

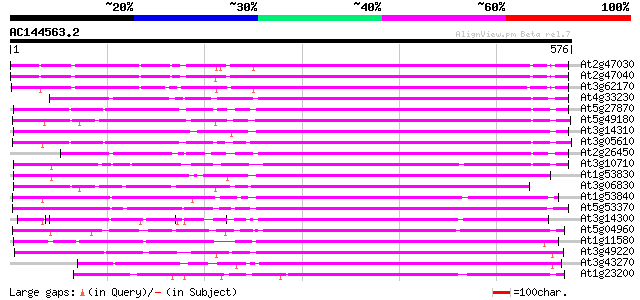
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144563.2 + phase: 0
(576 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At2g47030 putative pectinesterase 417 e-117
At2g47040 putative pectinesterase 402 e-112
At3g62170 PECTINESTERASE-like protein 390 e-108
At4g33230 pectinesterase - like protein 365 e-101
At5g27870 pectin methyl-esterase - like protein 360 e-100
At5g49180 pectin methylesterase 360 2e-99
At3g14310 pectin methylesterase like protein 359 3e-99
At3g05610 putative pectinesterase 355 4e-98
At2g26450 putative pectinesterase 350 1e-96
At3g10710 putative pectinesterase 342 3e-94
At1g53830 unknown protein 338 6e-93
At3g06830 pectin methylesterase like protein 331 6e-91
At1g53840 unknown protein 327 8e-90
At5g53370 pectinesterase 326 2e-89
At3g14300 putative pectin methylesterase 323 1e-88
At5g04960 pectinesterase 322 4e-88
At1g11580 unknown protein 322 4e-88
At3g49220 pectinesterase - like protein 311 6e-85
At3g43270 pectinesterase like protein 310 2e-84
At1g23200 putative pectinesterase 300 1e-81
>At2g47030 putative pectinesterase
Length = 588
Score = 417 bits (1072), Expect = e-117
Identities = 240/608 (39%), Positives = 349/608 (56%), Gaps = 56/608 (9%)
Query: 1 MSGKVIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRNLRIMCQNSQDQKLCHETLS 60
M GKV++S S++L+VGVAIGVV + KNG D + Q + ++ +CQ++ D+ C +TL
Sbjct: 1 MIGKVVVSVASILLIVGVAIGVVAFINKNG-DANLSPQMKAVQGICQSTSDKASCVKTLE 59
Query: 61 SVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGA--KMALNDCKDLMQF 118
V + DP I A + A D + K+ N + + G K L+ CK + +
Sbjct: 60 PVK---SEDPNKLIKAFMLATKDELTKSSNFTGQTEVNMGSSISPNNKAVLDYCKRVFMY 116
Query: 119 ALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFH 178
AL+ L + + ++ + + +I ++ WL V +Y+ C++ ++ +D K I E
Sbjct: 117 ALEDLATIIEEMGED-LSQIGSKIDQLKQWLIGVYNYQTDCLDDIEE-DDLRKAIGE--- 171
Query: 179 VQSLDSVQKVTAVALDIVTGLSDILQQFNLKFD-------------VKPA---------- 215
+ + + +T A+DI + + + N K D P
Sbjct: 172 --GIANSKILTTNAIDIFHTVVSAMAKINNKVDDLKNMTGGIPTPGAPPVVDESPVADPD 229
Query: 216 --SRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQR-----KNWRANIMPNAVVAKDGSGQ 268
+RRLL +D+ G P+W+S +DRKL+AK R + A + N VVAKDGSGQ
Sbjct: 230 GPARRLLED---IDETGIPTWVSGADRKLMAKAGRGRRGGRGGGARVRTNFVVAKDGSGQ 286
Query: 269 FKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFA 328
FKT+Q A+ + P+ N+GR +IY+KAG+Y E + +PK NI M+GDG +T+++ +S A
Sbjct: 287 FKTVQQAVDACPENNRGRCIIYIKAGLYREQVIIPKKKNNIFMFGDGARKTVISYNRSVA 346
Query: 329 A--GVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQD 386
G T +AT + GF+ K M F+NTAGP GHQA A R GD + + C GYQD
Sbjct: 347 LSRGTTTSLSATVQVESEGFMAKWMGFKNTAGPMGHQAAAIRVNGDRAVIFNCRFDGYQD 406
Query: 387 SLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDT-MNL 445
+LYV + RQ+YRNC+VSGTVDFIFG SAT+IQ++ I+VRK KGQ+NT+TADG++ + +
Sbjct: 407 TLYVNNGRQFYRNCVVSGTVDFIFGKSATVIQNTLIVVRKGSKGQYNTVTADGNELGLGM 466
Query: 446 NTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGE 505
GIV+Q+C I+P+ L PER T+ +YLGRPWK + TV+M + +GD I P+GW IW GE
Sbjct: 467 KIGIVLQNCRIVPDRKLTPERLTVATYLGRPWKKFSTTVIMSTEMGDLIRPEGWKIWDGE 526
Query: 506 QNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNG 565
H +C Y EY N GPGA RRV W S AE N FTA WL GP W+
Sbjct: 527 SFHKSCRYVEYNNRGPGAFANRRVNWAKV--ARSAAEVNGFTAANWL--GP---INWIQE 579
Query: 566 LHVPHYLG 573
+VP +G
Sbjct: 580 ANVPVTIG 587
>At2g47040 putative pectinesterase
Length = 595
Score = 402 bits (1033), Expect = e-112
Identities = 237/615 (38%), Positives = 343/615 (55%), Gaps = 63/615 (10%)
Query: 1 MSGKVIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRNLRIMCQNSQDQKLCHETLS 60
M GKV++S S++L+VGVAIGVV + KNG D + Q + +R +C+ + D+ C +TL
Sbjct: 1 MIGKVVVSVASILLIVGVAIGVVAYINKNG-DANLSPQMKAVRGICEATSDKASCVKTLE 59
Query: 61 SVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGK--ENGAKMALNDCKDLMQF 118
V + DP I A + A D + ++ N + + G K L+ CK + +
Sbjct: 60 PVK---SDDPNKLIKAFMLATRDAITQSSNFTGKTEGNLGSGISPNNKAVLDYCKKVFMY 116
Query: 119 ALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFH 178
AL+ L + + ++ + + +I ++ WL+ V +Y+ C++ ++ +D K I E
Sbjct: 117 ALEDLSTIVEEMGED-LNQIGSKIDQLKQWLTGVYNYQTDCLDDIEE-DDLRKTIGE--- 171
Query: 179 VQSLDSVQKVTAVALDIVTGLSDILQQFNLKF---------------------------- 210
+ S + +T+ A+DI + + + NLK
Sbjct: 172 --GIASSKILTSNAIDIFHTVVSAMAKLNLKVEDFKNMTGGIFAPSDKGAAPVNKGTPPV 229
Query: 211 -------DVKPASRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQR--KNWRANIMPNAVV 261
D +RRLL +D+ G P+W+S +DRKL+ K R + A I VV
Sbjct: 230 ADDSPVADPDGPARRLLED---IDETGIPTWVSGADRKLMTKAGRGSNDGGARIRATFVV 286
Query: 262 AKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIV 321
AKDGSGQFKT+Q A+ + P+ N GR +I++KAG+Y E + +PK NI M+GDG +T++
Sbjct: 287 AKDGSGQFKTVQQAVNACPEKNPGRCIIHIKAGIYREQVIIPKKKNNIFMFGDGARKTVI 346
Query: 322 TGRKS--FAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGC 379
+ +S + G T + T + GF+ K + F+NTAGP GHQAVA R GD + + C
Sbjct: 347 SYNRSVKLSPGTTTSLSGTVQVESEGFMAKWIGFKNTAGPMGHQAVAIRVNGDRAVIFNC 406
Query: 380 HIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADG 439
GYQD+LYV + RQ+YRN +VSGTVDFIFG SAT+IQ+S I+VRK KGQFNT+TADG
Sbjct: 407 RFDGYQDTLYVNNGRQFYRNIVVSGTVDFIFGKSATVIQNSLIVVRKGNKGQFNTVTADG 466
Query: 440 SDT-MNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDG 498
++ + + GIV+Q+C I+P+ L ER + SYLGRPWK + TV++ S IGD I P+G
Sbjct: 467 NEKGLAMKIGIVLQNCRIVPDKKLAAERLIVESYLGRPWKKFSTTVIINSEIGDVIRPEG 526
Query: 499 WTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKS 558
W IW GE H +C Y EY N GPGA RRV W S AE N FT WL GP
Sbjct: 527 WKIWDGESFHKSCRYVEYNNRGPGAITNRRVNWVKI--ARSAAEVNDFTVANWL--GP-- 580
Query: 559 AAEWLNGLHVPHYLG 573
W+ +VP LG
Sbjct: 581 -INWIQEANVPVTLG 594
>At3g62170 PECTINESTERASE-like protein
Length = 588
Score = 390 bits (1001), Expect = e-108
Identities = 234/604 (38%), Positives = 336/604 (54%), Gaps = 53/604 (8%)
Query: 3 GKVIISAVSLILVVGVAIGVVVAVRKNG-----EDPEVQTQQRNLRIMCQNSQDQKLCHE 57
GKV++S SL+LVVGVAIGV+ V K G + + + Q+ ++ +CQ++ DQ C +
Sbjct: 4 GKVVVSVASLLLVVGVAIGVITFVNKGGGANGDSNGPINSHQKAVQTICQSTTDQGSCAK 63
Query: 58 TLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGA--KMALNDCKDL 115
TL V + DP + A + A D + K+ N + G A K L+ CK +
Sbjct: 64 TLDPVK---SDDPSKLVKAFLMATKDAITKSSNFTASTEGGMGTNMNATSKAVLDYCKRV 120
Query: 116 MQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKE 175
+ +AL+ L+ + + ++ +Q ++ ++ WL+ V +Y+ C+ DD + E K
Sbjct: 121 LMYALEDLETIVEEMGED-LQQSGTKLDQLKQWLTGVFNYQTDCL---DDIEEVELK--- 173
Query: 176 QFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFD----------VKPASRRLLNSEVT 225
+ + + + + +T+ A+DI + + Q +K D A+RRLL
Sbjct: 174 KIMGEGISNSKVLTSNAIDIFHSVVTAMAQMGVKVDDMKNITMGAGAGGAARRLLEDN-- 231
Query: 226 VDDQGYPSWISSSDRKLLAKMQRK-------------NWRANIMPNAVVAKDGSGQFKTI 272
D +G P W S DRKL+AK R I VVAKDGSGQFKTI
Sbjct: 232 -DSKGLPKWFSGKDRKLMAKAGRGAPAGGDDGIGEGGGGGGKIKATHVVAKDGSGQFKTI 290
Query: 273 QAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKS--FAAG 330
A+ + P N GR +I++KAG+Y+E + +PK NI M+GDG +TI+T +S + G
Sbjct: 291 SEAVMACPDKNPGRCIIHIKAGIYNEQVRIPKKKNNIFMFGDGATQTIITFDRSVKLSPG 350
Query: 331 VKTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYV 390
T + T + GF+ K + F+NTAGP GHQAVA R GD + + C GYQD+LYV
Sbjct: 351 TTTSLSGTVQVESEGFMAKWIGFKNTAGPLGHQAVALRVNGDRAVIFNCRFDGYQDTLYV 410
Query: 391 QSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDT-MNLNTGI 449
+ RQ+YRN +VSGTVDFIFG SAT+IQ+S I+VRK GQ N +TADG++ + GI
Sbjct: 411 NNGRQFYRNIVVSGTVDFIFGKSATVIQNSLILVRKGSPGQSNYVTADGNEKGAAMKIGI 470
Query: 450 VIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHN 509
V+ +C IIP+ L ++ TI+SYLGRPWK A TV++ + IGD I P+GWT WQGEQNH
Sbjct: 471 VLHNCRIIPDKELEADKLTIKSYLGRPWKKFATTVIIGTEIGDLIKPEGWTEWQGEQNHK 530
Query: 510 TCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVP 569
T Y E+ N GPGA +R W S AE +T W+ GP A W+ +VP
Sbjct: 531 TAKYIEFNNRGPGAATTQRPPW--VKVAKSAAEVETYTVANWV--GP---ANWIQEANVP 583
Query: 570 HYLG 573
LG
Sbjct: 584 VQLG 587
>At4g33230 pectinesterase - like protein
Length = 609
Score = 365 bits (937), Expect = e-101
Identities = 210/536 (39%), Positives = 306/536 (56%), Gaps = 29/536 (5%)
Query: 42 LRIMCQNSQDQKLCHETLSSVHGADA--ADPKAYIAASVKAATDNVIKAFNMSERLTTEY 99
++ +C ++ + C TL + D DP++ + +++ A D++ + F L TE
Sbjct: 95 IQTLCNSTLYKPTCQNTLKNETKKDTPQTDPRSLLKSAIVAVNDDLDQVFKRVLSLKTEN 154
Query: 100 GKENGAKMALNDCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQAC 159
+ K A+ CK L+ A + L S K ++D+ + + D+ +WLSAV+SY++ C
Sbjct: 155 KDD---KDAIAQCKLLVDEAKEELGTSMKRINDSEVNNFAKIVPDLDSWLSAVMSYQETC 211
Query: 160 MEGFDDANDGEKKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRL 219
++GF+ E K+K + ++ +S Q +T+ +L ++ L L K +R L
Sbjct: 212 VDGFE-----EGKLKTEIR-KNFNSSQVLTSNSLAMIKSLDGYLSSVP-----KVKTRLL 260
Query: 220 LNSEVTVDDQGY-PSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALAS 278
L + + + + SW+S+ +R++L + K + PNA VAKDGSG F TI AAL +
Sbjct: 261 LEARSSAKETDHITSWLSNKERRMLKAVDVKA----LKPNATVAKDGSGNFTTINAALKA 316
Query: 279 YPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTAT 338
P +GRY IY+K G+YDE + + K N+ M GDG +TIVTG KS A ++T TAT
Sbjct: 317 MPAKYQGRYTIYIKHGIYDESVIIDKKKPNVTMVGDGSQKTIVTGNKSHAKKIRTFLTAT 376
Query: 339 FANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYR 398
F GF+ ++M F NTAGP+GHQAVA R Q D S + C GYQD+LY ++RQYYR
Sbjct: 377 FVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQSDRSVFLNCRFEGYQDTLYAYTHRQYYR 436
Query: 399 NCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIP 458
+C++ GTVDFIFG +A + Q+ I +RK GQ NT+TA G TG VI +C + P
Sbjct: 437 SCVIIGTVDFIFGDAAAIFQNCDIFIRKGLPGQKNTVTAQGRVDKFQTTGFVIHNCTVAP 496
Query: 459 EAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQ-GEQNHNTCYYAEYA 517
L P + +SYLGRPWK ++TVVMESTI D I P GW WQ + +T YAEY
Sbjct: 497 NEDLKPVKAQFKSYLGRPWKPHSRTVVMESTIEDVIDPVGWLRWQETDFAIDTLSYAEYK 556
Query: 518 NTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
N GP A RVKW G+ V+++ EA KFT G +LQ EW+ + P LG
Sbjct: 557 NDGPSGATAARVKWPGFR-VLNKEEAMKFTVGPFLQ------GEWIQAIGSPVKLG 605
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 360 bits (925), Expect = e-100
Identities = 212/572 (37%), Positives = 314/572 (54%), Gaps = 32/572 (5%)
Query: 5 VIISAVSLI-LVVGVAIGVVVAVRKNGEDPEVQTQQRNLRIMCQNSQDQKLCHETLSSVH 63
+ IS+V LI +VV V IGV V N D E+ T + ++ +C + ++ C +TL
Sbjct: 19 ISISSVLLISMVVAVTIGVSVNKSDNAGDEEITTSVKAIKDVCAPTDYKETCEDTLRK-D 77
Query: 64 GADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFALDSL 123
D +DP + + A + S+ + E K+ AKMAL+ CK+LM +A+ L
Sbjct: 78 AKDTSDPLELVKTAFNATMKQISDVAKKSQTMI-ELQKDPRAKMALDQCKELMDYAIGEL 136
Query: 124 DLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFD--DANDGEKKIKEQFHVQS 181
S + + V + + +R WLSA IS+ Q C++GF N GE K +
Sbjct: 137 SKSFEELGKFEFHKVDEALVKLRIWLSATISHEQTCLDGFQGTQGNAGETIKK------A 190
Query: 182 LDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWISSSDRK 241
L + ++T L +VT +S+ L Q + + SRRLL+ E +PSW+ + R+
Sbjct: 191 LKTAVQLTHNGLAMVTEMSNYLGQMQIP---EMNSRRLLSQE-------FPSWMDARARR 240
Query: 242 LLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYIT 301
LL + + P+ VVA+DGSGQ+KTI AL PK +V+++K G+Y EY+
Sbjct: 241 LLNAPM-----SEVKPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHIKEGIYKEYVQ 295
Query: 302 VPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDG 361
V + +++ GDGP +T+++G KS+ G+ T +TAT A FI K + FENTAG
Sbjct: 296 VNRSMTHLVFIGDGPDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNIAFENTAGAIK 355
Query: 362 HQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHST 421
HQAVA R D S C GYQD+LY S+RQ+YR+C +SGT+DF+FG +A + Q+ T
Sbjct: 356 HQAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFGDAAAVFQNCT 415
Query: 422 IIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLA 481
++VRKP Q ITA G +TG V+Q C I+ E + ++YLGRPWK +
Sbjct: 416 LLVRKPLLNQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKTYLGRPWKEYS 475
Query: 482 KTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRA 541
+T++M + I DF+ P+GW W GE NT +Y+E NTGPGA + +RV W G +S
Sbjct: 476 RTIIMNTFIPDFVPPEGWQPWLGEFGLNTLFYSEVQNTGPGAAITKRVTWPGIK-KLSDE 534
Query: 542 EANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
E KFT ++Q W+ G VP+ LG
Sbjct: 535 EILKFTPAQYIQGD-----AWIPGKGVPYILG 561
>At5g49180 pectin methylesterase
Length = 571
Score = 360 bits (923), Expect = 2e-99
Identities = 214/586 (36%), Positives = 322/586 (54%), Gaps = 39/586 (6%)
Query: 4 KVIISAVSLILVVGVAIGVVVAVRKNGEDPE------VQTQQRNLRIMCQNSQDQKLCHE 57
K II+ V L+V + + V + +N E ++T + +C + ++ C
Sbjct: 11 KCIIAGVITALLVLMVVAVGITTSRNTSHSEKIVPVQIKTATTAVEAVCAPTDYKETCVN 70
Query: 58 TLSSVHGADAADP----KAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCK 113
+L D+ P K ++++ D++ KA S LT + + K AL C+
Sbjct: 71 SLMKA-SPDSTQPLDLIKLGFNVTIRSIEDSIKKA---SVELTAKAANDKDTKGALELCE 126
Query: 114 DLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKI 173
LM A D L +I + D + D+R WLS I+Y+Q CM+ F++ N +
Sbjct: 127 KLMNDATDDLKKCLDNFDGFSIPQIEDFVEDLRVWLSGSIAYQQTCMDTFEETNSKLSQD 186
Query: 174 KEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKF---DVKPASRRLLNSEVTVDDQG 230
++ S +++T+ L ++T +S++L +FN+ D+ +R+LL++E G
Sbjct: 187 MQKIFKTS----RELTSNGLAMITNISNLLGEFNVTGVTGDLGKYARKLLSAE-----DG 237
Query: 231 YPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIY 290
PSW+ + R+L+A + + N VVA DGSGQ+KTI AL + PK N+ +VIY
Sbjct: 238 IPSWVGPNTRRLMAT------KGGVKANVVVAHDGSGQYKTINEALNAVPKANQKPFVIY 291
Query: 291 VKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAG-VKTMQTATFANTAMGFIGK 349
+K GVY+E + V K ++ GDGP +T +TG ++ G VKT TAT A F K
Sbjct: 292 IKQGVYNEKVDVTKKMTHVTFIGDGPTKTKITGSLNYYIGKVKTYLTATVAINGDNFTAK 351
Query: 350 AMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFI 409
+ FENTAGP+GHQAVA R D++ C I GYQD+LYV S+RQ++R+C VSGTVDFI
Sbjct: 352 NIGFENTAGPEGHQAVALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRDCTVSGTVDFI 411
Query: 410 FGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTI 469
FG ++Q+ I+VRKP K Q ITA G +TG+V+Q+C+I E A P +
Sbjct: 412 FGDGIVVLQNCNIVVRKPMKSQSCMITAQGRSDKRESTGLVLQNCHITGEPAYIPVKSIN 471
Query: 470 RSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRV 529
++YLGRPWK ++T++M +TI D I P GW W G+ NT YYAEY N GPG+N A+RV
Sbjct: 472 KAYLGRPWKEFSRTIIMGTTIDDVIDPAGWLPWNGDFALNTLYYAEYENNGPGSNQAQRV 531
Query: 530 KWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLGFK 575
KW G +S +A +FT +L+ W+ VP+ F+
Sbjct: 532 KWPGIK-KLSPKQALRFTPARFLRGN-----LWIPPNRVPYMGNFQ 571
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 359 bits (921), Expect = 3e-99
Identities = 215/589 (36%), Positives = 319/589 (53%), Gaps = 38/589 (6%)
Query: 5 VIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRN-LRIMCQNSQDQKLCHETLSSVH 63
V++SA +L V G+ K E + LR C +++ +LC + +
Sbjct: 21 VLLSAAVALLFVAAVAGISAGASKANEKRTLSPSSHAVLRSSCSSTRYPELCISAVVTAG 80
Query: 64 GADAADPKAYIAASVKAATDNVI-KAFNMSERLTTEYGKENGAKMALNDCKDLMQFALDS 122
G + K I ASV V F + + + G K AL+DC + + LD
Sbjct: 81 GVELTSQKDVIEASVNLTITAVEHNYFTVKKLIKKRKGLTPREKTALHDCLETIDETLDE 140
Query: 123 LDLSTKCVH-DNNIQAVHDQIADMRNWLSAVISYRQACMEGF--DDANDGEKK--IKEQF 177
L + + +H + + + D++ +S+ I+ ++ C++GF DDA+ +K +K Q
Sbjct: 141 LHETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSHDDADKQVRKALLKGQI 200
Query: 178 HVQSLDSVQKVTAVALDIVTGLSDI-LQQFNLKFDVKPASRRLL--NSEVTV-------- 226
HV+ + S AL ++ ++D + F K + +R+L N E TV
Sbjct: 201 HVEHMCSN------ALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQETTVAVDIAGAG 254
Query: 227 --DDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNK 284
D +G+P+W+S+ DR+LL + + +A VA DGSG FKT+ AA+A+ P+ +
Sbjct: 255 ELDSEGWPTWLSAGDRRLLQG-------SGVKADATVAADGSGTFKTVAAAVAAAPENSN 307
Query: 285 GRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAM 344
RYVI++KAGVY E + V K NI+ GDG RTI+TG ++ G T +AT A
Sbjct: 308 KRYVIHIKAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHSATVAAVGE 367
Query: 345 GFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSG 404
F+ + +TF+NTAGP HQAVA R D SA C ++ YQD+LYV SNRQ++ CL++G
Sbjct: 368 RFLARDITFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQFFVKCLIAG 427
Query: 405 TVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFP 464
TVDFIFG++A ++Q I R+P GQ N +TA G N NTGIVIQ C I + L
Sbjct: 428 TVDFIFGNAAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRIGATSDLQS 487
Query: 465 ERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGAN 524
+ + +YLGRPWK ++TV+M+S I D I P+GW+ W G NT Y EY+NTG GA
Sbjct: 488 VKGSFPTYLGRPWKEYSQTVIMQSAISDVIRPEGWSEWTGTFALNTLTYREYSNTGAGAG 547
Query: 525 VARRVKWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
A RVKW+G+ + + AEA K+TAG ++ G WL+ P LG
Sbjct: 548 TANRVKWRGFKVITAAAEAQKYTAGQFIGGG-----GWLSSTGFPFSLG 591
>At3g05610 putative pectinesterase
Length = 669
Score = 355 bits (911), Expect = 4e-98
Identities = 206/581 (35%), Positives = 315/581 (53%), Gaps = 35/581 (6%)
Query: 4 KVIISAVSLILVVGVAIGVVVAVRKNGED------PEVQTQQRNLRIMCQNSQDQKLCHE 57
+ I+ +S +L++ + + V V V N D EV + ++ +C + +K C +
Sbjct: 14 RYIVITISSVLLISMVVAVTVGVSLNKHDGDSKGKAEVNASVKAVKDVCAPTDYRKTCED 73
Query: 58 TLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQ 117
TL +G + DP + + + A S+ + E K++ +MAL+ CK+LM
Sbjct: 74 TLIK-NGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIM-ELQKDSRTRMALDQCKELMD 131
Query: 118 FALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFD--DANDGEKKIKE 175
+ALD L S + + + + + ++R WLSA IS+ + C+EGF N GE K
Sbjct: 132 YALDELSNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGFQGTQGNAGETMKK- 190
Query: 176 QFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWI 235
+L + ++T L I++ +S+ + Q + SRRLL +G+PSW+
Sbjct: 191 -----ALKTAIELTHNGLAIISEMSNFVGQMQIP---GLNSRRLLA-------EGFPSWV 235
Query: 236 SSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGV 295
RKLL Q +++ P+ VVA+DGSGQ+KTI AL PK +V+++KAG+
Sbjct: 236 DQRGRKLL---QAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVHIKAGL 292
Query: 296 YDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFEN 355
Y EY+ V K +++ GDGP +TI++G K++ G+ T +TAT A FI K + FEN
Sbjct: 293 YKEYVQVNKTMSHLVFIGDGPDKTIISGNKNYKDGITTYRTATVAIVGNYFIAKNIGFEN 352
Query: 356 TAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSAT 415
TAG HQAVA R Q D S C GYQD+LY S+RQ++R+C +SGT+DF+FG +A
Sbjct: 353 TAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFLFGDAAA 412
Query: 416 LIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGR 475
+ Q+ T++VRKP Q ITA G +TG V Q C I E + T ++YLGR
Sbjct: 413 VFQNCTLLVRKPLPNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSKAYLGR 472
Query: 476 PWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYH 535
PWK ++T++M + I DF+ P GW W G+ T +Y+E NTGPG+ +A RV W G
Sbjct: 473 PWKEYSRTIIMNTFIPDFVQPQGWQPWLGDFGLKTLFYSEVQNTGPGSALANRVTWAGIK 532
Query: 536 GVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLGFKA 576
+S + KFT ++Q +W+ G VP+ G A
Sbjct: 533 -TLSEEDILKFTPAQYIQGD-----DWIPGKGVPYTTGLLA 567
>At2g26450 putative pectinesterase
Length = 496
Score = 350 bits (898), Expect = 1e-96
Identities = 199/524 (37%), Positives = 298/524 (55%), Gaps = 36/524 (6%)
Query: 53 KLCHETLSSV--HGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALN 110
++C +TL + G +P ++ ++++A +++ L TE + K A+
Sbjct: 2 QICEKTLKNRTDKGFALDNPTTFLKSAIEAVNEDLDLVLEKVLSLKTENQDD---KDAIE 58
Query: 111 DCKDLMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGE 170
CK L++ A + S ++ + + + D+ +WLSAV+SY++ C++GF++ N
Sbjct: 59 QCKLLVEDAKEETVASLNKINVTEVNSFEKVVPDLESWLSAVMSYQETCLDGFEEGN--- 115
Query: 171 KKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQG 230
+K + S++S Q +T+ +L ++ ++ NL +K R LL+
Sbjct: 116 --LKSEVKT-SVNSSQVLTSNSLALIKTFTE-----NLSPVMKVVERHLLDD-------- 159
Query: 231 YPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIY 290
PSW+S+ DR++L + K + PNA VAKDGSG F TI AL + P+ +GRY+IY
Sbjct: 160 IPSWVSNDDRRMLRAVDVKA----LKPNATVAKDGSGDFTTINDALRAMPEKYEGRYIIY 215
Query: 291 VKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKA 350
VK G+YDEY+TV K N+ M GDG +TIVTG KS A ++T TATF GF+ ++
Sbjct: 216 VKQGIYDEYVTVDKKKANLTMVGDGSQKTIVTGNKSHAKKIRTFLTATFVAQGEGFMAQS 275
Query: 351 MTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIF 410
M F NTAGP+GHQAVA R Q D S + C GYQD+LY ++RQYYR+C++ GT+DFIF
Sbjct: 276 MGFRNTAGPEGHQAVAIRVQSDRSIFLNCRFEGYQDTLYAYTHRQYYRSCVIVGTIDFIF 335
Query: 411 GSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIR 470
G +A + Q+ I +RK GQ NT+TA G TG V+ +C I L P + +
Sbjct: 336 GDAAAIFQNCNIFIRKGLPGQKNTVTAQGRVDKFQTTGFVVHNCKIAANEDLKPVKEEYK 395
Query: 471 SYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQ-GEQNHNTCYYAEYANTGPGANVARRV 529
SYLGRPWK ++T++MES I + I P GW WQ + +T YYAEY N G + RV
Sbjct: 396 SYLGRPWKNYSRTIIMESKIENVIDPVGWLRWQETDFAIDTLYYAEYNNKGSSGDTTSRV 455
Query: 530 KWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
KW G+ VI++ EA +T G +LQ +W++ P LG
Sbjct: 456 KWPGFK-VINKEEALNYTVGPFLQ------GDWISASGSPVKLG 492
>At3g10710 putative pectinesterase
Length = 561
Score = 342 bits (877), Expect = 3e-94
Identities = 212/579 (36%), Positives = 309/579 (52%), Gaps = 54/579 (9%)
Query: 5 VIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRN-------LRIMCQNSQDQKLCHE 57
+ I AVSL+++ G+ IG V + + PE N ++ +C + ++ C E
Sbjct: 26 IAIIAVSLVILAGIVIGAVFGTMAHKKSPETVETNNNGDSISVSVKAVCDVTLHKEKCFE 85
Query: 58 TLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQ 117
TL S A + +P+ +VK V KA N ++ G E + +N C +L+
Sbjct: 86 TLGSAPNASSLNPEELFRYAVKITIAEVSKAINA---FSSSLGDEKN-NITMNACAELLD 141
Query: 118 FALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGF--DDANDGEKKIKE 175
+D+L+ +T N V + + D+R WLS+ +Y++ C+E D GE +K
Sbjct: 142 LTIDNLN-NTLTSSSNGDVTVPELVDDLRTWLSSAGTYQRTCVETLAPDMRPFGESHLKN 200
Query: 176 QFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNS-EVTVDDQGYPSW 234
++T+ AL I+T L I F L+ RRLL + +V VD
Sbjct: 201 S---------TELTSNALAIITWLGKIADSFKLR-------RRLLTTADVEVDFHAGRRL 244
Query: 235 ISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAG 294
+ S+D + +A + VVAKDGSG+++TI+ AL P+ ++ R +IYVK G
Sbjct: 245 LQSTDLRKVADI-------------VVAKDGSGKYRTIKRALQDVPEKSEKRTIIYVKKG 291
Query: 295 VYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFE 354
VY E + V K N+++ GDG +++IV+GR + G T +TATFA GF+ + M F
Sbjct: 292 VYFENVKVEKKMWNVIVVGDGESKSIVSGRLNVIDGTPTFKTATFAVFGKGFMARDMGFI 351
Query: 355 NTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSA 414
NTAGP HQAVA D++A C + YQD+LYV + RQ+YR C + GTVDFIFG+SA
Sbjct: 352 NTAGPSKHQAVALMVSADLTAFYRCTMNAYQDTLYVHAQRQFYRECTIIGTVDFIFGNSA 411
Query: 415 TLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLG 474
+++Q I+ R+P KGQ NTITA G N+NTGI I CNI P L + ++LG
Sbjct: 412 SVLQSCRILPRRPMKGQQNTITAQGRTDPNMNTGISIHRCNISPLGDL----TDVMTFLG 467
Query: 475 RPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGY 534
RPWK + TV+M+S + FI GW W G+ +T +Y EY NTGPGA+ RVKWKG
Sbjct: 468 RPWKNFSTTVIMDSYLHGFIDRKGWLPWTGDSAPDTIFYGEYKNTGPGASTKNRVKWKGL 527
Query: 535 HGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
+S EAN+FT ++ G WL VP G
Sbjct: 528 R-FLSTKEANRFTVKPFIDGG-----RWLPATKVPFRSG 560
>At1g53830 unknown protein
Length = 587
Score = 338 bits (866), Expect = 6e-93
Identities = 201/569 (35%), Positives = 307/569 (53%), Gaps = 34/569 (5%)
Query: 5 VIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRN----LRIMCQNSQDQKLCHETLS 60
++ SA +L++ +G+ ++ ++ T L+ +C ++ +LC ++
Sbjct: 21 ILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHAILKSVCSSTLYPELCFSAVA 80
Query: 61 SVHGADAADPKAYIAASVKAATDNVI-KAFNMSERLTTEYGKENGAKMALNDCKDLMQFA 119
+ G + K I AS+ T V F + + + G AL+DC + +
Sbjct: 81 ATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREVTALHDCLETIDET 140
Query: 120 LDSLDLSTKCVHDNNIQ-AVHDQIADMRNWLSAVISYRQACMEGF--DDANDGEKK--IK 174
LD L ++ + +H Q ++ D++ +S+ I+ + C++GF DDA+ +K +K
Sbjct: 141 LDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGFSYDDADRKVRKALLK 200
Query: 175 EQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNS------EVT--V 226
Q HV+ + S A+A+ + ++ + F+++ S N+ EVT +
Sbjct: 201 GQVHVEHMCS----NALAM-----IKNMTETDIANFELRDKSSTFTNNNNRKLKEVTGDL 251
Query: 227 DDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGR 286
D G+P W+S DR+LL + I +A VA DGSG F T+ AA+A+ P+ + R
Sbjct: 252 DSDGWPKWLSVGDRRLLQG-------STIKADATVADDGSGDFTTVAAAVAAAPEKSNKR 304
Query: 287 YVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGF 346
+VI++KAGVY E + V K NI+ GDG +TI+TG ++ G T +AT A F
Sbjct: 305 FVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFHSATVAAVGERF 364
Query: 347 IGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTV 406
+ + +TF+NTAGP HQAVA R D SA C + YQD+LYV SNRQ++ C ++GTV
Sbjct: 365 LARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQFFVKCHITGTV 424
Query: 407 DFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPER 466
DFIFG++A ++Q I R+P GQ N +TA G N NTGIVIQ+C I + L +
Sbjct: 425 DFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCRIGGTSDLLAVK 484
Query: 467 FTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVA 526
T +YLGRPWK ++TV+M+S I D I P+GW W G +T Y EY N G GA A
Sbjct: 485 GTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEWSGSFALDTLTYREYLNRGGGAGTA 544
Query: 527 RRVKWKGYHGVISRAEANKFTAGIWLQAG 555
RVKWKGY + S EA FTAG ++ G
Sbjct: 545 NRVKWKGYKVITSDTEAQPFTAGQFIGGG 573
>At3g06830 pectin methylesterase like protein
Length = 568
Score = 331 bits (849), Expect = 6e-91
Identities = 195/540 (36%), Positives = 309/540 (57%), Gaps = 29/540 (5%)
Query: 5 VIISAVSLILVVGVAIGVVVAVRKNGEDPE--VQTQQRNLRIMCQNSQDQKLCHETLSSV 62
++ +VS LV+ V VV + + +D E ++ + ++ +C + + C +L
Sbjct: 12 IVAGSVSGFLVIMVVSVAVVTSKHSPKDDENHIRKTTKAVQAVCAPTDFKDTCVNSLMGA 71
Query: 63 HGADAADP----KAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQF 118
D+ DP K ++K+ +++ KA S + + K AK A C+ LM
Sbjct: 72 -SPDSDDPVDLIKLGFKVTIKSINESLEKA---SGDIKAKADKNPEAKGAFELCEKLM-- 125
Query: 119 ALDSLDLSTKCV-HDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQF 177
+D++D KC+ H ++ + + D+R WLS I+++Q CM+ F + K Q
Sbjct: 126 -IDAIDDLKKCMDHGFSVDQIEVFVEDLRVWLSGSIAFQQTCMDSFGEI----KSNLMQD 180
Query: 178 HVQSLDSVQKVTAVALDIVTGLSDILQQFNLKF---DVKPASRRLLNSEVTVDDQGYPSW 234
++ + +++++ +L +VT +S ++ NL + +R+LL++E ++ P+W
Sbjct: 181 MLKIFKTSRELSSNSLAMVTRISTLIPNSNLTGLTGALAKYARKLLSTEDSI-----PTW 235
Query: 235 ISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAG 294
+ R+L+A + + NAVVA+DG+GQFKTI AL + PKGNK ++I++K G
Sbjct: 236 VGPEARRLMAA--QGGGPGPVKANAVVAQDGTGQFKTITDALNAVPKGNKVPFIIHIKEG 293
Query: 295 VYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAG-VKTMQTATFANTAMGFIGKAMTF 353
+Y E +TV K ++ GDGP +T++TG +F G VKT TAT F K +
Sbjct: 294 IYKEKVTVTKKMPHVTFIGDGPNKTLITGSLNFGIGKVKTFLTATITIEGDHFTAKNIGI 353
Query: 354 ENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSS 413
ENTAGP+G QAVA R D + C I G+QD+LYV S+RQ+YR+C VSGTVDFIFG +
Sbjct: 354 ENTAGPEGGQAVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTVSGTVDFIFGDA 413
Query: 414 ATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYL 473
++Q+ I+VRKP KGQ +TA G + +TG+V+ C+I + A P + ++YL
Sbjct: 414 KCILQNCKIVVRKPNKGQTCMVTAQGRSNVRESTGLVLHGCHITGDPAYIPMKSVNKAYL 473
Query: 474 GRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKG 533
GRPWK ++T++M++TI D I P GW W G+ T YYAE+ NTGPG+N A+RVKW G
Sbjct: 474 GRPWKEFSRTIIMKTTIDDVIDPAGWLPWSGDFALKTLYYAEHMNTGPGSNQAQRVKWPG 533
>At1g53840 unknown protein
Length = 586
Score = 327 bits (839), Expect = 8e-90
Identities = 199/574 (34%), Positives = 301/574 (51%), Gaps = 39/574 (6%)
Query: 4 KVIISAVSLILVVGVAIGVVVA--VRKNGED-----PEVQTQQRNLRIMCQNSQDQKLCH 56
++++ ++S+++++ V I VVA V KN + P T +L+ +C ++ + C
Sbjct: 28 RLLLLSISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAICSVTRFPESCI 87
Query: 57 ETLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLM 116
++S + ++ DP+ S+K D + ++ E+L+ E E K AL C DL+
Sbjct: 88 SSISKLPSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDER-IKSALRVCGDLI 146
Query: 117 QFALDSLDLSTKCVHDNNIQAV--HDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIK 174
+ ALD L+ + + D + +I D++ WLSA ++ + C + D+ + +
Sbjct: 147 EDALDRLNDTVSAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCFDSLDELKQNKTEYA 206
Query: 175 EQFHVQSLDSVQ----KVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQG 230
Q+L S + T+ +L IV+ + L + RR L S
Sbjct: 207 NSTITQNLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIH----RRRRLMSHHHQQSVD 262
Query: 231 YPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIY 290
+ W + R+LL A + P+ VA DG+G T+ A+A PK + +VIY
Sbjct: 263 FEKW---ARRRLLQT-------AGLKPDVTVAGDGTGDVLTVNEAVAKVPKKSLKMFVIY 312
Query: 291 VKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKA 350
VK+G Y E + + K N+++YGDG +TI++G K+F G T +TATFA GFI K
Sbjct: 313 VKSGTYVENVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFAIQGKGFIMKD 372
Query: 351 MTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIF 410
+ NTAG HQAVAFR+ D S C G+QD+LY SNRQ+YR+C V+GT+DFIF
Sbjct: 373 IGIINTAGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDCDVTGTIDFIF 432
Query: 411 GSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIR 470
GS+A + Q I+ R+P QFNTITA G N ++G+ IQ C I +
Sbjct: 433 GSAAVVFQGCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTISANGNVIAP----- 487
Query: 471 SYLGRPWKYLAKTVVMESTIGDFIHPDGWTIW-QGEQNHNTCYYAEYANTGPGANVARRV 529
+YLGRPWK + TV+ME+ IG + P GW W G + Y EY NTGPG++V +RV
Sbjct: 488 TYLGRPWKEFSTTVIMETVIGAVVRPSGWMSWVSGVDPPASIVYGEYKNTGPGSDVTQRV 547
Query: 530 KWKGYHGVISRAEANKFTAGIWLQAGPKSAAEWL 563
KW GY V+S AEA KFT L A+W+
Sbjct: 548 KWAGYKPVMSDAEAAKFTVATLLH-----GADWI 576
>At5g53370 pectinesterase
Length = 587
Score = 326 bits (836), Expect = 2e-89
Identities = 202/575 (35%), Positives = 301/575 (52%), Gaps = 28/575 (4%)
Query: 4 KVIISAVSLILVVGVAIGVVVAVRK--NGEDPEVQTQQRNLRIM--CQNSQDQKLCHETL 59
K+I+ +++++V V G+ +R +G+ T++ I C S LC +TL
Sbjct: 33 KLILFTLAVLVVGVVCFGIFAGIRAVDSGKTEPKLTRKPTQAISRTCSKSLYPNLCIDTL 92
Query: 60 SSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFA 119
G+ AD I S A KA S +T + A + C +L+
Sbjct: 93 LDFPGSLTADENELIHISFNATLQKFSKALYTSSTITYTQMPPR-VRSAYDSCLELLD-- 149
Query: 120 LDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFHV 179
DS+D T+ + + + + +D+ WLS+ ++ C +GFD+ ++K+Q +
Sbjct: 150 -DSVDALTRALSSVVVVSGDESHSDVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQV-I 207
Query: 180 QSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWISSSD 239
++ + ++ + L I G L + +R+LL +E T + P+W+ D
Sbjct: 208 GAVKDLSEMVSNCLAIFAGKVKDLSGVPVV-----NNRKLLGTEETEE---LPNWLKRED 259
Query: 240 RKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEY 299
R+LL + I + V+KDGSG FKTI A+ P+ + R+VIYVKAG Y+E
Sbjct: 260 RELLGTPT-----SAIQADITVSKDGSGTFKTIAEAIKKAPEHSSRRFVIYVKAGRYEEE 314
Query: 300 -ITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAG 358
+ V + N++ GDG +T++TG KS A + T TATFA T GFI + MTFEN AG
Sbjct: 315 NLKVGRKKTNLMFIGDGKGKTVITGGKSIADDLTTFHTATFAATGAGFIVRDMTFENYAG 374
Query: 359 PDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQ 418
P HQAVA R GD + + C+I+GYQD+LYV SNRQ++R C + GTVDFIFG++A ++Q
Sbjct: 375 PAKHQAVALRVGGDHAVVYRCNIIGYQDALYVHSNRQFFRECEIYGTVDFIFGNAAVILQ 434
Query: 419 HSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWK 478
I RKP Q TITA N NTGI I C ++ L + + +YLGRPWK
Sbjct: 435 SCNIYARKPMAQQKITITAQNRKDPNQNTGISIHACKLLATPDLEASKGSYPTYLGRPWK 494
Query: 479 YLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVI 538
++ V M S +GD I P GW W G ++ YY EY N G G+ + +RVKW GYH +
Sbjct: 495 LYSRVVYMMSDMGDHIDPRGWLEWNGPFALDSLYYGEYMNKGLGSGIGQRVKWPGYHVIT 554
Query: 539 SRAEANKFTAGIWLQAGPKSAAEWLNGLHVPHYLG 573
S EA+KFT ++ S + WL V + G
Sbjct: 555 STVEASKFTVAQFI-----SGSSWLPSTGVSFFSG 584
>At3g14300 putative pectin methylesterase
Length = 968
Score = 323 bits (829), Expect = 1e-88
Identities = 194/524 (37%), Positives = 277/524 (52%), Gaps = 42/524 (8%)
Query: 42 LRIMCQNSQDQKLCHETLSSVH-GADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYG 100
LR +C + C ++S + DPK S++ D + + ++L E
Sbjct: 459 LRTVCNVTNYPASCISSISKLPLSKTTTDPKVLFRLSLQVTFDELNSIVGLPKKLAEETN 518
Query: 101 KENGAKMALNDCKDLMQFALDSLDLSTKCVHD------NNIQAVHDQIADMRNWLSAVIS 154
E G K AL+ C D+ A+DS++ + + + N+ + I D+ WLS+ ++
Sbjct: 519 DE-GLKSALSVCADVFDLAVDSVNDTISSLDEVISGGKKNLNS--STIGDLITWLSSAVT 575
Query: 155 YRQACMEGFDDANDGE---KKIKEQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFD 211
C + D+ N +K+K V S + A+ ++ S K
Sbjct: 576 DIGTCGDTLDEDNYNSPIPQKLKSAM-VNSTEFTSNSLAIVAQVLKKPS--------KSR 626
Query: 212 VKPASRRLLNSEVTVDDQGYPSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKT 271
+ RRLLNS +P+W+ R+LL Q KN + P+ VA DGSG +T
Sbjct: 627 IPVQGRRLLNSN------SFPNWVRPGVRRLL---QAKN----LTPHVTVAADGSGDVRT 673
Query: 272 IQAALASYPKGNKGRYVIYVKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGV 331
+ A+ PK K +VIYVKAG Y E + + KD N+ +YGDG +TI++G + GV
Sbjct: 674 VNEAVWRVPKKGKTMFVIYVKAGTYVENVLMKKDKWNVFIYGDGRDKTIISGSTNMVDGV 733
Query: 332 KTMQTATFANTAMGFIGKAMTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQ 391
+T T+TFA GF+ K M NTAGP+ HQAVAFR+ D S C GYQD+LY
Sbjct: 734 RTFNTSTFATEGKGFMMKDMGIINTAGPEKHQAVAFRSDSDRSVYYRCSFDGYQDTLYTH 793
Query: 392 SNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVI 451
SNRQYYRNC V+GTVDFIFG+ + Q +I R+P QFNTITA+G+ N NTGI I
Sbjct: 794 SNRQYYRNCDVTGTVDFIFGAGTVVFQGCSIRPRQPLPNQFNTITAEGTQEANQNTGISI 853
Query: 452 QDCNIIPEAALFPERFTIRSYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQG--EQNHN 509
C I P T +YLGRPWK +KTV+M+S IG F++P GW W +
Sbjct: 854 HQCTISPNG-----NVTATTYLGRPWKLFSKTVIMQSVIGSFVNPAGWIAWNSTYDPPPR 908
Query: 510 TCYYAEYANTGPGANVARRVKWKGYHGVISRAEANKFTAGIWLQ 553
T +Y EY N+GPG+++++RVKW GY + S EA +FT +L+
Sbjct: 909 TIFYREYKNSGPGSDLSKRVKWAGYKPISSDDEAARFTVKYFLR 952
Score = 61.2 bits (147), Expect = 1e-09
Identities = 52/226 (23%), Positives = 97/226 (42%), Gaps = 22/226 (9%)
Query: 9 AVSLILVVGVAIGVVVAVRKNGED-------PEVQTQQRNLRIMCQNSQDQKLCHETLSS 61
AV + +++ + + + RK PE+ T +L+ +C + C ++S
Sbjct: 38 AVLVAIIISSTVTIAIHSRKGNSPHPTPSSVPEL-TPAASLKTVCSVTNYPVSCFSSISK 96
Query: 62 VHGADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFALD 121
+ ++ DP+ S++ D + + ++L E E G K AL+ C+ L+ A+D
Sbjct: 97 LPLSNTTDPEVIFRLSLQVVIDELNSIVELPKKLAEETDDE-GLKSALSVCEHLLDLAID 155
Query: 122 SLD--LSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFH- 178
++ +S V D I D+ WLSA ++Y C++ D+ + I +
Sbjct: 156 RVNETVSAMEVVDGKKILNAATIDDLLTWLSAAVTYHGTCLDALDEISHTNSAIPLKLKS 215
Query: 179 --VQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNS 222
V S + A+ I++ +SD F + RRLLNS
Sbjct: 216 GMVNSTEFTSNSLAIVAKILSTISD--------FGIPIHGRRLLNS 253
Score = 44.3 bits (103), Expect = 2e-04
Identities = 30/137 (21%), Positives = 65/137 (46%), Gaps = 4/137 (2%)
Query: 37 TQQRNLRIMCQNSQDQKLCHETLSSVHGADAADPKAYIAASVKAATDNVIKAFNMSERLT 96
T +LR +C ++ C ++S + ++ DP+A S++ + + + ++L
Sbjct: 266 TPAASLRNVCSVTRYPASCVSSISKLPSSNTTDPEALFRLSLQVVINELNSIAGLPKKLA 325
Query: 97 TEYGKENGAKMALNDCKDLMQFALDSLD--LST-KCVHDNNIQAVHDQIADMRNWLSAVI 153
E E K +L+ C D+ A+D ++ +ST + V D I +++ WLSA +
Sbjct: 326 EETDDER-LKSSLSVCGDVFNDAIDIVNDTISTMEEVGDGKKILKSSTIDEIQTWLSAAV 384
Query: 154 SYRQACMEGFDDANDGE 170
+ C++ D+ + +
Sbjct: 385 TDHDTCLDALDELSQNK 401
>At5g04960 pectinesterase
Length = 564
Score = 322 bits (825), Expect = 4e-88
Identities = 208/578 (35%), Positives = 304/578 (51%), Gaps = 54/578 (9%)
Query: 4 KVIISAVSLILVVGVAIGVVVAV--RKNGEDPEVQTQQR----NLRIMCQNSQDQKLCHE 57
++ I A+S I++V + +G VV R N + P + +++ +C + ++ C E
Sbjct: 24 RIAIIAISSIVLVCIVVGAVVGTTARDNSKKPPTENNGEPISVSVKALCDVTLHKEKCFE 83
Query: 58 TLSSVHGADAADPKAYIAASVKAAT---DNVIKAFNMSERLTTEYGKENGAKMALNDCKD 114
TL S A + P+ +VK V+ F+ E + +N A+ C +
Sbjct: 84 TLGSAPNASRSSPEELFKYAVKVTITELSKVLDGFSNGEHM------DNATSAAMGACVE 137
Query: 115 LMQFALDSLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIK 174
L+ A+D L+ + N D+R WLS+V +Y++ CM+ +AN K
Sbjct: 138 LIGLAVDQLNETMTSSLKN--------FDDLRTWLSSVGTYQETCMDALVEAN---KPSL 186
Query: 175 EQFHVQSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLL---NSEVTVDDQGY 231
F L + ++T+ AL I+T L I VK RRLL N++V V D
Sbjct: 187 TTFGENHLKNSTEMTSNALAIITWLGKIADT------VKFRRRRLLETGNAKVVVADLPM 240
Query: 232 PSWISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYV 291
R+LL K +A I VVAKDGSG+++TI ALA + N+ +IYV
Sbjct: 241 -----MEGRRLLESGDLKK-KATI----VVAKDGSGKYRTIGEALAEVEEKNEKPTIIYV 290
Query: 292 KAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAM 351
K GVY E + V K N++M GDG ++TIV+ +F G T +TATFA GF+ + M
Sbjct: 291 KKGVYLENVRVEKTKWNVVMVGDGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMARDM 350
Query: 352 TFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFG 411
F NTAGP HQAVA D+S C + +QD++Y + RQ+YR+C++ GTVDFIFG
Sbjct: 351 GFINTAGPAKHQAVALMVSADLSVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFIFG 410
Query: 412 SSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRS 471
++A + Q I+ R+P KGQ NTITA G N NTGI I +C I P I++
Sbjct: 411 NAAVVFQKCEILPRRPMKGQQNTITAQGRKDPNQNTGISIHNCTIKP----LDNLTDIQT 466
Query: 472 YLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKW 531
+LGRPWK + TV+M+S + FI+P GW W G+ +T +YAEY N+GPGA+ RVKW
Sbjct: 467 FLGRPWKDFSTTVIMKSFMDKFINPKGWLPWTGDTAPDTIFYAEYLNSGPGASTKNRVKW 526
Query: 532 KGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVP 569
+G +++ EANKFT ++ WL VP
Sbjct: 527 QGLKTSLTKKEANKFTVKPFIDGN-----NWLPATKVP 559
>At1g11580 unknown protein
Length = 557
Score = 322 bits (825), Expect = 4e-88
Identities = 187/568 (32%), Positives = 294/568 (50%), Gaps = 41/568 (7%)
Query: 5 VIISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQQRNLRIMCQNSQDQKLCHETLSSVHG 64
+++S V+++ V +++V N D + T + +C + DQ C LS
Sbjct: 22 LVLSFVAILGSVAFFTAQLISVNTNNNDDSLLTTSQ----ICHGAHDQDSCQALLSEFTT 77
Query: 65 ADAADPKAYIAASVKAATDNVIKAFNMSERLTTEYG-KENGA--KMALNDCKDLMQFALD 121
+ K + N + + + +E + NG K DC+++M + D
Sbjct: 78 LSLS--KLNRLDLLHVFLKNSVWRLESTMTMVSEARIRSNGVRDKAGFADCEEMMDVSKD 135
Query: 122 SLDLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFHVQS 181
+ S + + N + +++ WLS+V++ C+E D + K+I +
Sbjct: 136 RMMSSMEELRGGNYNL--ESYSNVHTWLSSVLTNYMTCLESISDVSVNSKQIVKPQLEDL 193
Query: 182 LDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGYPSWISSSDRK 241
+ + A+ + ++ D+ + +F PSW+++ DRK
Sbjct: 194 VSRARVALAIFVSVLPARDDLKMIISNRF---------------------PSWLTALDRK 232
Query: 242 LLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYIT 301
LL + + N VVAKDG+G+FKT+ A+A+ P+ + RYVIYVK GVY E I
Sbjct: 233 LLESSPKT---LKVTANVVVAKDGTGKFKTVNEAVAAAPENSNTRYVIYVKKGVYKETID 289
Query: 302 VPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDG 361
+ K N+++ GDG TI+TG + G T ++AT A GF+ + + F+NTAGP
Sbjct: 290 IGKKKKNLMLVGDGKDATIITGSLNVIDGSTTFRSATVAANGDGFMAQDIWFQNTAGPAK 349
Query: 362 HQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHST 421
HQAVA R D + + C I YQD+LY + RQ+YR+ ++GTVDFIFG+SA + Q+
Sbjct: 350 HQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYITGTVDFIFGNSAVVFQNCD 409
Query: 422 IIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLA 481
I+ R PG GQ N +TA G + N NT I IQ C I + L P + +++++LGRPWK +
Sbjct: 410 IVARNPGAGQKNMLTAQGREDQNQNTAISIQKCKITASSDLAPVKGSVKTFLGRPWKLYS 469
Query: 482 KTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRA 541
+TV+M+S I + I P GW W GE +T YY EYANTGPGA+ ++RV WKG+ +
Sbjct: 470 RTVIMQSFIDNHIDPAGWFPWDGEFALSTLYYGEYANTGPGADTSKRVNWKGFKVIKDSK 529
Query: 542 EANKFT------AGIWLQAGPKSAAEWL 563
EA +FT G+WL+ + EWL
Sbjct: 530 EAEQFTVAKLIQGGLWLKPTGVTFQEWL 557
>At3g49220 pectinesterase - like protein
Length = 598
Score = 311 bits (797), Expect = 6e-85
Identities = 195/575 (33%), Positives = 301/575 (51%), Gaps = 38/575 (6%)
Query: 6 IISAVSLILVVGVAIGVVVAVRKNGEDPEVQTQ-QRNLRIMCQNSQDQKLCHETLSSVHG 64
I+ A+SLIL + GV ++ N P + + + + C+ ++ +LC ++L G
Sbjct: 50 IVLAISLILAAAIFAGVRSRLKLNQSVPGLARKPSQAISKACELTRFPELCVDSLMDFPG 109
Query: 65 ADAADP-KAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFALDSL 123
+ AA K I +V + A S L+ A+ A + C +L+ ++D+L
Sbjct: 110 SLAASSSKDLIHVTVNMTLHHFSHALYSSASLSF-VDMPPRARSAYDSCVELLDDSVDAL 168
Query: 124 DLSTKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFHVQSLD 183
+ V ++ + D+ WLSA ++ C EGFD +DG K
Sbjct: 169 SRALSSVVSSSAKP-----QDVTTWLSAALTNHDTCTEGFDGVDDGGVK----------- 212
Query: 184 SVQKVTAVALDIVTGLSDILQQFNLKFD------VKPASRRLLNSEVTVDDQGYPSWISS 237
+TA ++ +S+ L F+ D V +RRLL E ++ +P W+
Sbjct: 213 --DHMTAALQNLSELVSNCLAIFSASHDGDDFAGVPIQNRRLLGVEER--EEKFPRWMRP 268
Query: 238 SDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYD 297
+R++L + I + +V+KDG+G KTI A+ P+ + R +IYVKAG Y+
Sbjct: 269 KEREILEMPV-----SQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYVKAGRYE 323
Query: 298 EY-ITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENT 356
E + V + +N++ GDG +T+++G KS + T TA+FA T GFI + +TFEN
Sbjct: 324 ENNLKVGRKKINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARDITFENW 383
Query: 357 AGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATL 416
AGP HQAVA R D + + C+I+GYQD+LYV SNRQ++R C + GTVDFIFG++A +
Sbjct: 384 AGPAKHQAVALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIFGNAAVV 443
Query: 417 IQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRP 476
+Q+ +I RKP Q NTITA N NTGI I ++ + L + ++YLGRP
Sbjct: 444 LQNCSIYARKPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQTYLGRP 503
Query: 477 WKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHG 536
WK ++TV M S IG +H GW W +T YY EY N+GPG+ + +RV W GY
Sbjct: 504 WKLFSRTVYMMSYIGGHVHTRGWLEWNTTFALDTLYYGEYLNSGPGSGLGQRVSWPGYRV 563
Query: 537 VISRAEANKFTAGIWLQAG---PKSAAEWLNGLHV 568
+ S AEAN+FT ++ P + +L GL +
Sbjct: 564 INSTAEANRFTVAEFIYGSSWLPSTGVSFLAGLSI 598
>At3g43270 pectinesterase like protein
Length = 527
Score = 310 bits (793), Expect = 2e-84
Identities = 186/505 (36%), Positives = 268/505 (52%), Gaps = 30/505 (5%)
Query: 70 PKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFALDSLD--LST 127
P A + K D + KA + + + GK + A+ DC DL+ A + L +S
Sbjct: 43 PPLEFAEAAKTVVDAITKAVAIVSKFDKKAGKSRVSN-AIVDCVDLLDSAAEELSWIISA 101
Query: 128 KCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDDANDGEKKIKEQFHVQSLDSVQK 187
+ + D +D+R W+SA +S + C++GF+ N KKI + K
Sbjct: 102 SQSPNGKDNSTGDVGSDLRTWISAALSNQDTCLDGFEGTNGIIKKIVA-------GGLSK 154
Query: 188 VTAVALDIVTGLSDILQQFNLKFDVKPASRRLLNSEVTVDDQGY---PSWISSSDRKLLA 244
V +++T + KP + + +T G+ PSW+ DRKLL
Sbjct: 155 VGTTVRNLLTMVHSPPS--------KPKPKPIKAQTMTKAHSGFSKFPSWVKPGDRKLL- 205
Query: 245 KMQRKNWRANIMPNAVVAKDGSGQFKTIQAALASYPKGNKGRYVIYVKAGVYDEYITVPK 304
Q N + +AVVA DG+G F TI A+ + P + RYVI+VK GVY E + + K
Sbjct: 206 --QTDNIT---VADAVVAADGTGNFTTISDAVLAAPDYSTKRYVIHVKRGVYVENVEIKK 260
Query: 305 DAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKAMTFENTAGPDGHQA 364
NI+M GDG T++TG +SF G T ++ATFA + GFI + +TF+NTAGP+ HQA
Sbjct: 261 KKWNIMMVGDGIDATVITGNRSFIDGWTTFRSATFAVSGRGFIARDITFQNTAGPEKHQA 320
Query: 365 VAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIFGSSATLIQHSTIIV 424
VA R+ D+ C + GYQD+LY S RQ++R C+++GTVDFIFG + + Q I
Sbjct: 321 VAIRSDTDLGVFYRCAMRGYQDTLYAHSMRQFFRECIITGTVDFIFGDATAVFQSCQIKA 380
Query: 425 RKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIRSYLGRPWKYLAKTV 484
++ Q N+ITA G N TG IQ NI + L T +YLGRPWK ++TV
Sbjct: 381 KQGLPNQKNSITAQGRKDPNEPTGFTIQFSNIAADTDLLLNLNTTATYLGRPWKLYSRTV 440
Query: 485 VMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVKWKGYHGVISRAEAN 544
M++ + D I+P GW W G +T YY EY N+GPGA++ RRVKW GYH + + AEAN
Sbjct: 441 FMQNYMSDAINPVGWLEWNGNFALDTLYYGEYMNSGPGASLDRRVKWPGYHVLNTSAEAN 500
Query: 545 KFTAGIWLQAG---PKSAAEWLNGL 566
FT +Q P + ++ GL
Sbjct: 501 NFTVSQLIQGNLWLPSTGITFIAGL 525
>At1g23200 putative pectinesterase
Length = 554
Score = 300 bits (768), Expect = 1e-81
Identities = 188/519 (36%), Positives = 260/519 (49%), Gaps = 45/519 (8%)
Query: 66 DAADPKAYIAASVKAATDNVIKAFNMSERLTTEYGKENGAKMALNDCKDLMQFALDSLDL 125
D D + V + D ++ + L + A AL DC +L + +D L+
Sbjct: 61 DQTDGFTFHDLVVSSTMDQAVQLHRLVSSLKQHHSLHKHATSALFDCLELYEDTIDQLN- 119
Query: 126 STKCVHDNNIQAVHDQIADMRNWLSAVISYRQACMEGFDD---ANDGEKKIKEQFH---V 179
H + D + LSA I+ + C GF D + K QFH
Sbjct: 120 -----HSRRSYGQYSSPHDRQTSLSAAIANQDTCRNGFRDFKLTSSYSKYFPVQFHRNLT 174
Query: 180 QSLDSVQKVTAVALDIVTGLSDILQQFNLKFDVKPAS------RRLLNSEVTVDDQGYPS 233
+S+ + VT A + KF + +S RRLL D+ +PS
Sbjct: 175 KSISNSLAVTKAAAEAEAVAEKYPSTGFTKFSKQRSSAGGGSHRRLL----LFSDEKFPS 230
Query: 234 WISSSDRKLLAKMQRKNWRANIMPNAVVAKDGSGQFKTIQAAL---ASYPKGNKGRYVIY 290
W SDRKLL ++ + + VVAKDGSG + +IQ A+ A P+ N+ R VIY
Sbjct: 231 WFPLSDRKLL-----EDSKTTAKADLVVAKDGSGHYTSIQQAVNAAAKLPRRNQ-RLVIY 284
Query: 291 VKAGVYDEYITVPKDAVNILMYGDGPARTIVTGRKSFAAGVKTMQTATFANTAMGFIGKA 350
VKAGVY E + + K N+++ GDG TIVTG ++ G T ++ATFA + GFI +
Sbjct: 285 VKAGVYRENVVIKKSIKNVMVIGDGIDSTIVTGNRNVQDGTTTFRSATFAVSGNGFIAQG 344
Query: 351 MTFENTAGPDGHQAVAFRNQGDMSALVGCHIVGYQDSLYVQSNRQYYRNCLVSGTVDFIF 410
+TFENTAGP+ HQAVA R+ D S C GYQD+LY+ S+RQ+ RNC + GTVDFIF
Sbjct: 345 ITFENTAGPEKHQAVALRSSSDFSVFYACSFKGYQDTLYLHSSRQFLRNCNIYGTVDFIF 404
Query: 411 GSSATLIQHSTIIVRKPGKGQFNTITADGSDTMNLNTGIVIQDCNIIPEAALFPERFTIR 470
G + ++Q+ I RKP GQ NTITA + TG VIQ + +
Sbjct: 405 GDATAILQNCNIYARKPMSGQKNTITAQSRKEPDETTGFVIQSSTVATAS---------E 455
Query: 471 SYLGRPWKYLAKTVVMESTIGDFIHPDGWTIWQGEQNHNTCYYAEYANTGPGANVARRVK 530
+YLGRPW+ ++TV M+ +G + P GW W G +T YY EY NTG GA+V+ RVK
Sbjct: 456 TYLGRPWRSHSRTVFMKCNLGALVSPAGWLPWSGSFALSTLYYGEYGNTGAGASVSGRVK 515
Query: 531 WKGYHGVISRAEANKFTAGIWLQAGPKSAAEWLNGLHVP 569
W GYH + + EA KFT +L W+ VP
Sbjct: 516 WPGYHVIKTVTEAEKFTVENFLDGN-----YWITATGVP 549
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.133 0.394
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 12,477,192
Number of Sequences: 26719
Number of extensions: 512608
Number of successful extensions: 1463
Number of sequences better than 10.0: 93
Number of HSP's better than 10.0 without gapping: 74
Number of HSP's successfully gapped in prelim test: 19
Number of HSP's that attempted gapping in prelim test: 1177
Number of HSP's gapped (non-prelim): 113
length of query: 576
length of database: 11,318,596
effective HSP length: 105
effective length of query: 471
effective length of database: 8,513,101
effective search space: 4009670571
effective search space used: 4009670571
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144563.2


