

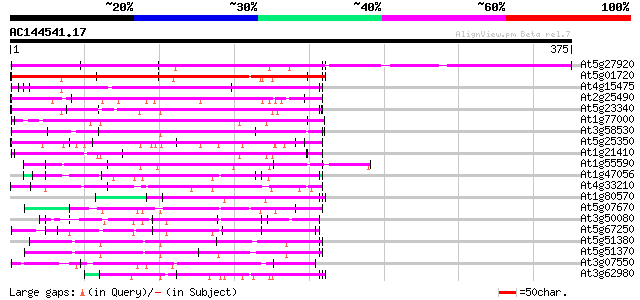
BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.17 - phase: 0 /pseudo
(375 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
Score E
Sequences producing significant alignments: (bits) Value
At5g27920 unknown protein 266 2e-71
At5g01720 putative F-box protein family, AtFBL3 173 1e-43
At4g15475 unknown protein 94 1e-19
At2g25490 putative glucose regulated repressor protein 91 1e-18
At5g23340 unknown protein 89 5e-18
At1g77000 unknown protein 82 4e-16
At3g58530 unknown protein 80 1e-15
At5g25350 leucine-rich repeats containing protein 76 3e-14
At1g21410 unknown protein 73 2e-13
At1g55590 unknown protein 72 6e-13
At1g47056 hypothetical protein 65 8e-11
At4g33210 unknown protein 63 3e-10
At1g80570 unknown protein 61 1e-09
At5g07670 unknown protein 60 1e-09
At3g50080 unknown protein 60 1e-09
At5g67250 unknown protein 59 6e-09
At5g51380 putative protein 58 7e-09
At5g51370 putative protein 57 1e-08
At3g07550 unknown protein 57 2e-08
At3g62980 transport inhibitor response 1 (TIR1) 55 6e-08
>At5g27920 unknown protein
Length = 642
Score = 266 bits (679), Expect = 2e-71
Identities = 157/402 (39%), Positives = 222/402 (55%), Gaps = 41/402 (10%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRV 61
+DV+RC+ VS SGL+S++ GH ++ + A HC+SE+S +K LKHL I IDG V
Sbjct: 253 VDVTRCDRVSLSGLISIVRGHPDIQLLKASHCVSEVSGSFLKYIKGLKHLKTIWIDGAHV 312
Query: 62 SDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAISTI 120
SD L + S+C+SL+E+GLS+C+ VT++G++ + C NL TL+L CC FVTD AIS +
Sbjct: 313 SDSSLVSLSSSCRSLMEIGLSRCVDVTDIGMISLARNCLNLKTLNLACCGFVTDVAISAV 372
Query: 121 ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVR 180
A SC NL LKLESC ++TE GL +G ++++ELDLTDC GVND L+Y+S+CS L R
Sbjct: 373 AQSCRNLGTLKLESCHLITEKGLQSLGCYSMLVQELDLTDCYGVNDRGLEYISKCSNLQR 432
Query: 181 LKLGL--------------------------CTNISDIGLAHIACNCPKLTELDLYRR*W 214
LKLGL C D GLA ++ C L L L
Sbjct: 433 LKLGLCTNISDKGIFHIGSKCSKLLELDLYRCAGFGDDGLAALSRGCKSLNRLILSY--C 490
Query: 215 ASSTDNRMQQVGHAQP-SILQ*DYRRRVEVYQQSR*TL*F*VAWAFKHHEHWYKSSCSQL 273
TD ++Q+ + S L+ + + + +A K + C +
Sbjct: 491 CELTDTGVEQIRQLELLSHLELRGLKNITGVGLAA------IASGCKKLGYLDVKLCENI 544
Query: 274 QEIG*FRFEALRKT**YGFSGTCFLFTKPTADHVLWLLMSNLKRLQDAKLVYLVNVTIQG 333
+ G + K L +D L +LMSNL R+QD LV+L VT++G
Sbjct: 545 DDSGFWALAYFSKN-----LRQINLCNCSVSDTALCMLMSNLSRVQDVDLVHLSRVTVEG 599
Query: 334 LELALISCCGRIKKVKLQRSLEFSISSEILETIHERGCKVRW 375
E AL +CC R+KK+KL L F +SSE+LET+H RGC++RW
Sbjct: 600 FEFALRACCNRLKKLKLLAPLRFLLSSELLETLHARGCRIRW 641
Score = 67.0 bits (162), Expect = 2e-11
Identities = 58/235 (24%), Positives = 97/235 (40%), Gaps = 29/235 (12%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDG-VR 60
+DVS C + +S GL ++ CLS L + +L+ I + +
Sbjct: 126 VDVSHCWGFGDREA-AALSSATGLRELKMDKCLSLSDVGLARIVVGCSNLNKISLKWCME 184
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTI 120
+SD + ++ CK L L +S + +TN I + L LD+ C + D + +
Sbjct: 185 ISDLGIDLLCKICKGLKSLDVSY-LKITNDSIRSIALLVKLEVLDMVSCPLIDDGGLQFL 243
Query: 121 ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDC-SGVNDIALKYL------- 172
N P+L + + CD V+ GL I ++ L + C S V+ LKY+
Sbjct: 244 ENGSPSLQEVDVTRCDRVSLSGLISIVRGHPDIQLLKASHCVSEVSGSFLKYIKGLKHLK 303
Query: 173 ------------------SRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
S C L+ + L C +++DIG+ +A NC L L+L
Sbjct: 304 TIWIDGAHVSDSSLVSLSSSCRSLMEIGLSRCVDVTDIGMISLARNCLNLKTLNL 358
Score = 60.1 bits (144), Expect = 2e-09
Identities = 39/119 (32%), Positives = 55/119 (45%), Gaps = 14/119 (11%)
Query: 100 NLTTLDLTCC---------RFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSC 150
NL++LDL+ C R D AIST+ + L L V GL + C
Sbjct: 66 NLSSLDLSVCPKLDDDVVLRLALDGAISTLG-----IKSLNLSRSTAVRARGLETLARMC 120
Query: 151 LMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
LE +D++ C G D LS + L LK+ C ++SD+GLA I C L ++ L
Sbjct: 121 HALERVDVSHCWGFGDREAAALSSATGLRELKMDKCLSLSDVGLARIVVGCSNLNKISL 179
Score = 56.6 bits (135), Expect = 2e-08
Identities = 41/164 (25%), Positives = 76/164 (46%), Gaps = 3/164 (1%)
Query: 48 LKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLT 107
+K L++ R VR L+ + C +L + +S C G + + L L +
Sbjct: 97 IKSLNLSRSTAVRARG--LETLARMCHALERVDVSHCWGFGDREAAALSSATGLRELKMD 154
Query: 108 CCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDI 167
C ++D ++ I C NL + L+ C ++++G+ + C L+ LD++ ND
Sbjct: 155 KCLSLSDVGLARIVVGCSNLNKISLKWCMEISDLGIDLLCKICKGLKSLDVSYLKITND- 213
Query: 168 ALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYR 211
+++ ++ KL L + C I D GL + P L E+D+ R
Sbjct: 214 SIRSIALLVKLEVLDMVSCPLIDDGGLQFLENGSPSLQEVDVTR 257
>At5g01720 putative F-box protein family, AtFBL3
Length = 665
Score = 173 bits (439), Expect = 1e-43
Identities = 93/211 (44%), Positives = 133/211 (62%), Gaps = 2/211 (0%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRV 61
+D S C ++ GL S++SG L++++ HC S +S + LK + L IR+DG V
Sbjct: 257 LDASSCQNLTHRGLTSLLSGAGYLQRLDLSHCSSVISLDFASSLKKVSALQSIRLDGCSV 316
Query: 62 SDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQ-VVGCCNLTTLDLTCCRFVTDAAISTI 120
+ L+ IG+ C SL E+ LSKC+ VT+ G+ V+ +L LD+TCCR ++ +I+ I
Sbjct: 317 TPDGLKAIGTLCNSLKEVSLSKCVSVTDEGLSSLVMKLKDLRKLDITCCRKLSRVSITQI 376
Query: 121 ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVR 180
ANSCP L LK+ESC +V+ + IG C +LEELDLTD + ++D LK +S C L
Sbjct: 377 ANSCPLLVSLKMESCSLVSREAFWLIGQKCRLLEELDLTD-NEIDDEGLKSISSCLSLSS 435
Query: 181 LKLGLCTNISDIGLAHIACNCPKLTELDLYR 211
LKLG+C NI+D GL++I C L ELDLYR
Sbjct: 436 LKLGICLNITDKGLSYIGMGCSNLRELDLYR 466
Score = 94.7 bits (234), Expect = 7e-20
Identities = 60/209 (28%), Positives = 105/209 (49%), Gaps = 2/209 (0%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR- 60
+ +S+C V+ GL S++ + L +++ C +T + L ++++
Sbjct: 334 VSLSKCVSVTDEGLSSLVMKLKDLRKLDITCCRKLSRVSITQIANSCPLLVSLKMESCSL 393
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTI 120
VS +IG C+ L EL L+ + + G+ + C +L++L L C +TD +S I
Sbjct: 394 VSREAFWLIGQKCRLLEELDLTDN-EIDDEGLKSISSCLSLSSLKLGICLNITDKGLSYI 452
Query: 121 ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVR 180
C NL L L +T++G+ I C+ LE ++++ C + D +L LS+CS L
Sbjct: 453 GMGCSNLRELDLYRSVGITDVGISTIAQGCIHLETINISYCQDITDKSLVSLSKCSLLQT 512
Query: 181 LKLGLCTNISDIGLAHIACNCPKLTELDL 209
+ C NI+ GLA IA C +L ++DL
Sbjct: 513 FESRGCPNITSQGLAAIAVRCKRLAKVDL 541
Score = 76.6 bits (187), Expect = 2e-14
Identities = 43/145 (29%), Positives = 78/145 (53%), Gaps = 5/145 (3%)
Query: 59 VRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAI 117
+ ++D L IG C +L EL L + +G+T++GI + GC +L T++++ C+ +TD ++
Sbjct: 442 LNITDKGLSYIGMGCSNLRELDLYRSVGITDVGISTIAQGCIHLETINISYCQDITDKSL 501
Query: 118 STIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVND---IALKYLSR 174
+++ C L + C +T GL I C L ++DL C +ND +AL + S+
Sbjct: 502 VSLSK-CSLLQTFESRGCPNITSQGLAAIAVRCKRLAKVDLKKCPSINDAGLLALAHFSQ 560
Query: 175 CSKLVRLKLGLCTNISDIGLAHIAC 199
K + + T + + LA+I C
Sbjct: 561 NLKQINVSDTAVTEVGLLSLANIGC 585
Score = 68.2 bits (165), Expect = 7e-12
Identities = 40/111 (36%), Positives = 58/111 (52%), Gaps = 1/111 (0%)
Query: 100 NLTTLDLTCCRFVTDAAISTIAN-SCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDL 158
N T LDLT C VTD A+S + S P L L L + GL ++ C+ L E+DL
Sbjct: 73 NTTDLDLTFCPRVTDYALSVVGCLSGPTLRSLDLSRSGSFSAAGLLRLALKCVNLVEIDL 132
Query: 159 TDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
++ + + D ++ L RLKLG C ++D+G+ IA C KL + L
Sbjct: 133 SNATEMRDADAAVVAEARSLERLKLGRCKMLTDMGIGCIAVGCKKLNTVSL 183
Score = 64.3 bits (155), Expect = 1e-10
Identities = 61/262 (23%), Positives = 110/262 (41%), Gaps = 55/262 (20%)
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHC--LSELSAPLTNGLKNLKHLSVIRIDG 58
++D+SR S +GLL + L +I+ + + + A + ++L+ L + R
Sbjct: 103 SLDLSRSGSFSAAGLLRLALKCVNLVEIDLSNATEMRDADAAVVAEARSLERLKLGRCK- 161
Query: 59 VRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGI-MQVVGCCNLTTLDLTC--------- 108
++D + I CK L + L C+GV ++G+ + V C ++ TLDL+
Sbjct: 162 -MLTDMGIGCIAVGCKKLNTVSLKWCVGVGDLGVGLLAVKCKDIRTLDLSYLPITGKCLH 220
Query: 109 ---------------CRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLML 153
C V D ++ ++ + C +L L SC +T GL + S L
Sbjct: 221 DILKLQHLEELLLEGCFGVDDDSLKSLRHDCKSLKKLDASSCQNLTHRGLTSLLSGAGYL 280
Query: 154 EELDLTDCSGVNDI----ALKYLSR----------------------CSKLVRLKLGLCT 187
+ LDL+ CS V + +LK +S C+ L + L C
Sbjct: 281 QRLDLSHCSSVISLDFASSLKKVSALQSIRLDGCSVTPDGLKAIGTLCNSLKEVSLSKCV 340
Query: 188 NISDIGLAHIACNCPKLTELDL 209
+++D GL+ + L +LD+
Sbjct: 341 SVTDEGLSSLVMKLKDLRKLDI 362
Score = 34.7 bits (78), Expect = 0.087
Identities = 19/66 (28%), Positives = 33/66 (49%), Gaps = 1/66 (1%)
Query: 310 LLMSNLKRLQDAKLVYLVNVTIQGLELALISCCGRIKKVKLQRSLEFSISSEILETIHER 369
L ++N+ LQ+ +V + G+ AL+ C G ++K KL SL + ++ + R
Sbjct: 578 LSLANIGCLQNIAVVNSSGLRPSGVAAALLGC-GGLRKAKLHASLRSLLPLSLIHHLEAR 636
Query: 370 GCKVRW 375
GC W
Sbjct: 637 GCAFLW 642
>At4g15475 unknown protein
Length = 610
Score = 94.0 bits (232), Expect = 1e-19
Identities = 58/205 (28%), Positives = 102/205 (49%), Gaps = 6/205 (2%)
Query: 7 CNCVSPSGLLSVISGHEGLEQINAGHC--LSELS-APLTNGLKNLKHLSVIRIDGVRVSD 63
C + S L + G + LE ++ C + +++ + G +NLK L + R +
Sbjct: 382 CQRIGNSALQEIGKGCKSLEILHLVDCSGIGDIAMCSIAKGCRNLKKLHIRRCYEIGNKG 441
Query: 64 FILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANS 123
I IG +CKSL EL L C V N ++ + C+L L+++ C ++DA I+ IA
Sbjct: 442 IIS--IGKHCKSLTELSLRFCDKVGNKALIAIGKGCSLQQLNVSGCNQISDAGITAIARG 499
Query: 124 CPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYL-SRCSKLVRLK 182
CP L L + + ++ L ++G C ML++L L+ C + D L +L +C L
Sbjct: 500 CPQLTHLDISVLQNIGDMPLAELGEGCPMLKDLVLSHCHHITDNGLNHLVQKCKLLETCH 559
Query: 183 LGLCTNISDIGLAHIACNCPKLTEL 207
+ C I+ G+A + +CP + ++
Sbjct: 560 MVYCPGITSAGVATVVSSCPHIKKV 584
Score = 92.0 bits (227), Expect = 5e-19
Identities = 60/198 (30%), Positives = 91/198 (45%), Gaps = 2/198 (1%)
Query: 14 GLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR-VSDFILQIIGSN 72
G+ ++ G + L+ + C L K L + I+G + ++ IG +
Sbjct: 311 GMRAIGKGSKKLKDLTLSDCYFVSCKGLEAIAHGCKELERVEINGCHNIGTRGIEAIGKS 370
Query: 73 CKSLVELGLSKCIGVTNMGIMQV-VGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLK 131
C L EL L C + N + ++ GC +L L L C + D A+ +IA C NL L
Sbjct: 371 CPRLKELALLYCQRIGNSALQEIGKGCKSLEILHLVDCSGIGDIAMCSIAKGCRNLKKLH 430
Query: 132 LESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISD 191
+ C + G+ IG C L EL L C V + AL + + L +L + C ISD
Sbjct: 431 IRRCYEIGNKGIISIGKHCKSLTELSLRFCDKVGNKALIAIGKGCSLQQLNVSGCNQISD 490
Query: 192 IGLAHIACNCPKLTELDL 209
G+ IA CP+LT LD+
Sbjct: 491 AGITAIARGCPQLTHLDI 508
Score = 77.0 bits (188), Expect = 2e-14
Identities = 57/203 (28%), Positives = 99/203 (48%), Gaps = 5/203 (2%)
Query: 10 VSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQII 69
++ +GL ++ +G +E ++ C + S L + + L + + G V D L +
Sbjct: 127 LTDTGLTALANGFPRIENLSLIWCPNVSSVGLCSLAQKCTSLKSLDLQGCYVGDQGLAAV 186
Query: 70 GSNCKSLVELGLSKCIGVTNMGIMQ-VVGCC-NLTTLDLTCCRFVTDAAISTIANSCPNL 127
G CK L EL L C G+T++G++ VVGC +L ++ + +TD ++ + + C L
Sbjct: 187 GKFCKQLEELNLRFCEGLTDVGVIDLVVGCSKSLKSIGVAASAKITDLSLEAVGSHCKLL 246
Query: 128 ACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSR-CSKLVRLKLGLC 186
L L+S + + + GL + C L+ L L C V D+A + C+ L RL L
Sbjct: 247 EVLYLDS-EYIHDKGLIAVAQGCHRLKNLKL-QCVSVTDVAFAAVGELCTSLERLALYSF 304
Query: 187 TNISDIGLAHIACNCPKLTELDL 209
+ +D G+ I KL +L L
Sbjct: 305 QHFTDKGMRAIGKGSKKLKDLTL 327
Score = 50.4 bits (119), Expect = 2e-06
Identities = 38/148 (25%), Positives = 65/148 (43%), Gaps = 27/148 (18%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRV 61
++VS CN +S +G+ ++ G L HL + + +
Sbjct: 480 LNVSGCNQISDAGITAI-----------------------ARGCPQLTHLDISVLQ--NI 514
Query: 62 SDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCN-LTTLDLTCCRFVTDAAISTI 120
D L +G C L +L LS C +T+ G+ +V C L T + C +T A ++T+
Sbjct: 515 GDMPLAELGEGCPMLKDLVLSHCHHITDNGLNHLVQKCKLLETCHMVYCPGITSAGVATV 574
Query: 121 ANSCPNLACLKLESCDMVTEIGLYQIGS 148
+SCP++ + +E VTE + GS
Sbjct: 575 VSSCPHIKKVLIEKW-KVTERTTRRAGS 601
>At2g25490 putative glucose regulated repressor protein
Length = 628
Score = 90.5 bits (223), Expect = 1e-18
Identities = 60/172 (34%), Positives = 91/172 (52%), Gaps = 6/172 (3%)
Query: 42 TNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCN 100
T G L LS+ + +VSD L+ IG +C SL L L +T+ G++++ GC
Sbjct: 145 TAGRGGLGKLSIRGSNSAKVSDLGLRSIGRSCPSLGSLSLWNVSTITDNGLLEIAEGCAQ 204
Query: 101 LTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTD 160
L L+L C +TD + IA SCPNL L LE+C + + GL I SC L+ + + +
Sbjct: 205 LEKLELNRCSTITDKGLVAIAKSCPNLTELTLEACSRIGDEGLLAIARSCSKLKSVSIKN 264
Query: 161 CSGVNDIALKYL---SRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
C V D + L + CS L +LKL + N++D+ LA + +T+L L
Sbjct: 265 CPLVRDQGIASLLSNTTCS-LAKLKLQM-LNVTDVSLAVVGHYGLSITDLVL 314
Score = 75.1 bits (183), Expect = 6e-14
Identities = 59/241 (24%), Positives = 102/241 (41%), Gaps = 33/241 (13%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLK-HLSVIRIDGVR 60
+ + C+ + GLL++ L+ ++ +C + + L N L+ +++ +
Sbjct: 234 LTLEACSRIGDEGLLAIARSCSKLKSVSIKNCPLVRDQGIASLLSNTTCSLAKLKLQMLN 293
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQV---VGCCNLTTLDLTCCRFVTDAAI 117
V+D L ++G S+ +L L+ V+ G + VG L +L +T C+ VTD +
Sbjct: 294 VTDVSLAVVGHYGLSITDLVLAGLSHVSEKGFWVMGNGVGLQKLNSLTITACQGVTDMGL 353
Query: 118 STIANSCPN--------------------------LACLKLESCDMVTEIGLYQIGSSC- 150
++ CPN L L+LE C VT+ G + +C
Sbjct: 354 ESVGKGCPNMKKAIISKSPLLSDNGLVSFAKASLSLESLQLEECHRVTQFGFFGSLLNCG 413
Query: 151 LMLEELDLTDCSGVNDI--ALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELD 208
L+ L +C + D+ L S CS L L + C D LA I CP+L ++D
Sbjct: 414 EKLKAFSLVNCLSIRDLTTGLPASSHCSALRSLSIRNCPGFGDANLAAIGKLCPQLEDID 473
Query: 209 L 209
L
Sbjct: 474 L 474
Score = 68.6 bits (166), Expect = 5e-12
Identities = 55/205 (26%), Positives = 100/205 (47%), Gaps = 12/205 (5%)
Query: 1 AIDVSRCNCVSPSGLL-SVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV 59
++ + C+ V+ G S+++ E L+ + +CLS LT GL H S +R +
Sbjct: 391 SLQLEECHRVTQFGFFGSLLNCGEKLKAFSLVNCLS--IRDLTTGLPASSHCSALRSLSI 448
Query: 60 R----VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDA 115
R D L IG C L ++ L G+T G + ++ +L ++ + C +TD
Sbjct: 449 RNCPGFGDANLAAIGKLCPQLEDIDLCGLKGITESGFLHLIQS-SLVKINFSGCSNLTDR 507
Query: 116 AISTI-ANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSR 174
IS I A + L L ++ C +T+ L I ++C +L +LD++ C+ ++D ++ L+
Sbjct: 508 VISAITARNGWTLEVLNIDGCSNITDASLVSIAANCQILSDLDISKCA-ISDSGIQALAS 566
Query: 175 CSKLVR--LKLGLCTNISDIGLAHI 197
KL L + C+ ++D L I
Sbjct: 567 SDKLKLQILSVAGCSMVTDKSLPAI 591
Score = 56.2 bits (134), Expect = 3e-08
Identities = 57/219 (26%), Positives = 102/219 (46%), Gaps = 16/219 (7%)
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQ--INAGHCLSELSAPLTNGLKNLKHLSVIRIDG 58
++ ++ C V+ GL SV G +++ I+ LS+ L + K L ++++
Sbjct: 339 SLTITACQGVTDMGLESVGKGCPNMKKAIISKSPLLSDNG--LVSFAKASLSLESLQLEE 396
Query: 59 V-RVSDFILQIIGS--NC-KSLVELGLSKCIGVTNM--GIMQVVGCCNLTTLDLTCCRFV 112
RV+ F GS NC + L L C+ + ++ G+ C L +L + C
Sbjct: 397 CHRVTQF--GFFGSLLNCGEKLKAFSLVNCLSIRDLTTGLPASSHCSALRSLSIRNCPGF 454
Query: 113 TDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYL 172
DA ++ I CP L + L +TE G + S L+ +++ + CS + D + +
Sbjct: 455 GDANLAAIGKLCPQLEDIDLCGLKGITESGFLHLIQSSLV--KINFSGCSNLTDRVISAI 512
Query: 173 SRCS--KLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
+ + L L + C+NI+D L IA NC L++LD+
Sbjct: 513 TARNGWTLEVLNIDGCSNITDASLVSIAANCQILSDLDI 551
>At5g23340 unknown protein
Length = 405
Score = 88.6 bits (218), Expect = 5e-18
Identities = 59/213 (27%), Positives = 103/213 (47%), Gaps = 5/213 (2%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR- 60
+++ C ++ +GL S+ L+ ++ +C L+ + L + + G R
Sbjct: 103 LNLHNCKGITDTGLASIGRCLSLLQFLDVSYCRKLSDKGLSAVAEGCHDLRALHLAGCRF 162
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAIST 119
++D L+ + C+ L LGL C +T+ G+ +V GC + +LD+ C V DA +S+
Sbjct: 163 ITDESLKSLSERCRDLEALGLQGCTNITDSGLADLVKGCRKIKSLDINKCSNVGDAGVSS 222
Query: 120 IANSCPN-LACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSK- 177
+A +C + L LKL C V + + C LE L + C ++D ++ L+ K
Sbjct: 223 VAKACASSLKTLKLLDCYKVGNESISSLAQFCKNLETLIIGGCRDISDESIMLLADSCKD 282
Query: 178 -LVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
L L++ C NISD L+ I C L LD+
Sbjct: 283 SLKNLRMDWCLNISDSSLSCILKQCKNLEALDI 315
Score = 79.3 bits (194), Expect = 3e-15
Identities = 53/173 (30%), Positives = 91/173 (51%), Gaps = 7/173 (4%)
Query: 39 APLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVE-LGLSKCIGVTNMGIMQVV- 96
A ++ G K L+ L++ G+ +D L IG C SL++ L +S C +++ G+ V
Sbjct: 91 AVISEGFKFLRVLNLHNCKGI--TDTGLASIG-RCLSLLQFLDVSYCRKLSDKGLSAVAE 147
Query: 97 GCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEEL 156
GC +L L L CRF+TD ++ +++ C +L L L+ C +T+ GL + C ++ L
Sbjct: 148 GCHDLRALHLAGCRFITDESLKSLSERCRDLEALGLQGCTNITDSGLADLVKGCRKIKSL 207
Query: 157 DLTDCSGVNDIALKYLSR--CSKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
D+ CS V D + +++ S L LKL C + + ++ +A C L L
Sbjct: 208 DINKCSNVGDAGVSSVAKACASSLKTLKLLDCYKVGNESISSLAQFCKNLETL 260
Score = 72.0 bits (175), Expect = 5e-13
Identities = 52/158 (32%), Positives = 77/158 (47%), Gaps = 8/158 (5%)
Query: 60 RVSDFILQIIGSNCKSLVELGLSKCI------GVTNMGIMQVV-GCCNLTTLDLTCCRFV 112
R +L+ + S +VEL LS+ I GVT+ + + G L L+L C+ +
Sbjct: 52 RAGPHMLRRLASRFTQIVELDLSQSISRSFYPGVTDSDLAVISEGFKFLRVLNLHNCKGI 111
Query: 113 TDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYL 172
TD +++I L L + C +++ GL + C L L L C + D +LK L
Sbjct: 112 TDTGLASIGRCLSLLQFLDVSYCRKLSDKGLSAVAEGCHDLRALHLAGCRFITDESLKSL 171
Query: 173 S-RCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
S RC L L L CTNI+D GLA + C K+ LD+
Sbjct: 172 SERCRDLEALGLQGCTNITDSGLADLVKGCRKIKSLDI 209
Score = 70.9 bits (172), Expect = 1e-12
Identities = 48/219 (21%), Positives = 102/219 (45%), Gaps = 16/219 (7%)
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHC-------LSELSAPLTNGLKNLKHLSV 53
A+ + C ++ SGL ++ G ++ ++ C +S ++ + LK LK L
Sbjct: 180 ALGLQGCTNITDSGLADLVKGCRKIKSLDINKCSNVGDAGVSSVAKACASSLKTLKLLDC 239
Query: 54 IRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCC--NLTTLDLTCCRF 111
++ +S + CK+L L + C +++ IM + C +L L + C
Sbjct: 240 YKVGNESISS-----LAQFCKNLETLIIGGCRDISDESIMLLADSCKDSLKNLRMDWCLN 294
Query: 112 VTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGS-SCLMLEELDLTDCSGVNDIAL- 169
++D+++S I C NL L + C+ VT+ +GS L L+ L +++C+ + +
Sbjct: 295 ISDSSLSCILKQCKNLEALDIGCCEEVTDTAFRDLGSDDVLGLKVLKVSNCTKITVTGIG 354
Query: 170 KYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELD 208
K L +CS L + + +++++ + PK +++
Sbjct: 355 KLLDKCSSLEYIDVRSLPHVTEVRCSEAGLEFPKCCKVN 393
>At1g77000 unknown protein
Length = 360
Score = 82.4 bits (202), Expect = 4e-16
Identities = 60/226 (26%), Positives = 101/226 (44%), Gaps = 31/226 (13%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV-- 59
+D+S+ + ++ L S+ G L ++N C S L + + + L ++ + G
Sbjct: 122 LDLSKSSKITDHSLYSLARGCTNLTKLNLSGCTSFSDTALAHLTRFCRKLKILNLCGCVE 181
Query: 60 RVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAIS 118
VSD LQ IG NC L L L C +++ G+M + GC +L TLDL C +TD ++
Sbjct: 182 AVSDNTLQAIGENCNQLQSLNLGWCENISDDGVMSLAYGCPDLRTLDLCSCVLITDESVV 241
Query: 119 TIANSCPNLACLKLESCDMVTEIGLYQIGSSCLM------------------LEELDLTD 160
+AN C +L L L C +T+ +Y + S + L L+++
Sbjct: 242 ALANRCIHLRSLGLYYCRNITDRAMYSLAQSGVKNKHEMWRAVKKGKFDEEGLRSLNISQ 301
Query: 161 CSGVNDIALK-------YLSRCSKLVRLKLGLCTNISDIGLAHIAC 199
C+ + A++ L CS L + C N+ + H AC
Sbjct: 302 CTYLTPSAVQAVCDTFPALHTCSGRHSLVMSGCLNLQSV---HCAC 344
Score = 77.4 bits (189), Expect = 1e-14
Identities = 60/212 (28%), Positives = 102/212 (47%), Gaps = 10/212 (4%)
Query: 4 VSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLS--VIRIDGVRV 61
++ C C SG +S GL +++ C +++ + + L V+R D ++
Sbjct: 50 IASCIC---SGWRDAVS--LGLTRLSLSWCKKNMNSLVLSLAPKFVKLQTLVLRQDKPQL 104
Query: 62 SDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAISTI 120
D ++ I ++C L +L LSK +T+ + + GC NLT L+L+ C +D A++ +
Sbjct: 105 EDNAVEAIANHCHELQDLDLSKSSKITDHSLYSLARGCTNLTKLNLSGCTSFSDTALAHL 164
Query: 121 ANSCPNLACLKLESC-DMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLS-RCSKL 178
C L L L C + V++ L IG +C L+ L+L C ++D + L+ C L
Sbjct: 165 TRFCRKLKILNLCGCVEAVSDNTLQAIGENCNQLQSLNLGWCENISDDGVMSLAYGCPDL 224
Query: 179 VRLKLGLCTNISDIGLAHIACNCPKLTELDLY 210
L L C I+D + +A C L L LY
Sbjct: 225 RTLDLCSCVLITDESVVALANRCIHLRSLGLY 256
>At3g58530 unknown protein
Length = 353
Score = 80.5 bits (197), Expect = 1e-15
Identities = 52/151 (34%), Positives = 81/151 (53%), Gaps = 2/151 (1%)
Query: 60 RVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAIS 118
++SD ++ I S C L + + VT+ GI +V C ++T L+L+ C+ +TD ++
Sbjct: 123 KISDNGIEAITSICPKLKVFSIYWNVRVTDAGIRNLVKNCRHITDLNLSGCKSLTDKSMQ 182
Query: 119 TIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKL 178
+A S P+L L + C +T+ GL Q+ C L+ L+L SG D A +S + L
Sbjct: 183 LVAESYPDLESLNITRCVKITDDGLLQVLQKCFSLQTLNLYALSGFTDKAYMKISLLADL 242
Query: 179 VRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
L + NISD G+ HIA C KL L+L
Sbjct: 243 RFLDICGAQNISDEGIGHIA-KCNKLESLNL 272
Score = 59.7 bits (143), Expect = 3e-09
Identities = 47/216 (21%), Positives = 94/216 (42%), Gaps = 53/216 (24%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR- 60
++++ C +S +G+ ++ S L+ + + A + N +KN +H++ + + G +
Sbjct: 116 LNLNVCQKISDNGIEAITSICPKLKVFSIYWNVRVTDAGIRNLVKNCRHITDLNLSGCKS 175
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCC--------------------- 99
++D +Q++ + L L +++C+ +T+ G++QV+ C
Sbjct: 176 LTDKSMQLVAESYPDLESLNITRCVKITDDGLLQVLQKCFSLQTLNLYALSGFTDKAYMK 235
Query: 100 ------------------------------NLTTLDLTCCRFVTDAAISTIANSCPNLAC 129
L +L+LT C +TDA ++TIANSC +L
Sbjct: 236 ISLLADLRFLDICGAQNISDEGIGHIAKCNKLESLNLTWCVRITDAGVNTIANSCTSLEF 295
Query: 130 LKLESCDMVTEIGLYQIGSSC-LMLEELDLTDCSGV 164
L L VT+ L + +C L LD+ C+G+
Sbjct: 296 LSLFGIVGVTDRCLETLSQTCSTTLTTLDVNGCTGI 331
Score = 53.9 bits (128), Expect = 1e-07
Identities = 45/187 (24%), Positives = 81/187 (43%), Gaps = 28/187 (14%)
Query: 26 EQINAG-HCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKC 84
E NAG L+ LS P + +KH+++ GV D L+++ + C +
Sbjct: 63 EMTNAGDRLLAALSLPR---YRQVKHINLEFAQGV--VDSHLKLVKTECPDAL------- 110
Query: 85 IGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLY 144
+L L+L C+ ++D I I + CP L + VT+ G+
Sbjct: 111 --------------LSLEWLNLNVCQKISDNGIEAITSICPKLKVFSIYWNVRVTDAGIR 156
Query: 145 QIGSSCLMLEELDLTDCSGVNDIALKYLSRC-SKLVRLKLGLCTNISDIGLAHIACNCPK 203
+ +C + +L+L+ C + D +++ ++ L L + C I+D GL + C
Sbjct: 157 NLVKNCRHITDLNLSGCKSLTDKSMQLVAESYPDLESLNITRCVKITDDGLLQVLQKCFS 216
Query: 204 LTELDLY 210
L L+LY
Sbjct: 217 LQTLNLY 223
Score = 40.8 bits (94), Expect = 0.001
Identities = 38/162 (23%), Positives = 67/162 (40%), Gaps = 29/162 (17%)
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDG-- 58
+++++RC ++ GLL V+ L+ +N + LS + + L L + I G
Sbjct: 193 SLNITRCVKITDDGLLQVLQKCFSLQTLNL-YALSGFTDKAYMKISLLADLRFLDICGAQ 251
Query: 59 ------------------------VRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQ 94
VR++D + I ++C SL L L +GVT+ +
Sbjct: 252 NISDEGIGHIAKCNKLESLNLTWCVRITDAGVNTIANSCTSLEFLSLFGIVGVTDRCLET 311
Query: 95 VVGCCN--LTTLDLTCCRFVTDAAISTIANSCPNLACLKLES 134
+ C+ LTTLD+ C + + + P L C K+ S
Sbjct: 312 LSQTCSTTLTTLDVNGCTGIKRRSREELLQMFPRLTCFKVHS 353
>At5g25350 leucine-rich repeats containing protein
Length = 623
Score = 76.3 bits (186), Expect = 3e-14
Identities = 63/223 (28%), Positives = 99/223 (44%), Gaps = 34/223 (15%)
Query: 2 IDVSRCNCVSPSGLLSVIS----------------GHEGLEQINAGHCLSELSAPLTN-- 43
+D+SRC ++ SGL+++ G+EGL I A C++ S + +
Sbjct: 198 LDLSRCPGITDSGLVAIAENCVNLSDLTIDSCSGVGNEGLRAI-ARRCVNLRSISIRSCP 256
Query: 44 -----GLKNL-----KHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIM 93
G+ L +L+ +++ + VS L +IG ++ +L L GV G
Sbjct: 257 RIGDQGVAFLLAQAGSYLTKVKLQMLNVSGLSLAVIGHYGAAVTDLVLHGLQGVNEKGFW 316
Query: 94 ---QVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSC 150
G L +L + CR +TD + + N CP+L + L C +V+ GL + S
Sbjct: 317 VMGNAKGLKKLKSLSVMSCRGMTDVGLEAVGNGCPDLKHVSLNKCLLVSGKGLVALAKSA 376
Query: 151 LMLEELDLTDCSGVNDIALK-YLSRC-SKLVRLKLGLCTNISD 191
L LE L L +C +N L +L C SKL L C ISD
Sbjct: 377 LSLESLKLEECHRINQFGLMGFLMNCGSKLKAFSLANCLGISD 419
Score = 72.0 bits (175), Expect = 5e-13
Identities = 55/217 (25%), Positives = 107/217 (48%), Gaps = 8/217 (3%)
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV- 59
++ V C ++ GL +V +G L+ ++ CL L K+ L ++++
Sbjct: 329 SLSVMSCRGMTDVGLEAVGNGCPDLKHVSLNKCLLVSGKGLVALAKSALSLESLKLEECH 388
Query: 60 RVSDFILQIIGSNCKS-LVELGLSKCIGVTNMGIMQVV---GCCNLTTLDLTCCRFVTDA 115
R++ F L NC S L L+ C+G+++ + C +L +L + CC DA
Sbjct: 389 RINQFGLMGFLMNCGSKLKAFSLANCLGISDFNSESSLPSPSCSSLRSLSIRCCPGFGDA 448
Query: 116 AISTIANSCPNLACLKLESCDMVTEIGLYQ-IGSSCLMLEELDLTDCSGVNDIALKYLSR 174
+++ + C L ++L + VT+ G+ + + S+ + L +++L++C V+D + +S
Sbjct: 449 SLAFLGKFCHQLQDVELCGLNGVTDAGVRELLQSNNVGLVKVNLSECINVSDNTVSAISV 508
Query: 175 C--SKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
C L L L C NI++ L +A NC + +LD+
Sbjct: 509 CHGRTLESLNLDGCKNITNASLVAVAKNCYSVNDLDI 545
Score = 68.9 bits (167), Expect = 4e-12
Identities = 47/150 (31%), Positives = 74/150 (49%), Gaps = 8/150 (5%)
Query: 56 IDGVRVSDFILQIIGSNCKSLVELGLSKCIG------VTNMGIMQVV-GCCNLTTLDLTC 108
++G + +D L I S LG + G VT++G+ V GC +L + L
Sbjct: 117 LEGKKATDLRLAAIAVGTSSRGGLGKLQIRGSGFESKVTDVGLGAVAHGCPSLRIVSLWN 176
Query: 109 CRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIA 168
V+D +S IA SCP + L L C +T+ GL I +C+ L +L + CSGV +
Sbjct: 177 LPAVSDLGLSEIARSCPMIEKLDLSRCPGITDSGLVAIAENCVNLSDLTIDSCSGVGNEG 236
Query: 169 LKYLS-RCSKLVRLKLGLCTNISDIGLAHI 197
L+ ++ RC L + + C I D G+A +
Sbjct: 237 LRAIARRCVNLRSISIRSCPRIGDQGVAFL 266
Score = 63.2 bits (152), Expect = 2e-10
Identities = 50/199 (25%), Positives = 86/199 (43%), Gaps = 32/199 (16%)
Query: 41 LTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCC 99
+ +G +L+ +S+ + V SD L I +C + +L LS+C G+T+ G++ + C
Sbjct: 162 VAHGCPSLRIVSLWNLPAV--SDLGLSEIARSCPMIEKLDLSRCPGITDSGLVAIAENCV 219
Query: 100 NLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMV----------------TEIGL 143
NL+ L + C V + + IA C NL + + SC + T++ L
Sbjct: 220 NLSDLTIDSCSGVGNEGLRAIARRCVNLRSISIRSCPRIGDQGVAFLLAQAGSYLTKVKL 279
Query: 144 YQIGSSCLMLE----------ELDLTDCSGVNDIALKYLSRCSKLVRLK---LGLCTNIS 190
+ S L L +L L GVN+ + L +LK + C ++
Sbjct: 280 QMLNVSGLSLAVIGHYGAAVTDLVLHGLQGVNEKGFWVMGNAKGLKKLKSLSVMSCRGMT 339
Query: 191 DIGLAHIACNCPKLTELDL 209
D+GL + CP L + L
Sbjct: 340 DVGLEAVGNGCPDLKHVSL 358
Score = 52.4 bits (124), Expect = 4e-07
Identities = 52/205 (25%), Positives = 98/205 (47%), Gaps = 19/205 (9%)
Query: 1 AIDVSRCNCVSPSGLLSVISG-HEGLEQINAGHCL------SELSAPLTNGLKNLKHLSV 53
++ + C+ ++ GL+ + L+ + +CL SE S P + +L+ LS+
Sbjct: 381 SLKLEECHRINQFGLMGFLMNCGSKLKAFSLANCLGISDFNSESSLP-SPSCSSLRSLSI 439
Query: 54 IRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCN--LTTLDLTCCRF 111
G D L +G C L ++ L GVT+ G+ +++ N L ++L+ C
Sbjct: 440 RCCPGF--GDASLAFLGKFCHQLQDVELCGLNGVTDAGVRELLQSNNVGLVKVNLSECIN 497
Query: 112 VTDAAISTIANSCPN--LACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIAL 169
V+D +S I+ C L L L+ C +T L + +C + +LD+++ + V+D +
Sbjct: 498 VSDNTVSAIS-VCHGRTLESLNLDGCKNITNASLVAVAKNCYSVNDLDISN-TLVSDHGI 555
Query: 170 KYLSRCS---KLVRLKLGLCTNISD 191
K L+ L L +G C++I+D
Sbjct: 556 KALASSPNHLNLQVLSIGGCSSITD 580
Score = 50.8 bits (120), Expect = 1e-06
Identities = 30/98 (30%), Positives = 47/98 (47%), Gaps = 25/98 (25%)
Query: 112 VTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKY 171
VTD + +A+ CP+L + L + V+++GL +I SC M+E+LDL+
Sbjct: 154 VTDVGLGAVAHGCPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSR----------- 202
Query: 172 LSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
C I+D GL IA NC L++L +
Sbjct: 203 --------------CPGITDSGLVAIAENCVNLSDLTI 226
Score = 32.7 bits (73), Expect = 0.33
Identities = 19/51 (37%), Positives = 29/51 (56%), Gaps = 1/51 (1%)
Query: 162 SGVNDIALKYLSR-CSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYR 211
S V D+ L ++ C L + L +SD+GL+ IA +CP + +LDL R
Sbjct: 152 SKVTDVGLGAVAHGCPSLRIVSLWNLPAVSDLGLSEIARSCPMIEKLDLSR 202
>At1g21410 unknown protein
Length = 360
Score = 73.2 bits (178), Expect = 2e-13
Identities = 55/226 (24%), Positives = 101/226 (44%), Gaps = 31/226 (13%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV-- 59
+D+S+ ++ L ++ G L ++N C S + + + L V+ + G
Sbjct: 122 LDLSKSLKITDRSLYALAHGCPDLTKLNLSGCTSFSDTAIAYLTRFCRKLKVLNLCGCVK 181
Query: 60 RVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAIS 118
V+D L+ IG+NC + L L C +++ G+M + GC +L TLDL C +TD ++
Sbjct: 182 AVTDNALEAIGNNCNQMQSLNLGWCENISDDGVMSLAYGCPDLRTLDLCGCVLITDESVV 241
Query: 119 TIANSCPNLACLKLESCDMVTEIGLYQIGSSCLM------------------LEELDLTD 160
+A+ C +L L L C +T+ +Y + S + L L+++
Sbjct: 242 ALADWCVHLRSLGLYYCRNITDRAMYSLAQSGVKNKPGSWKSVKKGKYDEEGLRSLNISQ 301
Query: 161 CSGVNDIALK-------YLSRCSKLVRLKLGLCTNISDIGLAHIAC 199
C+ + A++ L CS L + C N++ + H AC
Sbjct: 302 CTALTPSAVQAVCDSFPALHTCSGRHSLVMSGCLNLTTV---HCAC 344
Score = 66.6 bits (161), Expect = 2e-11
Identities = 54/201 (26%), Positives = 91/201 (44%), Gaps = 7/201 (3%)
Query: 4 VSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRID---GVR 60
+S CN S +LS++ L+ +N +L + N H + +D ++
Sbjct: 71 LSWCNNNMNSLVLSLVPKFVKLQTLNLRQDKPQLEDNAVEAIANHCH-ELQELDLSKSLK 129
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCN-LTTLDLT-CCRFVTDAAIS 118
++D L + C L +L LS C ++ I + C L L+L C + VTD A+
Sbjct: 130 ITDRSLYALAHGCPDLTKLNLSGCTSFSDTAIAYLTRFCRKLKVLNLCGCVKAVTDNALE 189
Query: 119 TIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSR-CSK 177
I N+C + L L C+ +++ G+ + C L LDL C + D ++ L+ C
Sbjct: 190 AIGNNCNQMQSLNLGWCENISDDGVMSLAYGCPDLRTLDLCGCVLITDESVVALADWCVH 249
Query: 178 LVRLKLGLCTNISDIGLAHIA 198
L L L C NI+D + +A
Sbjct: 250 LRSLGLYYCRNITDRAMYSLA 270
Score = 63.9 bits (154), Expect = 1e-10
Identities = 50/164 (30%), Positives = 72/164 (43%), Gaps = 30/164 (18%)
Query: 76 LVELGLSKCIGVTNMGIMQVVG-CCNLTTLDLTCCR-FVTDAAISTIANSCPNLACLKLE 133
L L LS C N ++ +V L TL+L + + D A+ IAN C L L L
Sbjct: 66 LTRLRLSWCNNNMNSLVLSLVPKFVKLQTLNLRQDKPQLEDNAVEAIANHCHELQELDLS 125
Query: 134 SCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSR------------------- 174
+T+ LY + C L +L+L+ C+ +D A+ YL+R
Sbjct: 126 KSLKITDRSLYALAHGCPDLTKLNLSGCTSFSDTAIAYLTRFCRKLKVLNLCGCVKAVTD 185
Query: 175 ---------CSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
C+++ L LG C NISD G+ +A CP L LDL
Sbjct: 186 NALEAIGNNCNQMQSLNLGWCENISDDGVMSLAYGCPDLRTLDL 229
>At1g55590 unknown protein
Length = 607
Score = 71.6 bits (174), Expect = 6e-13
Identities = 58/207 (28%), Positives = 99/207 (47%), Gaps = 10/207 (4%)
Query: 10 VSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV-RVSDFILQ- 67
++ G+ + +GLE + G A + L + ++L + G +SD
Sbjct: 298 INDMGIFLLSEACKGLESVRLGGFPKVSDAGFASLLHSCRNLKKFEVRGAFLLSDLAFHD 357
Query: 68 IIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNL 127
+ GS+C SL E+ LS C +T+ + ++ C NL LDL C+ ++D+ ++++ ++ L
Sbjct: 358 VTGSSC-SLQEVRLSTCPLITSEAVKKLGLCGNLEVLDLGSCKSISDSCLNSV-SALRKL 415
Query: 128 ACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYL-----SRCSKLVRLK 182
L L D VT+ G+ +G S + + +L L C V+D + YL + L L
Sbjct: 416 TSLNLAGAD-VTDSGMLALGKSDVPITQLSLRGCRRVSDRGISYLLNNEGTISKTLSTLD 474
Query: 183 LGLCTNISDIGLAHIACNCPKLTELDL 209
LG ISD + I C LTEL +
Sbjct: 475 LGHMPGISDRAIHTITHCCKALTELSI 501
Score = 59.3 bits (142), Expect = 3e-09
Identities = 55/187 (29%), Positives = 89/187 (47%), Gaps = 16/187 (8%)
Query: 66 LQIIGSNCKSLVELGLSK-CIG-------VTNMGIMQVVGCCN-LTTLDLTCCRFVTDAA 116
LQ +G C+ L L L + C + +MGI + C L ++ L V+DA
Sbjct: 270 LQALGF-CQQLTSLSLVRTCYNRKISFKRINDMGIFLLSEACKGLESVRLGGFPKVSDAG 328
Query: 117 ISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCS 176
+++ +SC NL ++ +++++ + + S L+E+ L+ C + A+K L C
Sbjct: 329 FASLLHSCRNLKKFEVRGAFLLSDLAFHDVTGSSCSLQEVRLSTCPLITSEAVKKLGLCG 388
Query: 177 KLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYRR*WASSTDNRMQQVGHAQPSILQ*D 236
L L LG C +ISD L ++ KLT L+L A TD+ M +G + I Q
Sbjct: 389 NLEVLDLGSCKSISDSCLNSVSA-LRKLTSLNLA---GADVTDSGMLALGKSDVPITQLS 444
Query: 237 YR--RRV 241
R RRV
Sbjct: 445 LRGCRRV 451
Score = 55.8 bits (133), Expect = 4e-08
Identities = 44/164 (26%), Positives = 79/164 (47%), Gaps = 13/164 (7%)
Query: 25 LEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKC 84
LE ++ G C S +S N + L+ L+ + + G V+D + +G + + +L L C
Sbjct: 390 LEVLDLGSCKS-ISDSCLNSVSALRKLTSLNLAGADVTDSGMLALGKSDVPITQLSLRGC 448
Query: 85 IGVTNMGIMQVVG-----CCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVT 139
V++ GI ++ L+TLDL ++D AI TI + C L L + SC VT
Sbjct: 449 RRVSDRGISYLLNNEGTISKTLSTLDLGHMPGISDRAIHTITHCCKALTELSIRSCFHVT 508
Query: 140 EIGLYQIGS-------SCLMLEELDLTDCSGVNDIALKYLSRCS 176
+ + + + L +L++ +C + AL++LS+ S
Sbjct: 509 DSSIESLATWERQAEGGSKQLRKLNVHNCVSLTTGALRWLSKPS 552
Score = 31.6 bits (70), Expect = 0.74
Identities = 26/94 (27%), Positives = 39/94 (40%), Gaps = 8/94 (8%)
Query: 97 GCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSC-----L 151
GC L++L L C R + + P+L L L C +++ L IG+ C L
Sbjct: 65 GCIGLSSLTLNCLRLNAASVRGVLG---PHLRELHLLRCSLLSSTVLTYIGTLCPNLRVL 121
Query: 152 MLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGL 185
LE DL + L+ C L L+L +
Sbjct: 122 TLEMADLDSPDVFQSNLTQMLNGCPYLESLQLNI 155
>At1g47056 hypothetical protein
Length = 518
Score = 64.7 bits (156), Expect = 8e-11
Identities = 54/188 (28%), Positives = 87/188 (45%), Gaps = 15/188 (7%)
Query: 25 LEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKC 84
L+++ G C P+ G KNLK L + R G D +LQ + +VE+ L +
Sbjct: 216 LKELYNGQCFG----PVIVGAKNLKSLKLFRCSGDW--DLLLQEMSGKDHGVVEIHLER- 268
Query: 85 IGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLES--CDMVTEIG 142
+ V+++ + + C +L +L L T+ ++ IA C L L ++ +++ + G
Sbjct: 269 MQVSDVALSAISYCSSLESLHLVKTPECTNFGLAAIAEKCKRLRKLHIDGWKANLIGDEG 328
Query: 143 LYQIGSSCLMLEELDLTDCSGVNDIALKY---LSRCSKLVRLKLGLCTNISDIGLAHIAC 199
L + C L+EL L GVN L ++C L RL L C D L+ IA
Sbjct: 329 LVAVAKFCSQLQELVLI---GVNPTTLSLGMLAAKCLNLERLALCGCDTFGDPELSCIAA 385
Query: 200 NCPKLTEL 207
CP L +L
Sbjct: 386 KCPALRKL 393
Score = 50.8 bits (120), Expect = 1e-06
Identities = 42/152 (27%), Positives = 66/152 (42%), Gaps = 7/152 (4%)
Query: 16 LSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKS 75
+S S E L + C + A + K L+ L + + D L + C
Sbjct: 279 ISYCSSLESLHLVKTPECTNFGLAAIAEKCKRLRKLHIDGWKANLIGDEGLVAVAKFCSQ 338
Query: 76 LVELGLSKCIGV--TNMGI-MQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKL 132
L EL L IGV T + + M C NL L L C D +S IA CP L L +
Sbjct: 339 LQELVL---IGVNPTTLSLGMLAAKCLNLERLALCGCDTFGDPELSCIAAKCPALRKLCI 395
Query: 133 ESCDMVTEIGLYQIGSSCLMLEELDLTDCSGV 164
++C ++++G+ + + C L ++ + C GV
Sbjct: 396 KNCP-ISDVGIENLANGCPGLTKVKIKKCKGV 426
Score = 45.4 bits (106), Expect = 5e-05
Identities = 33/113 (29%), Positives = 54/113 (47%), Gaps = 7/113 (6%)
Query: 62 SDFILQI--IGSNCKSLVELGLS---KCIGVTNMGIMQV-VGCCNLTTLDLTCCRFVTDA 115
SD I I + S S+ +L L + + + + ++++ + C NL L L CR +TD
Sbjct: 87 SDLITSIPSLFSRFDSVTKLSLKCDRRSVSIGDEALVKISLRCRNLKRLKLRACRELTDV 146
Query: 116 AISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIA 168
++ A +C +L SCD + G+ + C LEEL + G DIA
Sbjct: 147 GMAAFAENCKDLKIFSCGSCDFGAK-GVKAVLDHCSNLEELSIKRLRGFTDIA 198
Score = 42.4 bits (98), Expect = 4e-04
Identities = 41/201 (20%), Positives = 80/201 (39%), Gaps = 2/201 (0%)
Query: 10 VSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQII 69
+ L+ + L+++ C + +N K L + ++ +
Sbjct: 117 IGDEALVKISLRCRNLKRLKLRACRELTDVGMAAFAENCKDLKIFSCGSCDFGAKGVKAV 176
Query: 70 GSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDA-AISTIANSCPNLA 128
+C +L EL + + G T++ + ++L C + + + + NL
Sbjct: 177 LDHCSNLEELSIKRLRGFTDIAPEMIGPGVAASSLKSICLKELYNGQCFGPVIVGAKNLK 236
Query: 129 CLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTN 188
LKL C ++ L ++ + E+ L V+D+AL +S CS L L L
Sbjct: 237 SLKLFRCSGDWDLLLQEMSGKDHGVVEIHLERMQ-VSDVALSAISYCSSLESLHLVKTPE 295
Query: 189 ISDIGLAHIACNCPKLTELDL 209
++ GLA IA C +L +L +
Sbjct: 296 CTNFGLAAIAEKCKRLRKLHI 316
>At4g33210 unknown protein
Length = 990
Score = 62.8 bits (151), Expect = 3e-10
Identities = 55/201 (27%), Positives = 93/201 (45%), Gaps = 17/201 (8%)
Query: 15 LLSVISGH------EGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQI 68
+L++ GH + L + N ++ A L NG + + HLS R+ ++++ +
Sbjct: 285 VLTIGKGHISESFFQALGECNMLRSVTVSDAILGNGAQEI-HLSHDRLRELKITKCRVMR 343
Query: 69 IGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLA 128
+ C L L L + +NM ++ C L LD+ C + DAAI + A SCP L
Sbjct: 344 LSIRCPQLRSLSLKR----SNMS-QAMLNCPLLQLLDIASCHKLLDAAIRSAAISCPQLE 398
Query: 129 CLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTN 188
L + +C V++ L +I +C L L+ + C ++ ++ L LKL C
Sbjct: 399 SLDVSNCSCVSDETLREIAQACANLHILNASYCPNISLESV----HLPMLTVLKLHSCEG 454
Query: 189 ISDIGLAHIACNCPKLTELDL 209
I+ + IA N P L L+L
Sbjct: 455 ITSASMTWIA-NSPALEVLEL 474
Score = 48.9 bits (115), Expect = 4e-06
Identities = 53/229 (23%), Positives = 97/229 (42%), Gaps = 29/229 (12%)
Query: 1 AIDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR 60
++DVS C+CVS L + L +NA +C + ++ +L L+V+++
Sbjct: 399 SLDVSNCSCVSDETLREIAQACANLHILNASYCPN-----ISLESVHLPMLTVLKLHSCE 453
Query: 61 VSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTI 120
+N +L L L C +T + + L ++ L CR TD + +I
Sbjct: 454 GITSASMTWIANSPALEVLELDNCNLLTTVSL----HLSRLQSISLVHCRKFTDLNLQSI 509
Query: 121 ------ANSCPNLACLKLES-----CDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIAL 169
++CP L + + S + + L + C L+E+DL+DC +++
Sbjct: 510 MLSSITVSNCPALRRITITSNALRRLALQKQENLTTLVLQCHSLQEVDLSDCESLSNSVC 569
Query: 170 KYLS---RCSKLVRLKLGLCTNISDI-----GLAHIA-CNCPKLTELDL 209
K S C L L L C +++ + LA ++ C +T L+L
Sbjct: 570 KIFSDDGGCPMLKSLILDNCESLTAVRFCNSSLASLSLVGCRAVTSLEL 618
Score = 45.8 bits (107), Expect = 4e-05
Identities = 45/147 (30%), Positives = 63/147 (42%), Gaps = 11/147 (7%)
Query: 66 LQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCP 125
L ++ +V L L C ++ IM C LT+LD + C + D +S SCP
Sbjct: 655 LSVLNIEAPYMVSLELKGCGVLSEASIM----CPLLTSLDASFCSQLRDDCLSATTASCP 710
Query: 126 NLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALK-YLSRCSKLVRLKLG 184
+ L L SC + GL SS L L + D S + L+ C +L LKL
Sbjct: 711 LIESLVLMSCPSIGSDGL----SSLNGLPNLTVLDLSYTFLMNLEPVFKSCIQLKVLKLQ 766
Query: 185 LCTNISDIGLAHIACN--CPKLTELDL 209
C ++D L + P L ELDL
Sbjct: 767 ACKYLTDSSLEPLYKEGALPALEELDL 793
Score = 38.5 bits (88), Expect = 0.006
Identities = 31/110 (28%), Positives = 42/110 (38%), Gaps = 31/110 (28%)
Query: 100 NLTTLDLTCCRFV-----TDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLE 154
NL +DLTC V ++ + CP LA L L+SC+M
Sbjct: 895 NLKEVDLTCSNLVLLNLSNCCSLEVLKLGCPRLASLFLQSCNM----------------- 937
Query: 155 ELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKL 204
D +GV +S CS L L L C IS + ++ CP L
Sbjct: 938 -----DEAGVEAA----ISGCSSLETLDLRFCPKISSVSMSKFRTVCPSL 978
Score = 35.8 bits (81), Expect = 0.039
Identities = 19/63 (30%), Positives = 31/63 (49%), Gaps = 2/63 (3%)
Query: 66 LQIIGSNCKSLVELGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAISTIANSC 124
L+++ C L L L C + G+ + GC +L TLDL C ++ ++S C
Sbjct: 917 LEVLKLGCPRLASLFLQSC-NMDEAGVEAAISGCSSLETLDLRFCPKISSVSMSKFRTVC 975
Query: 125 PNL 127
P+L
Sbjct: 976 PSL 978
>At1g80570 unknown protein
Length = 467
Score = 60.8 bits (146), Expect = 1e-09
Identities = 50/188 (26%), Positives = 73/188 (38%), Gaps = 36/188 (19%)
Query: 58 GVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAI 117
G +V D L ++ +NC SL +L LS C +T++GI + C L++L L +T +
Sbjct: 81 GKQVDDQGLLVLTTNCHSLTDLTLSFCTFITDVGIGHLSSCPELSSLKLNFAPRITGCGV 140
Query: 118 STIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIAL-------- 169
++A C L L L C V + + LEEL + +C + + L
Sbjct: 141 LSLAVGCKKLRRLHLIRCLNVASVEWLEYFGKLETLEELCIKNCRAIGEGDLIKLRNSWR 200
Query: 170 ----------------------------KYLSRCSKLVRLKLGLCTNISDIGLAHIACNC 201
K L C LV L LG C GLA + NC
Sbjct: 201 KLTSLQFEVDANYRYMKVYDQLDVERWPKQLVPCDSLVELSLGNCIIAPGRGLACVLRNC 260
Query: 202 PKLTELDL 209
L +L L
Sbjct: 261 KNLEKLHL 268
Score = 56.2 bits (134), Expect = 3e-08
Identities = 36/117 (30%), Positives = 54/117 (45%), Gaps = 2/117 (1%)
Query: 92 IMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCL 151
++ C +LT L L+ C F+TD I ++ SCP L+ LKL +T G+ + C
Sbjct: 90 LVLTTNCHSLTDLTLSFCTFITDVGIGHLS-SCPELSSLKLNFAPRITGCGVLSLAVGCK 148
Query: 152 MLEELDLTDCSGVNDIA-LKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
L L L C V + L+Y + L L + C I + L + + KLT L
Sbjct: 149 KLRRLHLIRCLNVASVEWLEYFGKLETLEELCIKNCRAIGEGDLIKLRNSWRKLTSL 205
Score = 50.1 bits (118), Expect = 2e-06
Identities = 30/116 (25%), Positives = 57/116 (48%), Gaps = 7/116 (6%)
Query: 103 TLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIG-------LYQIGSSCLMLEE 155
+L + C A+ ++ PNL+ +++ ++++G L + ++C L +
Sbjct: 42 SLRIGCGLVPASDALLSLCRRFPNLSKVEIIYSGWMSKLGKQVDDQGLLVLTTNCHSLTD 101
Query: 156 LDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYR 211
L L+ C+ + D+ + +LS C +L LKL I+ G+ +A C KL L L R
Sbjct: 102 LTLSFCTFITDVGIGHLSSCPELSSLKLNFAPRITGCGVLSLAVGCKKLRRLHLIR 157
Score = 39.7 bits (91), Expect = 0.003
Identities = 50/200 (25%), Positives = 87/200 (43%), Gaps = 19/200 (9%)
Query: 2 IDVSRCNCV-SPS-GLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGV 59
+++S NC+ +P GL V+ + LE+++ C + + ++ HL I +
Sbjct: 238 VELSLGNCIIAPGRGLACVLRNCKNLEKLHLDMCTGVSDSDIIALVQKASHLRSISL--- 294
Query: 60 RV-SDFILQIIGSNCKSLVELGLSK----CIGVTNMGIMQVVGCCNLTTLDLTCCRFVTD 114
RV SDF L ++ + L + LS C + + I G + T
Sbjct: 295 RVPSDFTLPLLNNITLRLTDESLSAIAQHCSKLESFKISFSDG-------EFPSLFSFTL 347
Query: 115 AAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSR 174
I T+ CP + L L+ + ++G+ + S LE L+L C V+D L +S+
Sbjct: 348 QGIITLIQKCP-VRELSLDHVCVFNDMGMEAL-CSAQKLEILELVHCQEVSDEGLILVSQ 405
Query: 175 CSKLVRLKLGLCTNISDIGL 194
L LKL C ++D G+
Sbjct: 406 FPSLNVLKLSKCLGVTDDGM 425
Score = 33.5 bits (75), Expect = 0.19
Identities = 43/206 (20%), Positives = 79/206 (37%), Gaps = 24/206 (11%)
Query: 10 VSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVR-VSDFILQI 68
++ G+LS+ G + L +++ CL+ S L+ L + I R + + L
Sbjct: 135 ITGCGVLSLAVGCKKLRRLHLIRCLNVASVEWLEYFGKLETLEELCIKNCRAIGEGDLIK 194
Query: 69 IGSNCKSLVELGLSKCIGVTNMGIM----------QVVGCCNLTTLDLTCCRFVTDAAIS 118
+ ++ + L L M + Q+V C +L L L C ++
Sbjct: 195 LRNSWRKLTSLQFEVDANYRYMKVYDQLDVERWPKQLVPCDSLVELSLGNCIIAPGRGLA 254
Query: 119 TIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKL 178
+ +C NL L L+ C V++ + + L + L S D L L+
Sbjct: 255 CVLRNCKNLEKLHLDMCTGVSDSDIIALVQKASHLRSISLRVPS---DFTLPLLN----- 306
Query: 179 VRLKLGLCTNISDIGLAHIACNCPKL 204
+ ++D L+ IA +C KL
Sbjct: 307 -----NITLRLTDESLSAIAQHCSKL 327
Score = 32.0 bits (71), Expect = 0.57
Identities = 21/56 (37%), Positives = 29/56 (51%)
Query: 68 IIGSNCKSLVELGLSKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANS 123
I+ S SL L LSKC+GVT+ G+ +VG L L + C V+ + A S
Sbjct: 401 ILVSQFPSLNVLKLSKCLGVTDDGMRPLVGSHKLELLVVEDCPQVSRRGVHGAATS 456
>At5g07670 unknown protein
Length = 476
Score = 60.5 bits (145), Expect = 1e-09
Identities = 54/177 (30%), Positives = 78/177 (43%), Gaps = 20/177 (11%)
Query: 41 LTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKC-----IGV---TNMGI 92
L G NL+ L V S+ L + C L EL L KC +G+ N+ I
Sbjct: 180 LAGGCSNLRKLVV-----TNTSELGLLNVAEECSRLQELELHKCSDSVLLGIGAFENLQI 234
Query: 93 MQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLM 152
+++VG +D V+D + +A C L L+L C+ + G+ +IG C M
Sbjct: 235 LRLVG-----NVDGLYNSLVSDIGLMILAQGCKRLVKLELVGCEGGFD-GIKEIGECCQM 288
Query: 153 LEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
LEEL + D + + L L C L LKL C I + ++C CP L L L
Sbjct: 289 LEELTVCD-NKMESGWLGGLRYCENLKTLKLVSCKKIDNDPDESLSCCCPALERLQL 344
Score = 57.0 bits (136), Expect = 2e-08
Identities = 59/236 (25%), Positives = 94/236 (39%), Gaps = 57/236 (24%)
Query: 11 SPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVI-RIDGVR---VSDFIL 66
S GLL+V L+++ C + + +NL+ L ++ +DG+ VSD L
Sbjct: 196 SELGLLNVAEECSRLQELELHKCSDSVLLGI-GAFENLQILRLVGNVDGLYNSLVSDIGL 254
Query: 67 QIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCC-------------------------NL 101
I+ CK LV+L L C G + GI ++ CC NL
Sbjct: 255 MILAQGCKRLVKLELVGCEGGFD-GIKEIGECCQMLEELTVCDNKMESGWLGGLRYCENL 313
Query: 102 TTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDC 161
TL L C+ + + +++ CP L L+LE C + + + + C E+ DC
Sbjct: 314 KTLKLVSCKKIDNDPDESLSCCCPALERLQLEKCQLRDKNTVKALFKMCEAAREIVFQDC 373
Query: 162 SGVND-----------IALKYLSRCS---------------KLVRLKLGLCTNISD 191
G+++ + L YL CS +L LK+ C NI D
Sbjct: 374 WGLDNDIFSLAMAFGRVKLLYLEGCSLLTTSGLESVILHWHELEHLKVVSCKNIKD 429
Score = 31.2 bits (69), Expect = 0.97
Identities = 15/37 (40%), Positives = 25/37 (67%), Gaps = 3/37 (8%)
Query: 175 CSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYR 211
CS L +L + TN S++GL ++A C +L EL+L++
Sbjct: 184 CSNLRKL---VVTNTSELGLLNVAEECSRLQELELHK 217
>At3g50080 unknown protein
Length = 522
Score = 60.5 bits (145), Expect = 1e-09
Identities = 53/187 (28%), Positives = 86/187 (45%), Gaps = 7/187 (3%)
Query: 25 LEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKC 84
L+++ CL L+ IR++ ++V+D L I S C +L L + K
Sbjct: 233 LKKVKIIRCLGNWDRVFEMNGNGNSSLTEIRLERLQVTDIGLFGI-SKCSNLETLHIVKT 291
Query: 85 IGVTNMGIMQVVGCCNLTT---LDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEI 141
+N+G+ VV C L +D + + D + ++A C NL L L D T +
Sbjct: 292 PDCSNLGLASVVERCKLLRKLHIDGWRVKRIGDQGLMSVAKHCLNLQELVLIGVD-ATYM 350
Query: 142 GLYQIGSSCLMLEELDLTDCSGVNDIALKYLS-RCSKLVRLKLGLCTNISDIGLAHIACN 200
L I S+C LE L L + D + ++ +C L + + C ISD+G+ +A
Sbjct: 351 SLSAIASNCKKLERLALCGSGTIGDAEIGCIAEKCVTLRKFCIKGCL-ISDVGVQALALG 409
Query: 201 CPKLTEL 207
CPKL +L
Sbjct: 410 CPKLVKL 416
Score = 48.1 bits (113), Expect = 8e-06
Identities = 39/121 (32%), Positives = 55/121 (45%), Gaps = 20/121 (16%)
Query: 21 GHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELG 80
G +GL + A HCL NL+ L +I +D +S L I SNCK L L
Sbjct: 323 GDQGLMSV-AKHCL------------NLQELVLIGVDATYMS---LSAIASNCKKLERLA 366
Query: 81 L--SKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMV 138
L S IG +G + C L + C ++D + +A CP L LK++ C +V
Sbjct: 367 LCGSGTIGDAEIGCI-AEKCVTLRKFCIKGC-LISDVGVQALALGCPKLVKLKVKKCSLV 424
Query: 139 T 139
T
Sbjct: 425 T 425
Score = 42.4 bits (98), Expect = 4e-04
Identities = 23/73 (31%), Positives = 38/73 (51%), Gaps = 1/73 (1%)
Query: 96 VGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEE 155
+ C NL + L CR +TD + + A +C +L L SC + G+ + C +LEE
Sbjct: 128 IRCSNLIRVKLRGCREITDLGMESFARNCKSLRKLSCGSCTFGAK-GINAMLEHCKVLEE 186
Query: 156 LDLTDCSGVNDIA 168
L L G++++A
Sbjct: 187 LSLKRIRGLHELA 199
Score = 42.0 bits (97), Expect = 5e-04
Identities = 40/138 (28%), Positives = 63/138 (44%), Gaps = 10/138 (7%)
Query: 41 LTNGLKNLKHLSVIRIDGVRVS---DFILQIIGSNCKSLVELGLSKCIGV--TNMGIMQV 95
L + ++ K L + IDG RV D L + +C +L EL L IGV T M + +
Sbjct: 299 LASVVERCKLLRKLHIDGWRVKRIGDQGLMSVAKHCLNLQELVL---IGVDATYMSLSAI 355
Query: 96 VGCCN-LTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLE 154
C L L L + DA I IA C L ++ C +++++G+ + C L
Sbjct: 356 ASNCKKLERLALCGSGTIGDAEIGCIAEKCVTLRKFCIKGC-LISDVGVQALALGCPKLV 414
Query: 155 ELDLTDCSGVNDIALKYL 172
+L + CS V ++L
Sbjct: 415 KLKVKKCSLVTGEVREWL 432
Score = 37.7 bits (86), Expect = 0.010
Identities = 20/45 (44%), Positives = 29/45 (64%), Gaps = 1/45 (2%)
Query: 164 VNDIALKYLS-RCSKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
++D AL +S RCS L+R+KL C I+D+G+ A NC L +L
Sbjct: 118 LSDEALFIVSIRCSNLIRVKLRGCREITDLGMESFARNCKSLRKL 162
Score = 30.0 bits (66), Expect = 2.2
Identities = 16/46 (34%), Positives = 25/46 (53%)
Query: 164 VNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
V DI L +S+CS L L + + S++GLA + C L +L +
Sbjct: 269 VTDIGLFGISKCSNLETLHIVKTPDCSNLGLASVVERCKLLRKLHI 314
>At5g67250 unknown protein
Length = 527
Score = 58.5 bits (140), Expect = 6e-09
Identities = 52/179 (29%), Positives = 81/179 (45%), Gaps = 10/179 (5%)
Query: 33 CLSELS-----APLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGV 87
CL EL PL + LK L +IR G D +LQ+I + SL E+ L + + V
Sbjct: 219 CLKELVNGQVFEPLLATTRTLKTLKIIRCLGDW--DKVLQMIANGKSSLSEIHLER-LQV 275
Query: 88 TNMGIMQVVGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLES--CDMVTEIGLYQ 145
+++G+ + C N+ TL + ++ + +A C L L ++ + + + GL
Sbjct: 276 SDIGLSAISKCSNVETLHIVKTPECSNFGLIYVAERCKLLRKLHIDGWRTNRIGDEGLLS 335
Query: 146 IGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKL 204
+ CL L+EL L + + S C KL RL L I D +A IA C L
Sbjct: 336 VAKHCLNLQELVLIGVNATHMSLAAIASNCEKLERLALCGSGTIGDTEIACIARKCGAL 394
Score = 56.2 bits (134), Expect = 3e-08
Identities = 51/187 (27%), Positives = 82/187 (43%), Gaps = 7/187 (3%)
Query: 25 LEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKC 84
L+ + CL + L LS I ++ ++VSD L I S C ++ L + K
Sbjct: 239 LKTLKIIRCLGDWDKVLQMIANGKSSLSEIHLERLQVSDIGLSAI-SKCSNVETLHIVKT 297
Query: 85 IGVTNMGIMQVVGCCNLTT---LDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEI 141
+N G++ V C L +D + D + ++A C NL L L + T +
Sbjct: 298 PECSNFGLIYVAERCKLLRKLHIDGWRTNRIGDEGLLSVAKHCLNLQELVLIGVN-ATHM 356
Query: 142 GLYQIGSSCLMLEELDLTDCSGVNDIALKYLSR-CSKLVRLKLGLCTNISDIGLAHIACN 200
L I S+C LE L L + D + ++R C L + + C +SD G+ +A
Sbjct: 357 SLAAIASNCEKLERLALCGSGTIGDTEIACIARKCGALRKFCIKGCP-VSDRGIEALAVG 415
Query: 201 CPKLTEL 207
CP L +L
Sbjct: 416 CPNLVKL 422
Score = 48.1 bits (113), Expect = 8e-06
Identities = 50/144 (34%), Positives = 63/144 (43%), Gaps = 31/144 (21%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRV 61
ID R N + GLLSV A HCL NL+ L +I GV
Sbjct: 320 IDGWRTNRIGDEGLLSV-----------AKHCL------------NLQELVLI---GVNA 353
Query: 62 SDFILQIIGSNCKSLVELGL--SKCIGVTNMGIMQVVGCCNLTTLDLTCCRFVTDAAIST 119
+ L I SNC+ L L L S IG T + + C L + C V+D I
Sbjct: 354 THMSLAAIASNCEKLERLALCGSGTIGDTEIACI-ARKCGALRKFCIKGCP-VSDRGIEA 411
Query: 120 IANSCPNLACLKLESCDMVT-EIG 142
+A CPNL LK++ C +VT EIG
Sbjct: 412 LAVGCPNLVKLKVKKCKVVTGEIG 435
Score = 46.2 bits (108), Expect = 3e-05
Identities = 25/73 (34%), Positives = 39/73 (53%), Gaps = 1/73 (1%)
Query: 96 VGCCNLTTLDLTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEE 155
V C NLT + L CR +TD + A +C NL L + SC+ + G+ + C +LEE
Sbjct: 130 VRCLNLTRVKLRGCREITDLGMEDFAKNCKNLKKLSVGSCNFGAK-GVNAMLEHCKLLEE 188
Query: 156 LDLTDCSGVNDIA 168
L + G+++ A
Sbjct: 189 LSVKRLRGIHEAA 201
Score = 38.5 bits (88), Expect = 0.006
Identities = 42/167 (25%), Positives = 74/167 (44%), Gaps = 8/167 (4%)
Query: 46 KNLKHLSVIRID-GVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMG-IMQVVGCCNLTT 103
KNLK LSV + G + + +L+ +CK L EL + + G+ ++ + + ++
Sbjct: 159 KNLKKLSVGSCNFGAKGVNAMLE----HCKLLEELSVKRLRGIHEAAELIHLPDDASSSS 214
Query: 104 LDLTCCR-FVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCS 162
L C + V + + L LK+ C + L I + L E+ L
Sbjct: 215 LRSICLKELVNGQVFEPLLATTRTLKTLKIIRCLGDWDKVLQMIANGKSSLSEIHLERLQ 274
Query: 163 GVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
V+DI L +S+CS + L + S+ GL ++A C L +L +
Sbjct: 275 -VSDIGLSAISKCSNVETLHIVKTPECSNFGLIYVAERCKLLRKLHI 320
Score = 35.8 bits (81), Expect = 0.039
Identities = 19/47 (40%), Positives = 28/47 (59%), Gaps = 1/47 (2%)
Query: 164 VNDIALKYLS-RCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
++D AL +S RC L R+KL C I+D+G+ A NC L +L +
Sbjct: 120 LSDEALAMISVRCLNLTRVKLRGCREITDLGMEDFAKNCKNLKKLSV 166
>At5g51380 putative protein
Length = 479
Score = 58.2 bits (139), Expect = 7e-09
Identities = 54/224 (24%), Positives = 97/224 (43%), Gaps = 32/224 (14%)
Query: 14 GLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVI-RIDGV---RVSDFILQII 69
GLLS+ L+++ C L + +NL+ L ++ +DG+ VSD L I+
Sbjct: 201 GLLSLAEDCSDLQELELHKCSDNLLRGIA-ACENLRGLRLVGSVDGLYSSSVSDIGLTIL 259
Query: 70 GSNCKSLVELGLSKCIGVTNMGIMQVVGCC-------------------------NLTTL 104
CK LV+L LS C G + GI + CC +L TL
Sbjct: 260 AQGCKRLVKLELSGCEGSFD-GIKAIGQCCEVLEELSICDHRMDDGWIAALSYFESLKTL 318
Query: 105 DLTCCRFVTDA-AISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSG 163
++ CR + + + SCP L L+L C + + G+ + C + ++++ DC G
Sbjct: 319 LISSCRKIDSSPGPGKLLGSCPALESLQLRRCCLNDKEGMRALFKVCDGVTKVNIQDCWG 378
Query: 164 VNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
++D + ++ L L C+ ++ GL + + +L +
Sbjct: 379 LDDDSFSLAKAFRRVRFLSLEGCSILTTSGLESVILHWEELESM 422
Score = 44.7 bits (104), Expect = 8e-05
Identities = 29/77 (37%), Positives = 44/77 (56%), Gaps = 9/77 (11%)
Query: 139 TEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLVRLKL-----GL-CTNISDI 192
TE+GL + C L+EL+L CS D L+ ++ C L L+L GL +++SDI
Sbjct: 198 TELGLLSLAEDCSDLQELELHKCS---DNLLRGIAACENLRGLRLVGSVDGLYSSSVSDI 254
Query: 193 GLAHIACNCPKLTELDL 209
GL +A C +L +L+L
Sbjct: 255 GLTILAQGCKRLVKLEL 271
>At5g51370 putative protein
Length = 446
Score = 57.4 bits (137), Expect = 1e-08
Identities = 55/227 (24%), Positives = 95/227 (41%), Gaps = 32/227 (14%)
Query: 11 SPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVI-RIDGV---RVSDFIL 66
S GLLS+ L+++ C L + KNLK L ++ +DG+ VSD L
Sbjct: 162 SELGLLSLAGDCSDLQELELHKCNDNLLHGIA-ACKNLKGLRLVGSVDGLYSSSVSDIGL 220
Query: 67 QIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCC-------------------------NL 101
+ C+SLV+L LS C G + GI + CC +L
Sbjct: 221 TFLAQGCRSLVKLELSGCEGSFD-GIKAIGQCCEVLEELSICDHRMDDGWIAALSYFESL 279
Query: 102 TTLDLTCCRFV-TDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTD 160
L ++ CR + + SCP + L+L+ C + + G+ + C E+++ D
Sbjct: 280 KILRISSCRKIDASPGPEKLLRSCPAMESLQLKRCCLNDKEGIKALFKVCDGATEVNIQD 339
Query: 161 CSGVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
C G++D ++ L L C+ ++ GL + + +L +
Sbjct: 340 CWGLSDDCFSLAKAFRRVRFLSLEGCSVLTSGGLESVILHWEELESM 386
Score = 43.5 bits (101), Expect = 2e-04
Identities = 33/96 (34%), Positives = 49/96 (50%), Gaps = 16/96 (16%)
Query: 127 LACLKLESCDMV-------TEIGLYQIGSSCLMLEELDLTDCSGVNDIALKYLSRCSKLV 179
L L ES D++ +E+GL + C L+EL+L C ND L ++ C L
Sbjct: 143 LRILSRESFDLLNLKVINASELGLLSLAGDCSDLQELELHKC---NDNLLHGIAACKNLK 199
Query: 180 RLKL-----GL-CTNISDIGLAHIACNCPKLTELDL 209
L+L GL +++SDIGL +A C L +L+L
Sbjct: 200 GLRLVGSVDGLYSSSVSDIGLTFLAQGCRSLVKLEL 235
Score = 30.0 bits (66), Expect = 2.2
Identities = 21/51 (41%), Positives = 31/51 (60%), Gaps = 4/51 (7%)
Query: 162 SGVNDIALKYLSRCS-KLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYR 211
S V D L+ LSR S L+ LK+ N S++GL +A +C L EL+L++
Sbjct: 136 SDVIDRGLRILSRESFDLLNLKV---INASELGLLSLAGDCSDLQELELHK 183
>At3g07550 unknown protein
Length = 395
Score = 56.6 bits (135), Expect = 2e-08
Identities = 37/131 (28%), Positives = 66/131 (50%), Gaps = 9/131 (6%)
Query: 48 LKHLSVIRIDGVRV-SDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCC-NLTTLD 105
L+HLS + G V +D L + L L L C G+++ GI + C NL+ +
Sbjct: 93 LEHLS---LSGCTVLNDSSLDSLRYPGARLHTLYLDCCFGISDDGISTIASFCPNLSVVS 149
Query: 106 LTCCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVN 165
L C ++D + T+A + +L C+ L C +V++ G+ + +CL LE + +++C +
Sbjct: 150 LYRCN-ISDIGLETLARASLSLKCVNLSYCPLVSDFGIKALSQACLQLESVKISNCKSIT 208
Query: 166 DIALKYLSRCS 176
+ S CS
Sbjct: 209 GVG---FSGCS 216
Score = 55.5 bits (132), Expect = 5e-08
Identities = 52/219 (23%), Positives = 93/219 (41%), Gaps = 33/219 (15%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTN--GLKNLKHLSVIRIDGV 59
+ + RCN +S GL ++ L+ +N +C PL + G+K L + ++++ V
Sbjct: 148 VSLYRCN-ISDIGLETLARASLSLKCVNLSYC------PLVSDFGIKALSQ-ACLQLESV 199
Query: 60 RVSDFILQIIGSNCKSLVELGLSKC---IGVTNM--------GIMQVVGCCNLTTLDLT- 107
++S NCKS+ +G S C +G + GI ++ + L+++
Sbjct: 200 KIS---------NCKSITGVGFSGCSPTLGYVDADSCQLEPKGITGIISGGGIEFLNISG 250
Query: 108 -CCRFVTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVND 166
C D + + L L L C V + + I C +L+E +L C V
Sbjct: 251 VSCYIRKDGLVPIGSGIASKLRILNLRMCRTVGDESIEAIAKGCPLLQEWNLALCHEVKI 310
Query: 167 IALKYLSR-CSKLVRLKLGLCTNISDIGLAHIACNCPKL 204
+ + + C L +L + C N+ D GL + C C L
Sbjct: 311 SGWEAVGKWCRNLKKLHVNRCRNLCDQGLLALRCGCMNL 349
Score = 41.2 bits (95), Expect = 0.001
Identities = 25/60 (41%), Positives = 35/60 (57%), Gaps = 1/60 (1%)
Query: 153 LEELDLTDCSGVNDIALKYLSRC-SKLVRLKLGLCTNISDIGLAHIACNCPKLTELDLYR 211
LE L L+ C+ +ND +L L ++L L L C ISD G++ IA CP L+ + LYR
Sbjct: 93 LEHLSLSGCTVLNDSSLDSLRYPGARLHTLYLDCCFGISDDGISTIASFCPNLSVVSLYR 152
Score = 33.5 bits (75), Expect = 0.19
Identities = 42/157 (26%), Positives = 62/157 (38%), Gaps = 28/157 (17%)
Query: 2 IDVSRCNCVSPSGLLSVISGHEGLEQINAGHCLSELSAPLTNGLKNLKHLSVIRIDGVRV 61
+D C + P G+ +ISG G+E +N +S +S IR DG
Sbjct: 222 VDADSCQ-LEPKGITGIISGG-GIEFLN----ISGVSC-------------YIRKDG--- 259
Query: 62 SDFILQIIGSNCKSLVE-LGLSKCIGVTNMGIMQVV-GCCNLTTLDLTCCRFVTDAAIST 119
L IGS S + L L C V + I + GC L +L C V +
Sbjct: 260 ----LVPIGSGIASKLRILNLRMCRTVGDESIEAIAKGCPLLQEWNLALCHEVKISGWEA 315
Query: 120 IANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEEL 156
+ C NL L + C + + GL + C+ L+ L
Sbjct: 316 VGKWCRNLKKLHVNRCRNLCDQGLLALRCGCMNLQIL 352
>At3g62980 transport inhibitor response 1 (TIR1)
Length = 594
Score = 55.1 bits (131), Expect = 6e-08
Identities = 45/167 (26%), Positives = 80/167 (46%), Gaps = 21/167 (12%)
Query: 61 VSDFILQIIGSNCKSLVELG--------LSKCIGVTNMGIMQV-VGCCNLTTLDLTCCRF 111
+ D L+++ S CK L EL + + +T G++ V +GC L ++ L CR
Sbjct: 325 IEDAGLEVLASTCKDLRELRVFPSEPFVMEPNVALTEQGLVSVSMGCPKLESV-LYFCRQ 383
Query: 112 VTDAAISTIANSCPNLACLKL-----ESCDMVT----EIGLYQIGSSCLMLEELDLTDCS 162
+T+AA+ TIA + PN+ +L ++ D +T +IG I C L L L+
Sbjct: 384 MTNAALITIARNRPNMTRFRLCIIEPKAPDYLTLEPLDIGFGAIVEHCKDLRRLSLSGL- 442
Query: 163 GVNDIALKYLSRCSKLVRLKLGLCTNISDIGLAHIACNCPKLTELDL 209
+ D +Y+ +K + + SD+G+ H+ C L +L++
Sbjct: 443 -LTDKVFEYIGTYAKKMEMLSVAFAGDSDLGMHHVLSGCDSLRKLEI 488
Score = 50.8 bits (120), Expect = 1e-06
Identities = 37/105 (35%), Positives = 53/105 (50%), Gaps = 6/105 (5%)
Query: 112 VTDAAISTIANSCPNLACLKLESCDMVTEIGLYQIGSSCLMLEELDLTDCSGVNDIALKY 171
VTD + IA S N L L SC+ + GL I ++C L+ELDL + S V+D++ +
Sbjct: 117 VTDDCLELIAKSFKNFKVLVLSSCEGFSTDGLAAIAATCRNLKELDLRE-SDVDDVSGHW 175
Query: 172 LSR----CSKLVRLKLG-LCTNISDIGLAHIACNCPKLTELDLYR 211
LS + LV L + L + +S L + CP L L L R
Sbjct: 176 LSHFPDTYTSLVSLNISCLASEVSFSALERLVTRCPNLKSLKLNR 220
Score = 44.7 bits (104), Expect = 8e-05
Identities = 51/214 (23%), Positives = 81/214 (37%), Gaps = 58/214 (27%)
Query: 51 LSVIRIDGVRVSDFILQIIGSNCKSLVELGLSKCIGVTNMGIMQVVGCC----------- 99
L IR+ + V+D L++I + K+ L LS C G + G+ + C
Sbjct: 107 LEEIRLKRMVVTDDCLELIAKSFKNFKVLVLSSCEGFSTDGLAAIAATCRNLKELDLRES 166
Query: 100 ------------------NLTTLDLTCCRF-VTDAAISTIANSCPNLACLKLESCDMVTE 140
+L +L+++C V+ +A+ + CPNL LKL + +
Sbjct: 167 DVDDVSGHWLSHFPDTYTSLVSLNISCLASEVSFSALERLVTRCPNLKSLKLNRAVPLEK 226
Query: 141 IG--------LYQIGSSCLMLE---------ELDLTDC------SGVNDIALKYL----S 173
+ L ++G+ E + L+ C SG D YL S
Sbjct: 227 LATLLQRAPQLEELGTGGYTAEVRPDVYSGLSVALSGCKELRCLSGFWDAVPAYLPAVYS 286
Query: 174 RCSKLVRLKLGLCTNISDIGLAHIACNCPKLTEL 207
CS+L L L T + L + C CPKL L
Sbjct: 287 VCSRLTTLNLSYAT-VQSYDLVKLLCQCPKLQRL 319
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.336 0.145 0.470
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 7,464,698
Number of Sequences: 26719
Number of extensions: 280779
Number of successful extensions: 1855
Number of sequences better than 10.0: 104
Number of HSP's better than 10.0 without gapping: 53
Number of HSP's successfully gapped in prelim test: 51
Number of HSP's that attempted gapping in prelim test: 1254
Number of HSP's gapped (non-prelim): 339
length of query: 375
length of database: 11,318,596
effective HSP length: 101
effective length of query: 274
effective length of database: 8,619,977
effective search space: 2361873698
effective search space used: 2361873698
T: 11
A: 40
X1: 15 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 39 (21.7 bits)
S2: 61 (28.1 bits)
Medicago: description of AC144541.17


