D K I E
Sbjct: 117 ALEDLSTIVEEMGED-LNQIGSKIDQLKQWLTGVYNYQTDCLDDIEE-DDLRKTIGE--- 171
Query: 179 VQSLYSVQKVTAVALDIVTGLSDILQQFNLNF---------------------------- 210
+ S + +T+ A+DI + + + NL
Sbjct: 172 --GIASSKILTSNAIDIFHTVVSAMAKLNLKVEDFKNMTGGIFAPSDKGAAPVNKGTPPV 229
Query: 211 -------DIKPPSRRLLNSEVTVDDQGYPSWISSSGRKLLAKMQR--KGWRANIRPNAVV 261
D P+RRLL +D+ G P+W+S + RKL+ K R A IR VV
Sbjct: 230 ADDSPVADPDGPARRLLED---IDETGIPTWVSGADRKLMTKAGRGSNDGGARIRATFVV 286
Query: 262 ANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIV 321
A DGSGQFKT+Q A+ + P+ N R +I++KAG+Y E + +PK+ NI M+GDG KT++
Sbjct: 287 AKDGSGQFKTVQQAVNACPEKNPGRCIIHIKAGIYREQVIIPKKKNNIFMFGDGARKTVI 346
Query: 322 TGRKNQMA--GTNTQNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGC 379
+ ++ GT T + T + GF+ K + F+NTAGP G QAVA R GD + + C
Sbjct: 347 SYNRSVKLSPGTTTSLSGTVQVESEGFMAKWIGFKNTAGPMGHQAVAIRVNGDRAVIFNC 406
Query: 380 HIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVA 439
GYQDTLYV RQFYRN V+SGT+DFIFG SAT+IQ+S I+VRKGN ++N + A
Sbjct: 407 RFDGYQDTLYVNNGRQFYRNIVVSGTVDFIFGKSATVIQNSLIVVRKGN--KGQFNTVTA 464
Query: 440 DGSPL-VNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQ 498
DG+ + M GIV+Q+C I+P+ L E+ V SYLGRPW+ S VI+ S IGD I
Sbjct: 465 DGNEKGLAMKIGIVLQNCRIVPDKKLAAERLIVESYLGRPWKKFSTTVIINSEIGDVIRP 524
Query: 499 DGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYTASI 558
+GW W E +C + EY N GPGA RRV W K S +E +T +
Sbjct: 525 EGWKIWDGES------FHKSCRYVEYNNRGPGAITNRRVNW--VKIARSAAEVNDFTVAN 576
Query: 559 WLDAGPKTAPKSAVEWLHDLHVPHYLG 585
WL GP + W+ + +VP LG
Sbjct: 577 WL--GP-------INWIQEANVPVTLG 594
>At3g62170 PECTINESTERASE-like protein
Length = 588
Score = 350 bits (897), Expect = 2e-96
Identities = 218/612 (35%), Positives = 321/612 (51%), Gaps = 57/612 (9%)
Query: 3 GKVIISVVSLILVVGVIIGVVVDIRKNG-----EDPKVQTQQRNLRIMCQNAQDQKLCHD 57
GKV++SV SL+LVVGV IGV+ + K G + + + Q+ ++ +CQ+ DQ C
Sbjct: 4 GKVVVSVASLLLVVGVAIGVITFVNKGGGANGDSNGPINSHQKAVQTICQSTTDQGSCAK 63
Query: 58 TLSSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGA--KMALDDCKDL 115
TL V+ + DP + A + A D + K+ N + G A K LD CK +
Sbjct: 64 TLDPVK---SDDPSKLVKAFLMATKDAITKSSNFTASTEGGMGTNMNATSKAVLDYCKRV 120
Query: 116 MQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKE 175
+ +AL+ L+ + ++ ++ + ++ WL+ V +Y+ C++ ++ KKI
Sbjct: 121 LMYALEDLETIVEEMGED-LQQSGTKLDQLKQWLTGVFNYQTDCLDDIEEVE--LKKIMG 177
Query: 176 QFHVQSLYSVQKVTAVALDIVTGLS------DILQQFNLNFDIKPPSRRLLNSEVTVDDQ 229
+ S + +VT ++ D ++ + +RRLL D +
Sbjct: 178 EGISNSKVLTSNAIDIFHSVVTAMAQMGVKVDDMKNITMGAGAGGAARRLLEDN---DSK 234
Query: 230 GYPSWISSSGRKLLAKMQRK-------------GWRANIRPNAVVANDGSGQFKTIQAAL 276
G P W S RKL+AK R G I+ VVA DGSGQFKTI A+
Sbjct: 235 GLPKWFSGKDRKLMAKAGRGAPAGGDDGIGEGGGGGGKIKATHVVAKDGSGQFKTISEAV 294
Query: 277 ASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMA--GTNTQ 334
+ P N R +I++KAG+Y+E + +PK+ NI M+GDG +TI+T ++ GT T
Sbjct: 295 MACPDKNPGRCIIHIKAGIYNEQVRIPKKKNNIFMFGDGATQTIITFDRSVKLSPGTTTS 354
Query: 335 NTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNR 394
+ T + GF+ K + F+NTAGP G QAVA R GD + + C GYQDTLYV R
Sbjct: 355 LSGTVQVESEGFMAKWIGFKNTAGPLGHQAVALRVNGDRAVIFNCRFDGYQDTLYVNNGR 414
Query: 395 QFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPL-VNMNTGIVI 453
QFYRN V+SGT+DFIFG SAT+IQ+S I+VRKG+ + Y + ADG+ M GIV+
Sbjct: 415 QFYRNIVVSGTVDFIFGKSATVIQNSLILVRKGSPGQSNY--VTADGNEKGAAMKIGIVL 472
Query: 454 QDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEK 513
+C IIP+ L +K T++SYLGRPW+ + VI+ + IGD I +GWT W EQN
Sbjct: 473 HNCRIIPDKELEADKLTIKSYLGRPWKKFATTVIIGTEIGDLIKPEGWTEWQGEQN---- 528
Query: 514 HHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVE 573
T + E+ N GPGA +R W K S +E YT + W+ GP
Sbjct: 529 --HKTAKYIEFNNRGPGAATTQRPPW--VKVAKSAAEVETYTVANWV--GP-------AN 575
Query: 574 WLHDLHVPHYLG 585
W+ + +VP LG
Sbjct: 576 WIQEANVPVQLG 587
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 333 bits (853), Expect = 2e-91
Identities = 211/589 (35%), Positives = 308/589 (51%), Gaps = 46/589 (7%)
Query: 2 KGKVIISVVSLIL---VVGVIIGVVVDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLCHDT 58
K VIIS+ S++L VV V IGV V+ N D ++ T + ++ +C ++ C DT
Sbjct: 14 KRYVIISISSVLLISMVVAVTIGVSVNKSDNAGDEEITTSVKAIKDVCAPTDYKETCEDT 73
Query: 59 LSSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQF 118
L D +DP + A A + S+ + E K AKMALD CK+LM +
Sbjct: 74 LRK-DAKDTSDPLELVKTAFNATMKQISDVAKKSQTMI-ELQKDPRAKMALDQCKELMDY 131
Query: 119 ALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFD--DANDGEKKIKEQ 176
A+ L S + V + +R WLSA IS++Q C++GF N GE K
Sbjct: 132 AIGELSKSFEELGKFEFHKVDEALVKLRIWLSATISHEQTCLDGFQGTQGNAGETIKK-- 189
Query: 177 FHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWIS 236
+L + ++T L +VT +S+ L Q + + SRRLL+ E +PSW+
Sbjct: 190 ----ALKTAVQLTHNGLAMVTEMSNYLGQMQIP---EMNSRRLLSQE-------FPSWMD 235
Query: 237 SSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVY 296
+ R+LL + ++P+ VVA DGSGQ+KTI AL PK +V+++K G+Y
Sbjct: 236 ARARRLLNAPM-----SEVKPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHIKEGIY 290
Query: 297 DEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENT 356
EY+ V + +++ GDGP KT+++G K+ G T TAT + FI K + FENT
Sbjct: 291 KEYVQVNRSMTHLVFIGDGPDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNIAFENT 350
Query: 357 AGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATL 416
AG QAVA R + D S C GYQDTLY ++RQFYR+C ISGTIDF+FG +A +
Sbjct: 351 AGAIKHQAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFGDAAAV 410
Query: 417 IQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLG 476
Q+ T++VRK N+ I A G +TG V+Q C I+ E + K ++YLG
Sbjct: 411 FQNCTLLVRKPLL--NQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKTYLG 468
Query: 477 RPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARR 536
RPW+ S+ +IM + I DF+ +GW W E NT +++E NTGPGA + +R
Sbjct: 469 RPWKEYSRTIIMNTFIPDFVPPEGWQPWLGEFGL------NTLFYSEVQNTGPGAAITKR 522
Query: 537 VKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLG 585
V W G K +S E K+T + ++ W+ VP+ LG
Sbjct: 523 VTWPGIK-KLSDEEILKFTPAQYIQGD---------AWIPGKGVPYILG 561
>At3g05610 putative pectinesterase
Length = 669
Score = 327 bits (838), Expect = 1e-89
Identities = 205/595 (34%), Positives = 306/595 (50%), Gaps = 47/595 (7%)
Query: 2 KGKVIISVVSLILVVGVIIGVVVDIRKNGED------PKVQTQQRNLRIMCQNAQDQKLC 55
K + I+ +S +L++ +++ V V + N D +V + ++ +C +K C
Sbjct: 12 KRRYIVITISSVLLISMVVAVTVGVSLNKHDGDSKGKAEVNASVKAVKDVCAPTDYRKTC 71
Query: 56 HDTLSSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDL 115
DTL G + DP + A + A S+ + E K + +MALD CK+L
Sbjct: 72 EDTLIK-NGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIM-ELQKDSRTRMALDQCKEL 129
Query: 116 MQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFD--DANDGEKKI 173
M +ALD L S + + + ++R WLSA IS+++ C+EGF N GE
Sbjct: 130 MDYALDELSNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGFQGTQGNAGETMK 189
Query: 174 KEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPS 233
K +L + ++T L I++ +S+ + Q + SRRLL +G+PS
Sbjct: 190 K------ALKTAIELTHNGLAIISEMSNFVGQMQIP---GLNSRRLLA-------EGFPS 233
Query: 234 WISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKA 293
W+ GRKLL Q ++++P+ VVA DGSGQ+KTI AL PK +V+++KA
Sbjct: 234 WVDQRGRKLL---QAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVHIKA 290
Query: 294 GVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTF 353
G+Y EY+ V K +++ GDGP KTI++G KN G T TAT + FI K + F
Sbjct: 291 GLYKEYVQVNKTMSHLVFIGDGPDKTIISGNKNYKDGITTYRTATVAIVGNYFIAKNIGF 350
Query: 354 ENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTS 413
ENTAG QAVA R D S C GYQDTLY ++RQF+R+C ISGTIDF+FG +
Sbjct: 351 ENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFLFGDA 410
Query: 414 ATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRS 473
A + Q+ T++VRK N+ I A G +TG V Q C I E + K T ++
Sbjct: 411 AAVFQNCTLLVRKPL--PNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSKA 468
Query: 474 YLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANV 533
YLGRPW+ S+ +IM + I DF+ GW W + T +++E NTGPG+ +
Sbjct: 469 YLGRPWKEYSRTIIMNTFIPDFVQPQGWQPWLGDFGL------KTLFYSEVQNTGPGSAL 522
Query: 534 ARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLGFKA 588
A RV W G K +S + K+T + ++ +W+ VP+ G A
Sbjct: 523 ANRVTWAGIK-TLSEEDILKFTPAQYIQGD---------DWIPGKGVPYTTGLLA 567
>At5g49180 pectin methylesterase
Length = 571
Score = 325 bits (833), Expect = 4e-89
Identities = 200/570 (35%), Positives = 309/570 (54%), Gaps = 43/570 (7%)
Query: 5 VIISVVSLILVVGVIIGVVVDIRKNGEDPKV-----QTQQRNLRIMCQNAQDQKLCHDTL 59
+I V++ +LV+ V+ + R K+ +T + +C ++ C ++L
Sbjct: 13 IIAGVITALLVLMVVAVGITTSRNTSHSEKIVPVQIKTATTAVEAVCAPTDYKETCVNSL 72
Query: 60 SSVRGADAADP----KAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDL 115
D+ P K +++ D++ KA S LT + K AL+ C+ L
Sbjct: 73 MKA-SPDSTQPLDLIKLGFNVTIRSIEDSIKKA---SVELTAKAANDKDTKGALELCEKL 128
Query: 116 MQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKE 175
M A D L + +I + D D+R WLS I+Y+Q CM+ F++ N K
Sbjct: 129 MNDATDDLKKCLDNFDGFSIPQIEDFVEDLRVWLSGSIAYQQTCMDTFEETNS-----KL 183
Query: 176 QFHVQSLYSVQK-VTAVALDIVTGLSDILQQFNLNF---DIKPPSRRLLNSEVTVDDQGY 231
+Q ++ + +T+ L ++T +S++L +FN+ D+ +R+LL++E G
Sbjct: 184 SQDMQKIFKTSRELTSNGLAMITNISNLLGEFNVTGVTGDLGKYARKLLSAE-----DGI 238
Query: 232 PSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYV 291
PSW+ + R+L+A + ++ N VVA+DGSGQ+KTI AL + PK N+ +VIY+
Sbjct: 239 PSWVGPNTRRLMAT------KGGVKANVVVAHDGSGQYKTINEALNAVPKANQKPFVIYI 292
Query: 292 KAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAG-TNTQNTATFSNTAMGFIGKA 350
K GVY+E + V K+ ++ GDGP KT +TG N G T TAT + F K
Sbjct: 293 KQGVYNEKVDVTKKMTHVTFIGDGPTKTKITGSLNYYIGKVKTYLTATVAINGDNFTAKN 352
Query: 351 MTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIF 410
+ FENTAGP G QAVA R D++ C I GYQDTLYV ++RQF+R+C +SGT+DFIF
Sbjct: 353 IGFENTAGPEGHQAVALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRDCTVSGTVDFIF 412
Query: 411 GTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFT 470
G ++Q+ I+VRK ++ +I A G +TG+V+Q+C+I E A +P K
Sbjct: 413 GDGIVVLQNCNIVVRKPM--KSQSCMITAQGRSDKRESTGLVLQNCHITGEPAYIPVKSI 470
Query: 471 VRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPG 530
++YLGRPW+ S+ +IM +TI D I GW W + NT Y+AEY N GPG
Sbjct: 471 NKAYLGRPWKEFSRTIIMGTTIDDVIDPAGWLPWNGD------FALNTLYYAEYENNGPG 524
Query: 531 ANVARRVKWKGYKGVISRSEATKYTASIWL 560
+N A+RVKW G K +S +A ++T + +L
Sbjct: 525 SNQAQRVKWPGIK-KLSPKQALRFTPARFL 553
>At4g33230 pectinesterase - like protein
Length = 609
Score = 325 bits (832), Expect = 6e-89
Identities = 198/547 (36%), Positives = 288/547 (52%), Gaps = 39/547 (7%)
Query: 42 LRIMCQNAQDQKLCHDTLSSVRGADA--ADPKAYIAAAVKAATDNVIKAFNMSERLTTEY 99
++ +C + + C +TL + D DP++ + +A+ A D++ + F L TE
Sbjct: 95 IQTLCNSTLYKPTCQNTLKNETKKDTPQTDPRSLLKSAIVAVNDDLDQVFKRVLSLKTE- 153
Query: 100 GKKNGAKMALDDCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGC 159
K A+ CK L+ A + L S + D+ + D+ +WLSAV+SY++ C
Sbjct: 154 --NKDDKDAIAQCKLLVDEAKEELGTSMKRINDSEVNNFAKIVPDLDSWLSAVMSYQETC 211
Query: 160 MEGFDDANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRL 219
++GF+ E K+K + ++ S Q +T+ +L ++ L L K +R L
Sbjct: 212 VDGFE-----EGKLKTEIR-KNFNSSQVLTSNSLAMIKSLDGYLSSVP-----KVKTRLL 260
Query: 220 LNSEVTVDDQGY-PSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALAS 278
L + + + + SW+S+ R++L + K ++PNA VA DGSG F TI AAL +
Sbjct: 261 LEARSSAKETDHITSWLSNKERRMLKAVDVKA----LKPNATVAKDGSGNFTTINAALKA 316
Query: 279 YPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTAT 338
P + RY IY+K G+YDE + + K+ N+ M GDG KTIVTG K+ T TAT
Sbjct: 317 MPAKYQGRYTIYIKHGIYDESVIIDKKKPNVTMVGDGSQKTIVTGNKSHAKKIRTFLTAT 376
Query: 339 FSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYR 398
F GF+ ++M F NTAGP G QAVA R D S + C GYQDTLY T+RQ+YR
Sbjct: 377 FVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQSDRSVFLNCRFEGYQDTLYAYTHRQYYR 436
Query: 399 NCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNI 458
+CVI GT+DFIFG +A + Q+ I +RKG + N + A G TG VI +C +
Sbjct: 437 SCVIIGTVDFIFGDAAAIFQNCDIFIRKGL--PGQKNTVTAQGRVDKFQTTGFVIHNCTV 494
Query: 459 IPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENT 518
P L P K +SYLGRPW+ S+ V+MESTI D I GW W E +T
Sbjct: 495 APNEDLKPVKAQFKSYLGRPWKPHSRTVVMESTIEDVIDPVGWLRWQETD-----FAIDT 549
Query: 519 CYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDL 578
+AEY N GP A RVKW G++ V+++ EA K+T +L EW+ +
Sbjct: 550 LSYAEYKNDGPSGATAARVKWPGFR-VLNKEEAMKFTVGPFLQG----------EWIQAI 598
Query: 579 HVPHYLG 585
P LG
Sbjct: 599 GSPVKLG 605
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 323 bits (827), Expect = 2e-88
Identities = 207/607 (34%), Positives = 316/607 (51%), Gaps = 56/607 (9%)
Query: 2 KGKVIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRN-LRIMCQNAQDQKLCHDTLS 60
K V++S +L V + G+ K E + LR C + + +LC +
Sbjct: 18 KKLVLLSAAVALLFVAAVAGISAGASKANEKRTLSPSSHAVLRSSCSSTRYPELCISAVV 77
Query: 61 SVRGADAADPKAYIAAAVKAATDNVI-KAFNMSERLTTEYGKKNGAKMALDDCKDLMQFA 119
+ G + K I A+V V F + + + G K AL DC + +
Sbjct: 78 TAGGVELTSQKDVIEASVNLTITAVEHNYFTVKKLIKKRKGLTPREKTALHDCLETIDET 137
Query: 120 LDSLDLSNNCVRDNNI----EAVHDQTADMRNWLSAVISYKQGCMEGF--DDANDGEKK- 172
LD L + V D ++ + + + D++ +S+ I+ ++ C++GF DDA+ +K
Sbjct: 138 LDEL---HETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSHDDADKQVRKA 194
Query: 173 -IKEQFHVQSLYSVQKVTAVALDIVTGLSDI-LQQFNLNFDIKPPSRRLL--NSEVTV-- 226
+K Q HV+ + S AL ++ ++D + F I +R+L N E TV
Sbjct: 195 LLKGQIHVEHMCSN------ALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQETTVAV 248
Query: 227 --------DDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALAS 278
D +G+P+W+S+ R+LL + ++ +A VA DGSG FKT+ AA+A+
Sbjct: 249 DIAGAGELDSEGWPTWLSAGDRRLLQG-------SGVKADATVAADGSGTFKTVAAAVAA 301
Query: 279 YPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTAT 338
P+ + RYVI++KAGVY E + V K+ NI+ GDG +TI+TG +N + G+ T ++AT
Sbjct: 302 APENSNKRYVIHIKAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSAT 361
Query: 339 FSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYR 398
+ F+ + +TF+NTAGP+ QAVA R D SA C ++ YQDTLYV +NRQF+
Sbjct: 362 VAAVGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFV 421
Query: 399 NCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNI 458
C+I+GT+DFIFG +A ++Q I R+ N + N++ A G N NTGIVIQ C I
Sbjct: 422 KCLIAGTVDFIFGNAAVVLQDCDIHARRPN--SGQKNMVTAQGRTDPNQNTGIVIQKCRI 479
Query: 459 IPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENT 518
+ L K + +YLGRPW+ S+ VIM+S I D I +GW+ W NT
Sbjct: 480 GATSDLQSVKGSFPTYLGRPWKEYSQTVIMQSAISDVIRPEGWSEW------TGTFALNT 533
Query: 519 CYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDL 578
+ EY+NTG GA A RVKW+G+K + + +EA KYTA ++ G WL
Sbjct: 534 LTYREYSNTGAGAGTANRVKWRGFKVITAAAEAQKYTAGQFIGGG---------GWLSST 584
Query: 579 HVPHYLG 585
P LG
Sbjct: 585 GFPFSLG 591
>At2g26450 putative pectinesterase
Length = 496
Score = 322 bits (824), Expect = 5e-88
Identities = 195/523 (37%), Positives = 283/523 (53%), Gaps = 36/523 (6%)
Query: 53 KLCHDTLSSV--RGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALD 110
++C TL + +G +P ++ +A++A +++ L TE K A++
Sbjct: 2 QICEKTLKNRTDKGFALDNPTTFLKSAIEAVNEDLDLVLEKVLSLKTE---NQDDKDAIE 58
Query: 111 DCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGE 170
CK L++ A + S N + + + D+ +WLSAV+SY++ C++GF++ N
Sbjct: 59 QCKLLVEDAKEETVASLNKINVTEVNSFEKVVPDLESWLSAVMSYQETCLDGFEEGN--- 115
Query: 171 KKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQG 230
+K + S+ S Q +T+ +L ++ ++ NL+ +K R LL+
Sbjct: 116 --LKSEVKT-SVNSSQVLTSNSLALIKTFTE-----NLSPVMKVVERHLLDD-------- 159
Query: 231 YPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIY 290
PSW+S+ R++L + K ++PNA VA DGSG F TI AL + P+ + RY+IY
Sbjct: 160 IPSWVSNDDRRMLRAVDVKA----LKPNATVAKDGSGDFTTINDALRAMPEKYEGRYIIY 215
Query: 291 VKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKA 350
VK G+YDEY+TV K+ N+ M GDG KTIVTG K+ T TATF GF+ ++
Sbjct: 216 VKQGIYDEYVTVDKKKANLTMVGDGSQKTIVTGNKSHAKKIRTFLTATFVAQGEGFMAQS 275
Query: 351 MTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIF 410
M F NTAGP G QAVA R D S + C GYQDTLY T+RQ+YR+CVI GTIDFIF
Sbjct: 276 MGFRNTAGPEGHQAVAIRVQSDRSIFLNCRFEGYQDTLYAYTHRQYYRSCVIVGTIDFIF 335
Query: 411 GTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFT 470
G +A + Q+ I +RKG + N + A G TG V+ +C I L P K
Sbjct: 336 GDAAAIFQNCNIFIRKGL--PGQKNTVTAQGRVDKFQTTGFVVHNCKIAANEDLKPVKEE 393
Query: 471 VRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPG 530
+SYLGRPW+N S+ +IMES I + I GW W E +T Y+AEY N G
Sbjct: 394 YKSYLGRPWKNYSRTIIMESKIENVIDPVGWLRWQETD-----FAIDTLYYAEYNNKGSS 448
Query: 531 ANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVE 573
+ RVKW G+K VI++ EA YT +L +A S V+
Sbjct: 449 GDTTSRVKWPGFK-VINKEEALNYTVGPFLQGDWISASGSPVK 490
>At1g53830 unknown protein
Length = 587
Score = 309 bits (791), Expect = 3e-84
Identities = 196/579 (33%), Positives = 310/579 (52%), Gaps = 43/579 (7%)
Query: 4 KVIISVVSL-ILVVGVIIGVVVDIRKNGEDPKVQTQQRN----LRIMCQNAQDQKLCHDT 58
K+I+S ++ +L++ I+G+ ++ K+ T L+ +C + +LC
Sbjct: 19 KLILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHAILKSVCSSTLYPELCFSA 78
Query: 59 LSSVRGADAADPKAYIAAAVKAATDNVI-KAFNMSERLTTEYGKKNGAKMALDDCKDLMQ 117
+++ G + K I A++ T V F + + + G AL DC + +
Sbjct: 79 VAATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREVTALHDCLETID 138
Query: 118 FALDSLDLSNNCVRDN-NIEAVHDQTADMRNWLSAVISYKQGCMEGF--DDANDGEKK-- 172
LD L ++ + +++ D++ +S+ I+ + C++GF DDA+ +K
Sbjct: 139 ETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFSYDDADRKVRKAL 198
Query: 173 IKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNS------EVT- 225
+K Q HV+ + S A+A+ + ++ + NF+++ S N+ EVT
Sbjct: 199 LKGQVHVEHMCS----NALAM-----IKNMTETDIANFELRDKSSTFTNNNNRKLKEVTG 249
Query: 226 -VDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNK 284
+D G+P W+S R+LL + I+ +A VA+DGSG F T+ AA+A+ P+ +
Sbjct: 250 DLDSDGWPKWLSVGDRRLLQG-------STIKADATVADDGSGDFTTVAAAVAAAPEKSN 302
Query: 285 DRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAM 344
R+VI++KAGVY E + V K+ NI+ GDG KTI+TG +N + G+ T ++AT +
Sbjct: 303 KRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGE 362
Query: 345 GFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISG 404
F+ + +TF+NTAGP+ QAVA R D SA C + YQDTLYV +NRQF+ C I+G
Sbjct: 363 RFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITG 422
Query: 405 TIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAAL 464
T+DFIFG +A ++Q I R+ N + N++ A G N NTGIVIQ+C I + L
Sbjct: 423 TVDFIFGNAAAVLQDCDINARRPN--SGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDL 480
Query: 465 VPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEY 524
+ K T +YLGRPW+ S+ VIM+S I D I +GW W +T + EY
Sbjct: 481 LAVKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEW------SGSFALDTLTYREY 534
Query: 525 ANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAG 563
N G GA A RVKWKGYK + S +EA +TA ++ G
Sbjct: 535 LNRGGGAGTANRVKWKGYKVITSDTEAQPFTAGQFIGGG 573
>At3g10710 putative pectinesterase
Length = 561
Score = 305 bits (780), Expect = 6e-83
Identities = 209/592 (35%), Positives = 303/592 (50%), Gaps = 68/592 (11%)
Query: 5 VIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRN-------LRIMCQNAQDQKLCHD 57
+ I VSL+++ G++IG V + + P+ N ++ +C ++ C +
Sbjct: 26 IAIIAVSLVILAGIVIGAVFGTMAHKKSPETVETNNNGDSISVSVKAVCDVTLHKEKCFE 85
Query: 58 TLSSVRGADAADPKAYIAAAVKAATDNVIKAFN-MSERLTTEYGKKNGAKMALDDCKDLM 116
TL S A + +P+ AVK V KA N S L E KN M + C +L+
Sbjct: 86 TLGSAPNASSLNPEELFRYAVKITIAEVSKAINAFSSSLGDE---KNNITM--NACAELL 140
Query: 117 QFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGF--DDANDGEKKIK 174
+D+L+ + + ++ V + D+R WLS+ +Y++ C+E D GE +K
Sbjct: 141 DLTIDNLNNTLTSSSNGDV-TVPELVDDLRTWLSSAGTYQRTCVETLAPDMRPFGESHLK 199
Query: 175 EQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNS-EVTVDDQGYPS 233
++T+ AL I+T L I F L RRLL + +V VD
Sbjct: 200 NS---------TELTSNALAIITWLGKIADSFKLR-------RRLLTTADVEVDFH---- 239
Query: 234 WISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKA 293
+GR+LL + + VVA DGSG+++TI+ AL P+ ++ R +IYVK
Sbjct: 240 ----AGRRLLQSTDLRKVA-----DIVVAKDGSGKYRTIKRALQDVPEKSEKRTIIYVKK 290
Query: 294 GVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTF 353
GVY E + V K+ N+++ GDG +K+IV+GR N + GT T TATF+ GF+ + M F
Sbjct: 291 GVYFENVKVEKKMWNVIVVGDGESKSIVSGRLNVIDGTPTFKTATFAVFGKGFMARDMGF 350
Query: 354 ENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTS 413
NTAGP+ QAVA D++A C + YQDTLYV RQFYR C I GT+DFIFG S
Sbjct: 351 INTAGPSKHQAVALMVSADLTAFYRCTMNAYQDTLYVHAQRQFYRECTIIGTVDFIFGNS 410
Query: 414 ATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRS 473
A+++QS I+ R+ + N I A G NMNTGI I CNI P L V +
Sbjct: 411 ASVLQSCRILPRRPM--KGQQNTITAQGRTDPNMNTGISIHRCNISPLGDLT----DVMT 464
Query: 474 YLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANV 533
+LGRPW+N S VIM+S + FI + GW W + +T ++ EY NTGPGA+
Sbjct: 465 FLGRPWKNFSTTVIMDSYLHGFIDRKGWLPWTGDS------APDTIFYGEYKNTGPGAST 518
Query: 534 ARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYLG 585
RVKWKG + +S EA ++T ++D G WL VP G
Sbjct: 519 KNRVKWKGLR-FLSTKEANRFTVKPFIDGG---------RWLPATKVPFRSG 560
>At1g53840 unknown protein
Length = 586
Score = 300 bits (768), Expect = 1e-81
Identities = 189/564 (33%), Positives = 292/564 (51%), Gaps = 37/564 (6%)
Query: 7 ISVVSLI-LVVGVIIGVVVDIRKNGEDPKVQ---TQQRNLRIMCQNAQDQKLCHDTLSSV 62
ISVV LI +++ ++ VV KN P T +L+ +C + + C ++S +
Sbjct: 34 ISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAICSVTRFPESCISSISKL 93
Query: 63 RGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFALDS 122
++ DP+ ++K D + ++ E+L+ E + K AL C DL++ ALD
Sbjct: 94 PSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDER-IKSALRVCGDLIEDALDR 152
Query: 123 LDLSNNCVRDNNIEAV--HDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQ 180
L+ + + + D + + D++ WLSA ++ + C + D+ + + Q
Sbjct: 153 LNDTVSAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCFDSLDELKQNKTEYANSTITQ 212
Query: 181 SLYSVQ----KVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWIS 236
+L S + T+ +L IV+ + L + RR L S + W
Sbjct: 213 NLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIH----RRRRLMSHHHQQSVDFEKW-- 266
Query: 237 SSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVY 296
+ R+LL A ++P+ VA DG+G T+ A+A PK + +VIYVK+G Y
Sbjct: 267 -ARRRLLQT-------AGLKPDVTVAGDGTGDVLTVNEAVAKVPKKSLKMFVIYVKSGTY 318
Query: 297 DEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENT 356
E + + K N+++YGDG KTI++G KN + GT T TATF+ GFI K + NT
Sbjct: 319 VENVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFAIQGKGFIMKDIGIINT 378
Query: 357 AGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATL 416
AG A QAVAFR+ D S C G+QDTLY +NRQFYR+C ++GTIDFIFG++A +
Sbjct: 379 AGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDVTGTIDFIFGSAAVV 438
Query: 417 IQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLG 476
Q I+ R+ N++N I A G N ++G+ IQ C I ++ +YLG
Sbjct: 439 FQGCKIMPRQPL--SNQFNTITAQGKKDPNQSSGMSIQRCTISANGNVI-----APTYLG 491
Query: 477 RPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARR 536
RPW+ S VIME+ IG + GW +W + P + + EY NTGPG++V +R
Sbjct: 492 RPWKEFSTTVIMETVIGAVVRPSGWMSWVSGVDPPA-----SIVYGEYKNTGPGSDVTQR 546
Query: 537 VKWKGYKGVISRSEATKYTASIWL 560
VKW GYK V+S +EA K+T + L
Sbjct: 547 VKWAGYKPVMSDAEAAKFTVATLL 570
>At3g06830 pectin methylesterase like protein
Length = 568
Score = 298 bits (764), Expect = 4e-81
Identities = 193/577 (33%), Positives = 312/577 (53%), Gaps = 31/577 (5%)
Query: 5 VIISVVSLILVVGVIIGVVVDIRKNGEDPK--VQTQQRNLRIMCQNAQDQKLCHDTLSSV 62
++ VS LV+ V+ VV + + +D + ++ + ++ +C + C ++L
Sbjct: 12 IVAGSVSGFLVIMVVSVAVVTSKHSPKDDENHIRKTTKAVQAVCAPTDFKDTCVNSLMGA 71
Query: 63 RGADAADPKAYIAAAVKAATDNVIKAFNM-SERLTTEYGKKNGAKMALDDCKDLMQFALD 121
D+ DP I K ++ ++ S + + K AK A + C+ LM A+D
Sbjct: 72 -SPDSDDPVDLIKLGFKVTIKSINESLEKASGDIKAKADKNPEAKGAFELCEKLMIDAID 130
Query: 122 SLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQS 181
DL +++ + D+R WLS I+++Q CM+ F + K Q ++
Sbjct: 131 --DLKKCMDHGFSVDQIEVFVEDLRVWLSGSIAFQQTCMDSFGEI----KSNLMQDMLKI 184
Query: 182 LYSVQKVTAVALDIVTGLSDILQQFNLNF---DIKPPSRRLLNSEVTVDDQGYPSWISSS 238
+ +++++ +L +VT +S ++ NL + +R+LL++E ++ P+W+
Sbjct: 185 FKTSRELSSNSLAMVTRISTLIPNSNLTGLTGALAKYARKLLSTEDSI-----PTWVGPE 239
Query: 239 GRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDE 298
R+L+A + G ++ NAVVA DG+GQFKTI AL + PKGNK ++I++K G+Y E
Sbjct: 240 ARRLMAA--QGGGPGPVKANAVVAQDGTGQFKTITDALNAVPKGNKVPFIIHIKEGIYKE 297
Query: 299 YITVPKEAVNILMYGDGPAKTIVTGRKNQMAG-TNTQNTATFSNTAMGFIGKAMTFENTA 357
+TV K+ ++ GDGP KT++TG N G T TAT + F K + ENTA
Sbjct: 298 KVTVTKKMPHVTFIGDGPNKTLITGSLNFGIGKVKTFLTATITIEGDHFTAKNIGIENTA 357
Query: 358 GPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLI 417
GP G QAVA R D + C I G+QDTLYV ++RQFYR+C +SGT+DFIFG + ++
Sbjct: 358 GPEGGQAVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTVSGTVDFIFGDAKCIL 417
Query: 418 QSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGR 477
Q+ I+VRK N + ++ A G V +TG+V+ C+I + A +P K ++YLGR
Sbjct: 418 QNCKIVVRKPN--KGQTCMVTAQGRSNVRESTGLVLHGCHITGDPAYIPMKSVNKAYLGR 475
Query: 478 PWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRV 537
PW+ S+ +IM++TI D I GW W + T Y+AE+ NTGPG+N A+RV
Sbjct: 476 PWKEFSRTIIMKTTIDDVIDPAGWLPWSGD------FALKTLYYAEHMNTGPGSNQAQRV 529
Query: 538 KWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEW 574
KW G K ++ +A YT +L G P++ V +
Sbjct: 530 KWPGIK-KLTPQDALLYTGDRFL-RGDTWIPQTQVPY 564
>At3g14300 putative pectin methylesterase
Length = 968
Score = 296 bits (757), Expect = 3e-80
Identities = 187/539 (34%), Positives = 282/539 (51%), Gaps = 42/539 (7%)
Query: 42 LRIMCQNAQDQKLCHDTLSSVR-GADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYG 100
LR +C C ++S + DPK +++ D + + ++L E
Sbjct: 459 LRTVCNVTNYPASCISSISKLPLSKTTTDPKVLFRLSLQVTFDELNSIVGLPKKLAEETN 518
Query: 101 KKNGAKMALDDCKDLMQFALDSLD---LSNNCVRDNNIEAVHDQT-ADMRNWLSAVISYK 156
+ G K AL C D+ A+DS++ S + V + ++ T D+ WLS+ ++
Sbjct: 519 DE-GLKSALSVCADVFDLAVDSVNDTISSLDEVISGGKKNLNSSTIGDLITWLSSAVTDI 577
Query: 157 QGCMEGFDDANDGE---KKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIK 213
C + D+ N +K+K ++ + + T+ +L IV + +L++ + + I
Sbjct: 578 GTCGDTLDEDNYNSPIPQKLKS-----AMVNSTEFTSNSLAIV---AQVLKKPSKS-RIP 628
Query: 214 PPSRRLLNSEVTVDDQGYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQ 273
RRLLNS +P+W+ R+LL Q K N+ P+ VA DGSG +T+
Sbjct: 629 VQGRRLLNSN------SFPNWVRPGVRRLL---QAK----NLTPHVTVAADGSGDVRTVN 675
Query: 274 AALASYPKGNKDRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNT 333
A+ PK K +VIYVKAG Y E + + K+ N+ +YGDG KTI++G N + G T
Sbjct: 676 EAVWRVPKKGKTMFVIYVKAGTYVENVLMKKDKWNVFIYGDGRDKTIISGSTNMVDGVRT 735
Query: 334 QNTATFSNTAMGFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTN 393
NT+TF+ GF+ K M NTAGP QAVAFR+ D S C GYQDTLY +N
Sbjct: 736 FNTSTFATEGKGFMMKDMGIINTAGPEKHQAVAFRSDSDRSVYYRCSFDGYQDTLYTHSN 795
Query: 394 RQFYRNCVISGTIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVI 453
RQ+YRNC ++GT+DFIFG + Q +I R+ N++N I A+G+ N NTGI I
Sbjct: 796 RQYYRNCDVTGTVDFIFGAGTVVFQGCSIRPRQPL--PNQFNTITAEGTQEANQNTGISI 853
Query: 454 QDCNIIPEAALVPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEK 513
C I P T +YLGRPW+ SK VIM+S IG F++ GW W + P +
Sbjct: 854 HQCTISPNG-----NVTATTYLGRPWKLFSKTVIMQSVIGSFVNPAGWIAWNSTYDPPPR 908
Query: 514 HHENTCYFAEYANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAV 572
T ++ EY N+GPG+++++RVKW GYK + S EA ++T +L PK+ +
Sbjct: 909 ----TIFYREYKNSGPGSDLSKRVKWAGYKPISSDDEAARFTVKYFLRGDDNWIPKAVM 963
Score = 53.1 bits (126), Expect = 4e-07
Identities = 49/228 (21%), Positives = 96/228 (41%), Gaps = 24/228 (10%)
Query: 10 VSLILVVGVIIGVVVDIR---KNGEDPKVQ-------TQQRNLRIMCQNAQDQKLCHDTL 59
+S+ ++V +II V I + G P T +L+ +C C ++
Sbjct: 35 ISVAVLVAIIISSTVTIAIHSRKGNSPHPTPSSVPELTPAASLKTVCSVTNYPVSCFSSI 94
Query: 60 SSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFA 119
S + ++ DP+ +++ D + + ++L E + G K AL C+ L+ A
Sbjct: 95 SKLPLSNTTDPEVIFRLSLQVVIDELNSIVELPKKLAEETDDE-GLKSALSVCEHLLDLA 153
Query: 120 LDSLDLSNNCVRDNNIEAVHDQTA--DMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQF 177
+D ++ + + + + + + + D+ WLSA ++Y C++ D+ + I +
Sbjct: 154 IDRVNETVSAMEVVDGKKILNAATIDDLLTWLSAAVTYHGTCLDALDEISHTNSAIPLKL 213
Query: 178 H---VQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNS 222
V S A+ I++ +SD F I RRLLNS
Sbjct: 214 KSGMVNSTEFTSNSLAIVAKILSTISD--------FGIPIHGRRLLNS 253
Score = 39.3 bits (90), Expect = 0.006
Identities = 31/146 (21%), Positives = 68/146 (46%), Gaps = 15/146 (10%)
Query: 33 PKVQTQQRNLRIMCQNAQDQKLCHDTLSSVRGADAADPKAYIAAAVKAATDNVIKAFNMS 92
PK+ T +LR +C + C ++S + ++ DP+A +++ + + +
Sbjct: 263 PKL-TPAASLRNVCSVTRYPASCVSSISKLPSSNTTDPEALFRLSLQVVINELNSIAGLP 321
Query: 93 ERLTTEYGKKNGAKMALDDCKDLMQFALDSLDLSNNCVRDNNIEAVHD-------QTAD- 144
++L E + K +L C D+ D++D+ N+ + + +E V D T D
Sbjct: 322 KKLAEETDDER-LKSSLSVCGDVFN---DAIDIVNDTI--STMEEVGDGKKILKSSTIDE 375
Query: 145 MRNWLSAVISYKQGCMEGFDDANDGE 170
++ WLSA ++ C++ D+ + +
Sbjct: 376 IQTWLSAAVTDHDTCLDALDELSQNK 401
>At1g11580 unknown protein
Length = 557
Score = 290 bits (743), Expect = 1e-78
Identities = 187/576 (32%), Positives = 285/576 (49%), Gaps = 45/576 (7%)
Query: 5 VIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQRNLRIMCQNAQDQKLCHDTLSSVRG 64
+++S V+++ V ++ + N D + T + +C A DQ C LS
Sbjct: 22 LVLSFVAILGSVAFFTAQLISVNTNNNDDSLLTTSQ----ICHGAHDQDSCQALLSEFTT 77
Query: 65 ADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYG-KKNGA--KMALDDCKDLMQFALD 121
+ K + N + + + +E + NG K DC+++M + D
Sbjct: 78 LSLS--KLNRLDLLHVFLKNSVWRLESTMTMVSEARIRSNGVRDKAGFADCEEMMDVSKD 135
Query: 122 SLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQS 181
+ S +R N + +++ WLS+V++ C+E D + K+I + ++
Sbjct: 136 RMMSSMEELRGGNYNL--ESYSNVHTWLSSVLTNYMTCLESISDVSVNSKQIVKP-QLED 192
Query: 182 LYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWISSSGRK 241
L S +V V D L+ N +PSW+++ RK
Sbjct: 193 LVSRARVALAIFVSVLPARDDLKMIISN--------------------RFPSWLTALDRK 232
Query: 242 LLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEYIT 301
LL + + N VVA DG+G+FKT+ A+A+ P+ + RYVIYVK GVY E I
Sbjct: 233 LLESSPKT---LKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKGVYKETID 289
Query: 302 VPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGPAG 361
+ K+ N+++ GDG TI+TG N + G+ T +AT + GF+ + + F+NTAGPA
Sbjct: 290 IGKKKKNLMLVGDGKDATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQNTAGPAK 349
Query: 362 MQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQSST 421
QAVA R D + + C I YQDTLY T RQFYR+ I+GT+DFIFG SA + Q+
Sbjct: 350 HQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSAVVFQNCD 409
Query: 422 IIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPWQN 481
I+ R N + N++ A G N NT I IQ C I + L P K +V+++LGRPW+
Sbjct: 410 IVAR--NPGAGQKNMLTAQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLGRPWKL 467
Query: 482 ESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKWKG 541
S+ VIM+S I + I GW W E +T Y+ EYANTGPGA+ ++RV WKG
Sbjct: 468 YSRTVIMQSFIDNHIDPAGWFPWDGE------FALSTLYYGEYANTGPGADTSKRVNWKG 521
Query: 542 YKGVISRSEATKYTASIWLDAGPKTAPKSAV--EWL 575
+K + EA ++T + + G P EWL
Sbjct: 522 FKVIKDSKEAEQFTVAKLIQGGLWLKPTGVTFQEWL 557
>At5g04960 pectinesterase
Length = 564
Score = 287 bits (735), Expect = 1e-77
Identities = 199/588 (33%), Positives = 294/588 (49%), Gaps = 58/588 (9%)
Query: 2 KGKVIISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQQR------NLRIMCQNAQDQKLC 55
K ++ I +S I++V +++G VV K T+ +++ +C ++ C
Sbjct: 22 KKRIAIIAISSIVLVCIVVGAVVGTTARDNSKKPPTENNGEPISVSVKALCDVTLHKEKC 81
Query: 56 HDTLSSVRGADAADPKAYIAAAVKAAT---DNVIKAFNMSERLTTEYGKKNGAKMALDDC 112
+TL S A + P+ AVK V+ F+ E + N A+ C
Sbjct: 82 FETLGSAPNASRSSPEELFKYAVKVTITELSKVLDGFSNGEHMD------NATSAAMGAC 135
Query: 113 KDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKK 172
+L+ A+D L+ + N D+R WLS+V +Y++ CM+ +AN K
Sbjct: 136 VELIGLAVDQLNETMTSSLKN--------FDDLRTWLSSVGTYQETCMDALVEAN---KP 184
Query: 173 IKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLL---NSEVTVDDQ 229
F L + ++T+ AL I+T L I +K RRLL N++V V D
Sbjct: 185 SLTTFGENHLKNSTEMTSNALAIITWLGKIADT------VKFRRRRLLETGNAKVVVADL 238
Query: 230 GYPSWISSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVI 289
GR+LL K + VVA DGSG+++TI ALA + N+ +I
Sbjct: 239 PM-----MEGRRLLESGDLKK-----KATIVVAKDGSGKYRTIGEALAEVEEKNEKPTII 288
Query: 290 YVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGK 349
YVK GVY E + V K N++M GDG +KTIV+ N + GT T TATF+ GF+ +
Sbjct: 289 YVKKGVYLENVRVEKTKWNVVMVGDGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMAR 348
Query: 350 AMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFI 409
M F NTAGPA QAVA D+S C + +QDT+Y RQFYR+CVI GT+DFI
Sbjct: 349 DMGFINTAGPAKHQAVALMVSADLSVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFI 408
Query: 410 FGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKF 469
FG +A + Q I+ R+ + N I A G N NTGI I +C I P L
Sbjct: 409 FGNAAVVFQKCEILPRRPM--KGQQNTITAQGRKDPNQNTGISIHNCTIKPLDNLT---- 462
Query: 470 TVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGP 529
++++LGRPW++ S VIM+S + FI+ GW W + +T ++AEY N+GP
Sbjct: 463 DIQTFLGRPWKDFSTTVIMKSFMDKFINPKGWLPWTGDT------APDTIFYAEYLNSGP 516
Query: 530 GANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHD 577
GA+ RVKW+G K +++ EA K+T ++D G P + V + D
Sbjct: 517 GASTKNRVKWQGLKTSLTKKEANKFTVKPFID-GNNWLPATKVPFNSD 563
>At5g53370 pectinesterase
Length = 587
Score = 286 bits (732), Expect = 2e-77
Identities = 194/584 (33%), Positives = 300/584 (51%), Gaps = 36/584 (6%)
Query: 2 KGKVIISVVSLILVVGVIIGVVVDIR---KNGEDPKVQTQQRN-LRIMCQNAQDQKLCHD 57
K K+I+ +++++V V G+ IR +PK+ + + C + LC D
Sbjct: 31 KTKLILFTLAVLVVGVVCFGIFAGIRAVDSGKTEPKLTRKPTQAISRTCSKSLYPNLCID 90
Query: 58 TLSSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLT-TEYGKKNGAKMALDDCKDLM 116
TL G+ AD I + A KA S +T T+ + + A D C +L+
Sbjct: 91 TLLDFPGSLTADENELIHISFNATLQKFSKALYTSSTITYTQMPPR--VRSAYDSCLELL 148
Query: 117 QFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQ 176
DS+D + + + + +D+ WLS+ ++ C +GFD+ ++K+Q
Sbjct: 149 D---DSVDALTRALSSVVVVSGDESHSDVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQ 205
Query: 177 FHVQSLYSVQKVTAVALDIVTG-LSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWI 235
+ ++ + ++ + L I G + D+ +N +R+LL +E T + P+W+
Sbjct: 206 V-IGAVKDLSEMVSNCLAIFAGKVKDLSGVPVVN------NRKLLGTEETEE---LPNWL 255
Query: 236 SSSGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGV 295
R+LL + I+ + V+ DGSG FKTI A+ P+ + R+VIYVKAG
Sbjct: 256 KREDRELLGTPT-----SAIQADITVSKDGSGTFKTIAEAIKKAPEHSSRRFVIYVKAGR 310
Query: 296 YDEY-ITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFE 354
Y+E + V ++ N++ GDG KT++TG K+ T +TATF+ T GFI + MTFE
Sbjct: 311 YEEENLKVGRKKTNLMFIGDGKGKTVITGGKSIADDLTTFHTATFAATGAGFIVRDMTFE 370
Query: 355 NTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSA 414
N AGPA QAVA R GD + + C+I+GYQD LYV +NRQF+R C I GT+DFIFG +A
Sbjct: 371 NYAGPAKHQAVALRVGGDHAVVYRCNIIGYQDALYVHSNRQFFRECEIYGTVDFIFGNAA 430
Query: 415 TLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSY 474
++QS I RK + I A N NTGI I C ++ L K + +Y
Sbjct: 431 VILQSCNIYARKPM--AQQKITITAQNRKDPNQNTGISIHACKLLATPDLEASKGSYPTY 488
Query: 475 LGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVA 534
LGRPW+ S+ V M S +GD I GW W N P ++ Y+ EY N G G+ +
Sbjct: 489 LGRPWKLYSRVVYMMSDMGDHIDPRGWLEW----NGP--FALDSLYYGEYMNKGLGSGIG 542
Query: 535 RRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDL 578
+RVKW GY + S EA+K+T + ++ +G P + V + L
Sbjct: 543 QRVKWPGYHVITSTVEASKFTVAQFI-SGSSWLPSTGVSFFSGL 585
>At3g49220 pectinesterase - like protein
Length = 598
Score = 278 bits (711), Expect = 6e-75
Identities = 186/584 (31%), Positives = 297/584 (50%), Gaps = 44/584 (7%)
Query: 6 IISVVSLILVVGVIIGVVVDIRKNGEDPKVQTQ-QRNLRIMCQNAQDQKLCHDTLSSVRG 64
I+ +SLIL + GV ++ N P + + + + C+ + +LC D+L G
Sbjct: 50 IVLAISLILAAAIFAGVRSRLKLNQSVPGLARKPSQAISKACELTRFPELCVDSLMDFPG 109
Query: 65 ADAADP-KAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFALDSL 123
+ AA K I V + A S L+ A+ A D C +L+ ++D+L
Sbjct: 110 SLAASSSKDLIHVTVNMTLHHFSHALYSSASLSF-VDMPPRARSAYDSCVELLDDSVDAL 168
Query: 124 DLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHVQSLY 183
+ + V ++ + D+ WLSA ++ C EGFD +DG K
Sbjct: 169 SRALSSVVSSSAKP-----QDVTTWLSAALTNHDTCTEGFDGVDDGGVK----------- 212
Query: 184 SVQKVTAVALDIVTGLSDILQQFNLNFD------IKPPSRRLLNSEVTVDDQGYPSWISS 237
+TA ++ +S+ L F+ + D + +RRLL E ++ +P W+
Sbjct: 213 --DHMTAALQNLSELVSNCLAIFSASHDGDDFAGVPIQNRRLLGVEER--EEKFPRWMRP 268
Query: 238 SGRKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYD 297
R++L + I+ + +V+ DG+G KTI A+ P+ + R +IYVKAG Y+
Sbjct: 269 KEREILEMPV-----SQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYVKAGRYE 323
Query: 298 EY-ITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENT 356
E + V ++ +N++ GDG KT+++G K+ T +TA+F+ T GFI + +TFEN
Sbjct: 324 ENNLKVGRKKINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARDITFENW 383
Query: 357 AGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATL 416
AGPA QAVA R D + + C+I+GYQDTLYV +NRQF+R C I GT+DFIFG +A +
Sbjct: 384 AGPAKHQAVALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGNAAVV 443
Query: 417 IQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLG 476
+Q+ +I RK + N I A N NTGI I ++ + L + ++YLG
Sbjct: 444 LQNCSIYARKPM--DFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTYLG 501
Query: 477 RPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARR 536
RPW+ S+ V M S IG +H GW W +T Y+ EY N+GPG+ + +R
Sbjct: 502 RPWKLFSRTVYMMSYIGGHVHTRGWLEW------NTTFALDTLYYGEYLNSGPGSGLGQR 555
Query: 537 VKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHV 580
V W GY+ + S +EA ++T + ++ G P + V +L L +
Sbjct: 556 VSWPGYRVINSTAEANRFTVAEFI-YGSSWLPSTGVSFLAGLSI 598
>At4g02300 putative pectinesterase
Length = 532
Score = 277 bits (709), Expect = 1e-74
Identities = 169/482 (35%), Positives = 262/482 (54%), Gaps = 41/482 (8%)
Query: 106 KMALDDCKDLMQFALDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDD 165
+ A +DC L+ + L+ + + +R +++E D+ L+ V++Y+ C++GF
Sbjct: 91 RCAFEDCLGLLDDTISDLETAVSDLRSSSLEF-----NDISMLLTNVMTYQDTCLDGFS- 144
Query: 166 ANDGEKKIKEQFHVQSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVT 225
+D E + + + + + LDI LS+ L + KP + +SEV
Sbjct: 145 TSDNENNNDMTYELP-----ENLKEIILDISNNLSNSLHMLQVISRKKPSPK---SSEVD 196
Query: 226 VDDQGYPSWISSSGRKLL-AKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNK 284
V+ YPSW+S + ++LL A +Q + N VA DG+G F TI A+ + P ++
Sbjct: 197 VE---YPSWLSENDQRLLEAPVQETNY------NLSVAIDGTGNFTTINDAVFAAPNMSE 247
Query: 285 DRYVIYVKAGVYDEYITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAM 344
R++IY+K G Y E + +PK+ I+ GDG KT++ ++++ G +T T T
Sbjct: 248 TRFIIYIKGGEYFENVELPKKKTMIMFIGDGIGKTVIKANRSRIDGWSTFQTPTVGVKGK 307
Query: 345 GFIGKAMTFENTAGPAGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISG 404
G+I K ++F N+AGPA QAVAFR+ D SA C GYQDTLYV + +QFYR C I G
Sbjct: 308 GYIAKDISFVNSAGPAKAQAVAFRSGSDHSAFYRCEFDGYQDTLYVHSAKQFYRECDIYG 367
Query: 405 TIDFIFGTSATLIQSSTIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAAL 464
TIDFIFG +A + Q+S++ RK N H A + TGI I +C I+ L
Sbjct: 368 TIDFIFGNAAVVFQNSSLYARKPNPGHK--IAFTAQSRNQSDQPTGISILNCRILAAPDL 425
Query: 465 VPEKFTVRSYLGRPWQNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEY 524
+P K ++YLGRPW+ S+ VI++S I D IH GW + K + E T Y+ EY
Sbjct: 426 IPVKENFKAYLGRPWRKYSRTVIIKSFIDDLIHPAGWL-----EGKKDFALE-TLYYGEY 479
Query: 525 ANTGPGANVARRVKWKGYKGVISRSEATKYTASIWLDAGPKTAPKSAVEWLHDLHVPHYL 584
N GPGAN+A+RV W G++ + +++EAT++T ++D WL+ +P L
Sbjct: 480 MNEGPGANMAKRVTWPGFRRIENQTEATQFTVGPFIDGS---------TWLNSTGIPFSL 530
Query: 585 GF 586
GF
Sbjct: 531 GF 532
>At3g59010 pectinesterase precursor-like protein
Length = 529
Score = 275 bits (704), Expect = 4e-74
Identities = 170/513 (33%), Positives = 265/513 (51%), Gaps = 42/513 (8%)
Query: 60 SSVRGADAADPKAYIAAAVKAATDNVIKAFNMSERLTTEYGKKNGAKMALDDCKDLMQFA 119
SS R + ++V+ + D+ + A +++ LT + + + LD D ++
Sbjct: 52 SSSRTKPSTSSNKGFLSSVQLSLDHALFARSLAFNLTLSH--RTSQTLMLDPVNDCLELL 109
Query: 120 LDSLDLSNNCVRDNNIEAVHDQTADMRNWLSAVISYKQGCMEGFDDANDGEKKIKEQFHV 179
D+LD+ V + V+D D+ WLSA ++ ++ C + + K F+
Sbjct: 110 DDTLDMLYRIVVIKRKDHVND---DVHTWLSAALTNQETCKQSLSE--------KSSFNK 158
Query: 180 QSLYSVQKVTAVALDIVTGLSDILQQFNLNFDIKPPSRRLLNSEVTVDDQGYPSWISSSG 239
+ + + + A ++ L++ L F + S L + D +P+W+SSS
Sbjct: 159 EGI----AIDSFARNLTGLLTNSLDMFVSDKQKSSSSSNLTGGRKLLSDHDFPTWVSSSD 214
Query: 240 RKLLAKMQRKGWRANIRPNAVVANDGSGQFKTIQAALASYPKGNKDRYVIYVKAGVYDEY 299
RKLL + +RP+AVVA DGSG ++ ALAS KG+ R VI++ AG Y E
Sbjct: 215 RKLLEASVEE-----LRPHAVVAADGSGTHMSVAEALASLEKGS-GRSVIHLTAGTYKEN 268
Query: 300 ITVPKEAVNILMYGDGPAKTIVTGRKNQMAGTNTQNTATFSNTAMGFIGKAMTFENTAGP 359
+ +P + N+++ GDG KT++ G ++ G NT +AT + GFI + +TF N+AGP
Sbjct: 269 LNIPSKQKNVMLVGDGKGKTVIVGSRSNRGGWNTYQSATVAAMGDGFIARDITFVNSAGP 328
Query: 360 AGMQAVAFRNIGDMSALVGCHIVGYQDTLYVQTNRQFYRNCVISGTIDFIFGTSATLIQS 419
QAVA R D S + C I GYQD+LY + RQFYR I+GT+DFIFG SA + QS
Sbjct: 329 NSEQAVALRVGSDRSVVYRCSIDGYQDSLYTLSKRQFYRETDITGTVDFIFGNSAVVFQS 388
Query: 420 STIIVRKGNYDHNEYNVIVADGSPLVNMNTGIVIQDCNIIPEAALVPEKFTVRSYLGRPW 479
++ RKG+ D N + A G N NTGI I +C I + ++YLGRPW
Sbjct: 389 CNLVSRKGSSDQ---NYVTAQGRSDPNQNTGISIHNCRITG---------STKTYLGRPW 436
Query: 480 QNESKAVIMESTIGDFIHQDGWTTWPEEQNKPEKHHENTCYFAEYANTGPGANVARRVKW 539
+ S+ V+M+S I IH GW+ W T Y+ E+ N+GPG++V+ RV W
Sbjct: 437 KQYSRTVVMQSFIDGSIHPSGWSPW------SSNFALKTLYYGEFGNSGPGSSVSGRVSW 490
Query: 540 KGYKGVISRSEATKYTASIWLDAGPKTAPKSAV 572
GY ++ +EA +T S ++D G P + V
Sbjct: 491 AGYHPALTLTEAQGFTVSGFID-GNSWLPSTGV 522
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.317 0.133 0.392
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,937,599
Number of Sequences: 26719
Number of extensions: 544116
Number of successful extensions: 1690
Number of sequences better than 10.0: 87
Number of HSP's better than 10.0 without gapping: 76
Number of HSP's successfully gapped in prelim test: 11
Number of HSP's that attempted gapping in prelim test: 1335
Number of HSP's gapped (non-prelim): 108
length of query: 588
length of database: 11,318,596
effective HSP length: 105
effective length of query: 483
effective length of database: 8,513,101
effective search space: 4111827783
effective search space used: 4111827783
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144563.4


