

BLAST2 result
BLASTP 2.2.2 [Dec-14-2001]
Reference: Altschul, Stephen F., Thomas L. Madden, Alejandro A. Schaffer,
Jinghui Zhang, Zheng Zhang, Webb Miller, and David J. Lipman (1997),
"Gapped BLAST and PSI-BLAST: a new generation of protein database search
programs", Nucleic Acids Res. 25:3389-3402.
Query= AC144541.15 - phase: 0
(593 letters)
Database: ara_mips
26,719 sequences; 11,318,596 total letters
Searching..................................................done
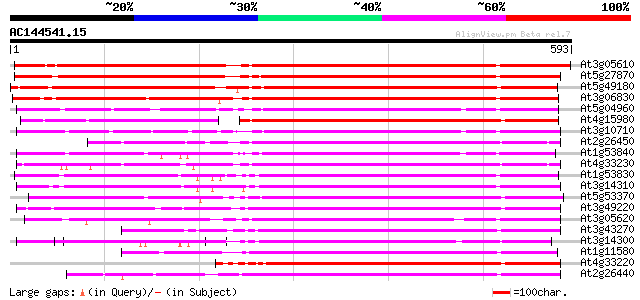
Score E
Sequences producing significant alignments: (bits) Value
At3g05610 putative pectinesterase 579 e-165
At5g27870 pectin methyl-esterase - like protein 560 e-160
At5g49180 pectin methylesterase 522 e-148
At3g06830 pectin methylesterase like protein 474 e-134
At5g04960 pectinesterase 391 e-109
At4g15980 pectinesterase like protein 382 e-106
At3g10710 putative pectinesterase 377 e-105
At2g26450 putative pectinesterase 370 e-103
At1g53840 unknown protein 369 e-102
At4g33230 pectinesterase - like protein 365 e-101
At1g53830 unknown protein 363 e-100
At3g14310 pectin methylesterase like protein 352 3e-97
At5g53370 pectinesterase 350 1e-96
At3g49220 pectinesterase - like protein 345 5e-95
At3g05620 putative pectinesterase 341 8e-94
At3g43270 pectinesterase like protein 338 5e-93
At3g14300 putative pectin methylesterase 337 8e-93
At1g11580 unknown protein 325 6e-89
At4g33220 pectinesterase like protein 314 8e-86
At2g26440 putative pectinesterase 313 2e-85
>At3g05610 putative pectinesterase
Length = 669
Score = 579 bits (1492), Expect = e-165
Identities = 288/590 (48%), Positives = 396/590 (66%), Gaps = 26/590 (4%)
Query: 6 DAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAP 65
++KR +R +I +S+ LL++MVVAVTV V N K G+ K + V+ SVKAVK +CAP
Sbjct: 8 ESKRKRRYIVITISSVLLISMVVAVTVGVSLN--KHDGDSKGK-AEVNASVKAVKDVCAP 64
Query: 66 TDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
TDY+K CED+LI + N T+P EL+K AFN+T+ +I++ KK+ + E +KD RT+ ALD
Sbjct: 65 TDYRKTCEDTLIKNGKNTTDPMELVKTAFNVTMKQITDAAKKSQTIMELQKDSRTRMALD 124
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDA 185
CK++M ++DE S E F+ + LD L +L++WLS AI+++ETCL+ F+ T +A
Sbjct: 125 QCKELMDYALDELSNSFEELGKFEFHLLDEALINLRIWLSAAISHEETCLEGFQGTQGNA 184
Query: 186 GKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPA--GRRLLKESVDGEEDVLGHGGDF 243
G+ MK+ L+T++ ++ NGL+II+++S +M+ P RRLL E
Sbjct: 185 GETMKKALKTAIELTHNGLAIISEMSNFVGQMQIPGLNSRRLLAEG-------------- 230
Query: 244 ELPEWVDDRAGVRKLLNKMTGRK-LQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIY 302
P WVD R RKLL ++ +VVA+DGSG + TI EAL+ VPKK FV++
Sbjct: 231 -FPSWVDQRG--RKLLQAAAAYSDVKPDIVVAQDGSGQYKTINEALQFVPKKRNTTFVVH 287
Query: 303 IKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
IK G+YKEYV+V KTM+H+VFIGDG KT I+GNKN+ DG+ T++TA+VAI G++F+
Sbjct: 288 IKAGLYKEYVQVNKTMSHLVFIGDGPDKTIISGNKNYKDGITTYRTATVAIVGNYFIAKN 347
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
IGFEN+AG KHQAVA+RVQSD SIF+ CR DGYQDTLY H+ RQF+RDC ISGTIDF+F
Sbjct: 348 IGFENTAGAIKHQAVAVRVQSDESIFFNCRFDGYQDTLYTHSHRQFFRDCTISGTIDFLF 407
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
GD+ AV QNCT +VRKPL NQ C +TA GRK+ + TG + QG +I +P Y V+ +K
Sbjct: 408 GDAAAVFQNCTLLVRKPLPNQACPITAHGRKDPRESTGFVFQGCTIAGEPDYLAVKETSK 467
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
AYL RPWK++SRTI ++T+I D + P+G+ PW G G T +Y E N GPGS +
Sbjct: 468 AYLGRPWKEYSRTIIMNTFIPDFVQPQGWQPW---LGDFGLKTLFYSEVQNTGPGSALAN 524
Query: 543 RVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPGATKTPPSRPT 592
RV W G+KT++ E F P ++ GDDWI VPY+ G P+ T
Sbjct: 525 RVTWAGIKTLSEEDILKFTPAQYIQGDDWIPGKGVPYTTGLLAGNPAAAT 574
>At5g27870 pectin methyl-esterase - like protein
Length = 732
Score = 560 bits (1444), Expect = e-160
Identities = 277/579 (47%), Positives = 388/579 (66%), Gaps = 28/579 (4%)
Query: 6 DAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAP 65
DAKR KR II +S+ LL++MVVAVT+ V N G+E ++ SVKA+K +CAP
Sbjct: 9 DAKRKKRYVIISISSVLLISMVVAVTIGVSVNKSDNAGDE-----EITTSVKAIKDVCAP 63
Query: 66 TDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
TDYK+ CED+L A + ++P EL+K AFN T+ +IS+ KK+ + E +KD R K ALD
Sbjct: 64 TDYKETCEDTLRKDAKDTSDPLELVKTAFNATMKQISDVAKKSQTMIELQKDPRAKMALD 123
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDA 185
CK++M +I E +S E F+ + +D L L++WLS I++++TCLD F+ T +A
Sbjct: 124 QCKELMDYAIGELSKSFEELGKFEFHKVDEALVKLRIWLSATISHEQTCLDGFQGTQGNA 183
Query: 186 GKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPA--GRRLLKESVDGEEDVLGHGGDF 243
G+ +K+ L+T++ ++ NGL+++ ++S +M+ P RRLL +
Sbjct: 184 GETIKKALKTAVQLTHNGLAMVTEMSNYLGQMQIPEMNSRRLLSQ--------------- 228
Query: 244 ELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYI 303
E P W+D RA R+LLN +++ +VVA+DGSG + TI EAL VPKK FV++I
Sbjct: 229 EFPSWMDARA--RRLLNAPMS-EVKPDIVVAQDGSGQYKTINEALNFVPKKKNTTFVVHI 285
Query: 304 KEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGI 363
KEG+YKEYV+V ++MTH+VFIGDG KT I+G+K++ DG+ T+KTA+VAI GD F+ I
Sbjct: 286 KEGIYKEYVQVNRSMTHLVFIGDGPDKTVISGSKSYKDGITTYKTATVAIVGDHFIAKNI 345
Query: 364 GFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFG 423
FEN+AG KHQAVA+RV +D SIFY C+ DGYQDTLYAH+ RQFYRDC ISGTIDF+FG
Sbjct: 346 AFENTAGAIKHQAVAIRVLADESIFYNCKFDGYQDTLYAHSHRQFYRDCTISGTIDFLFG 405
Query: 424 DSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKA 483
D+ AV QNCT +VRKPL NQ C +TA GRK+ + TG ++QG +IV +P Y V+ ++K
Sbjct: 406 DAAAVFQNCTLLVRKPLLNQACPITAHGRKDPRESTGFVLQGCTIVGEPDYLAVKEQSKT 465
Query: 484 YLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQR 543
YL RPWK++SRTI ++T+I D + PEG+ PW G G +T +Y E N GPG+ + +R
Sbjct: 466 YLGRPWKEYSRTIIMNTFIPDFVPPEGWQPW---LGEFGLNTLFYSEVQNTGPGAAITKR 522
Query: 544 VKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
V W G+K ++ E F P ++ GD WI VPY G
Sbjct: 523 VTWPGIKKLSDEEILKFTPAQYIQGDAWIPGKGVPYILG 561
>At5g49180 pectin methylesterase
Length = 571
Score = 522 bits (1345), Expect = e-148
Identities = 275/588 (46%), Positives = 372/588 (62%), Gaps = 31/588 (5%)
Query: 1 MSGGGDAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVK 60
M G+ K+ K+ I GV T LLV MVVAV + N E + + AV+
Sbjct: 1 MGVDGELKK-KKCIIAGVITALLVLMVVAVGITTSRNTSHS---EKIVPVQIKTATTAVE 56
Query: 61 TLCAPTDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKT--HLLQEAEKDE 118
+CAPTDYK+ C +SL+ + + T+P +LIK+ FN+TI I + +KK L +A D+
Sbjct: 57 AVCAPTDYKETCVNSLMKASPDSTQPLDLIKLGFNVTIRSIEDSIKKASVELTAKAANDK 116
Query: 119 RTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAF 178
TK AL+ C+++M + D+ ++ L+ F F + ++ + L+VWLSG+I YQ+TC+D F
Sbjct: 117 DTKGALELCEKLMNDATDDLKKCLDNFDGFSIPQIEDFVEDLRVWLSGSIAYQQTCMDTF 176
Query: 179 ENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLG 238
E T + + M+++ +TS ++SNGL++I +S E +V G LG
Sbjct: 177 EETNSKLSQDMQKIFKTSRELTSNGLAMITNISNLLGEF-----------NVTGVTGDLG 225
Query: 239 H------GGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVP 292
+ +P WV R+L+ G K A+VVVA DGSG + TI EAL VP
Sbjct: 226 KYARKLLSAEDGIPSWVGPNT--RRLMATKGGVK--ANVVVAHDGSGQYKTINEALNAVP 281
Query: 293 KKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDG-VGTFKTASV 351
K N KPFVIYIK+GVY E V+VTK MTHV FIGDG KT+ITG+ N+ G V T+ TA+V
Sbjct: 282 KANQKPFVIYIKQGVYNEKVDVTKKMTHVTFIGDGPTKTKITGSLNYYIGKVKTYLTATV 341
Query: 352 AITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRD 411
AI GD F IGFEN+AGPE HQAVALRV +D ++FY C++DGYQDTLY H+ RQF+RD
Sbjct: 342 AINGDNFTAKNIGFENTAGPEGHQAVALRVSADLAVFYNCQIDGYQDTLYVHSHRQFFRD 401
Query: 412 CIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVAD 471
C +SGT+DF+FGD I VLQNC VVRKP+++Q C++TAQGR +K + TGL++Q I +
Sbjct: 402 CTVSGTVDFIFGDGIVVLQNCNIVVRKPMKSQSCMITAQGRSDKRESTGLVLQNCHITGE 461
Query: 472 PKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEY 531
P Y PV+ NKAYL RPWK+FSRTI + T I D+I P G++PW G +T YY EY
Sbjct: 462 PAYIPVKSINKAYLGRPWKEFSRTIIMGTTIDDVIDPAGWLPWN---GDFALNTLYYAEY 518
Query: 532 NNRGPGSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPY 579
N GPGS+ QRVKW G+K ++ + A F P RF G+ WI RVPY
Sbjct: 519 ENNGPGSNQAQRVKWPGIKKLSPKQALRFTPARFLRGNLWIPPNRVPY 566
>At3g06830 pectin methylesterase like protein
Length = 568
Score = 474 bits (1220), Expect = e-134
Identities = 250/586 (42%), Positives = 367/586 (61%), Gaps = 31/586 (5%)
Query: 4 GGDAKRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLC 63
G D + K+ + G + LV MVV+V V +K P ++ E+H+ ++ KAV+ +C
Sbjct: 2 GSDGDKKKKFIVAGSVSGFLVIMVVSVAV---VTSKHSPKDD---ENHIRKTTKAVQAVC 55
Query: 64 APTDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTH--LLQEAEKDERTK 121
APTD+K C +SL+ + + +P +LIK+ F +TI I+E L+K + +A+K+ K
Sbjct: 56 APTDFKDTCVNSLMGASPDSDDPVDLIKLGFKVTIKSINESLEKASGDIKAKADKNPEAK 115
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENT 181
A + C+++M +ID+ ++ ++ F ++ ++ + L+VWLSG+I +Q+TC+D+F
Sbjct: 116 GAFELCEKLMIDAIDDLKKCMDH--GFSVDQIEVFVEDLRVWLSGSIAFQQTCMDSFGEI 173
Query: 182 TTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQP----AGRRLLKESVDGEEDVL 237
++ + M ++ +TS +SSN L+++ ++S A + ++ + E+ +
Sbjct: 174 KSNLMQDMLKIFKTSRELSSNSLAMVTRISTLIPNSNLTGLTGALAKYARKLLSTEDSI- 232
Query: 238 GHGGDFELPEWVDDRAGVRKLLNKMTGRK--LQAHVVVAKDGSGNFTTITEALKHVPKKN 295
P WV A R+L+ G ++A+ VVA+DG+G F TIT+AL VPK N
Sbjct: 233 --------PTWVGPEA--RRLMAAQGGGPGPVKANAVVAQDGTGQFKTITDALNAVPKGN 282
Query: 296 LKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNF-IDGVGTFKTASVAIT 354
PF+I+IKEG+YKE V VTK M HV FIGDG KT ITG+ NF I V TF TA++ I
Sbjct: 283 KVPFIIHIKEGIYKEKVTVTKKMPHVTFIGDGPNKTLITGSLNFGIGKVKTFLTATITIE 342
Query: 355 GDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCII 414
GD F IG EN+AGPE QAVALRV +D ++F+ C++DG+QDTLY H+ RQFYRDC +
Sbjct: 343 GDHFTAKNIGIENTAGPEGGQAVALRVSADYAVFHSCQIDGHQDTLYVHSHRQFYRDCTV 402
Query: 415 SGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKY 474
SGT+DF+FGD+ +LQNC VVRKP + Q C+VTAQGR + TGL++ G I DP Y
Sbjct: 403 SGTVDFIFGDAKCILQNCKIVVRKPNKGQTCMVTAQGRSNVRESTGLVLHGCHITGDPAY 462
Query: 475 YPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNR 534
P++ NKAYL RPWK+FSRTI + T I D+I P G++PW +G T YY E+ N
Sbjct: 463 IPMKSVNKAYLGRPWKEFSRTIIMKTTIDDVIDPAGWLPW---SGDFALKTLYYAEHMNT 519
Query: 535 GPGSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYS 580
GPGS+ QRVKW G+K +T + A + RF GD WI T+VPY+
Sbjct: 520 GPGSNQAQRVKWPGIKKLTPQDALLYTGDRFLRGDTWIPQTQVPYT 565
>At5g04960 pectinesterase
Length = 564
Score = 391 bits (1005), Expect = e-109
Identities = 234/579 (40%), Positives = 332/579 (56%), Gaps = 43/579 (7%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGF--NNKKEPGEETSKESHVSQSVKAVKTLCAP 65
K KR+AII +S+ +LV +VV V N+KK P E + VS VK LC
Sbjct: 20 KTKKRIAIIAISSIVLVCIVVGAVVGTTARDNSKKPPTENNGEPISVS-----VKALCDV 74
Query: 66 TDYKKECEDSL-IAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQAL 124
T +K++C ++L A + + P+EL K A +TI ++S+ L D T A+
Sbjct: 75 TLHKEKCFETLGSAPNASRSSPEELFKYAVKVTITELSKVLDG--FSNGEHMDNATSAAM 132
Query: 125 DTCKQVMQLSIDEFQRSL-ERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTT 183
C +++ L++D+ ++ NFD L+ WLS TYQETC+DA
Sbjct: 133 GACVELIGLAVDQLNETMTSSLKNFD---------DLRTWLSSVGTYQETCMDALVEANK 183
Query: 184 DAGKKMKEV-LQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGD 242
+ E L+ S M+SN L+II L K + +K RR L E+ + + V D
Sbjct: 184 PSLTTFGENHLKNSTEMTSNALAIITWLGKIADTVK--FRRRRLLETGNAKVVV----AD 237
Query: 243 FELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIY 302
+ E R+LL +K +A +VVAKDGSG + TI EAL V +KN KP +IY
Sbjct: 238 LPMMEG-------RRLLESGDLKK-KATIVVAKDGSGKYRTIGEALAEVEEKNEKPTIIY 289
Query: 303 IKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
+K+GVY E V V KT +VV +GDG KT ++ NFIDG TF+TA+ A+ G F+
Sbjct: 290 VKKGVYLENVRVEKTKWNVVMVGDGQSKTIVSAGLNFIDGTPTFETATFAVFGKGFMARD 349
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
+GF N+AGP KHQAVAL V +D S+FYKC MD +QDT+YAH RQFYRDC+I GT+DF+F
Sbjct: 350 MGFINTAGPAKHQAVALMVSADLSVFYKCTMDAFQDTMYAHAQRQFYRDCVILGTVDFIF 409
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
G++ V Q C + R+P++ QQ +TAQGRK+ NQ TG+ I +I + +
Sbjct: 410 GNAAVVFQKCEILPRRPMKGQQNTITAQGRKDPNQNTGISIHNCTIKPLDNLTDI----Q 465
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
+L RPWKDFS T+ + +++ I P+G++PW G T DT +Y EY N GPG+ K
Sbjct: 466 TFLGRPWKDFSTTVIMKSFMDKFINPKGWLPW---TGDTAPDTIFYAEYLNSGPGASTKN 522
Query: 543 RVKWQGVKT-ITSEGAASFVPIRFFHGDDWIRVTRVPYS 580
RVKWQG+KT +T + A F F G++W+ T+VP++
Sbjct: 523 RVKWQGLKTSLTKKEANKFTVKPFIDGNNWLPATKVPFN 561
>At4g15980 pectinesterase like protein
Length = 701
Score = 382 bits (981), Expect = e-106
Identities = 186/338 (55%), Positives = 241/338 (71%), Gaps = 5/338 (1%)
Query: 244 ELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYI 303
E P WV + R L + ++A+VVVAKDGSG TI +AL VP KN K FVI+I
Sbjct: 366 EFPPWVTPHSR-RLLARRPRNNGIKANVVVAKDGSGKCKTIAQALAMVPMKNTKKFVIHI 424
Query: 304 KEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFI-DGVGTFKTASVAITGDFFVGIG 362
KEGVYKE VEVTK M HV+F+GDG KT ITG+ F+ D VGT++TASVA+ GD+F+
Sbjct: 425 KEGVYKEKVEVTKKMLHVMFVGDGPTKTVITGDIAFLPDQVGTYRTASVAVNGDYFMAKD 484
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
IGFEN+AG +HQAVALRV +D ++F+ C M+GYQDTLY HT RQFYR+C +SGTIDFVF
Sbjct: 485 IGFENTAGAARHQAVALRVSADFAVFFNCHMNGYQDTLYVHTHRQFYRNCRVSGTIDFVF 544
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
GD+ AV QNC FV+R+P+E+QQCIVTAQGRK++ + TG++I I D Y PV+ KN+
Sbjct: 545 GDAKAVFQNCEFVIRRPMEHQQCIVTAQGRKDRRETTGIVIHNSRITGDASYLPVKAKNR 604
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
A+L RPWK+FSRTI ++T I D+I PEG++ W + +T +Y EY NRG GS +
Sbjct: 605 AFLGRPWKEFSRTIIMNTEIDDVIDPEGWLKWNETFAL---NTLFYTEYRNRGRGSGQGR 661
Query: 543 RVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYS 580
RV+W+G+K I+ A F P F G+ WI TR+PY+
Sbjct: 662 RVRWRGIKRISDRAAREFAPGNFLRGNTWIPQTRIPYN 699
Score = 135 bits (339), Expect = 8e-32
Identities = 74/211 (35%), Positives = 122/211 (57%), Gaps = 6/211 (2%)
Query: 12 RLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTDYKKE 71
+ ++GV T L++AMV+ V N G N+ EE +ES + + V +CA TDYK++
Sbjct: 3 KYVLLGV-TALIMAMVICVEANDG-NSPSRKMEEVHRESKLMITKTTVSIICASTDYKQD 60
Query: 72 CEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKK--THLLQEAEKDERTKQALDTCKQ 129
C SL +P+ LI+ AF++ I I G+ + L A+ D T++AL+TC++
Sbjct: 61 CTTSLATVRS--PDPRNLIRSAFDLAIISIRSGIDRGMIDLKSRADADMHTREALNTCRE 118
Query: 130 VMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKM 189
+M +ID+ +++ ++F F L + L VWLSG+ITYQ+TC+D FE ++A M
Sbjct: 119 LMDDAIDDLRKTRDKFRGFLFTRLSDFVEDLCVWLSGSITYQQTCIDGFEGIDSEAAVMM 178
Query: 190 KEVLQTSMHMSSNGLSIINQLSKTFEEMKQP 220
+ V++ H++SNGL+I L K + + P
Sbjct: 179 ERVMRKGQHLTSNGLAIAANLDKLLKAFRIP 209
>At3g10710 putative pectinesterase
Length = 561
Score = 377 bits (969), Expect = e-105
Identities = 223/577 (38%), Positives = 329/577 (56%), Gaps = 39/577 (6%)
Query: 8 KRNKRLAIIGVSTFLLVAMVV-AVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPT 66
K K +AII VS +L +V+ AV + E E + +S SVKAV C T
Sbjct: 21 KTRKNIAIIAVSLVILAGIVIGAVFGTMAHKKSPETVETNNNGDSISVSVKAV---CDVT 77
Query: 67 DYKKECEDSL-IAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
+K++C ++L A + P+EL + A ITIA++S+ + + + DE+ ++
Sbjct: 78 LHKEKCFETLGSAPNASSLNPEELFRYAVKITIAEVSKAI---NAFSSSLGDEKNNITMN 134
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDA 185
C +++ L+ID +L SN D+ ++ ++ L+ WLS A TYQ TC++
Sbjct: 135 ACAELLDLTIDNLNNTLTSSSNGDV-TVPELVDDLRTWLSSAGTYQRTCVETLAPDMRPF 193
Query: 186 GKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFEL 245
G+ L+ S ++SN L+II L K + K RR L + D E D H G
Sbjct: 194 GESH---LKNSTELTSNALAIITWLGKIADSFKL---RRRLLTTADVEVDF--HAG---- 241
Query: 246 PEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKE 305
R+LL RK+ A +VVAKDGSG + TI AL+ VP+K+ K +IY+K+
Sbjct: 242 ----------RRLLQSTDLRKV-ADIVVAKDGSGKYRTIKRALQDVPEKSEKRTIIYVKK 290
Query: 306 GVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGF 365
GVY E V+V K M +V+ +GDG K+ ++G N IDG TFKTA+ A+ G F+ +GF
Sbjct: 291 GVYFENVKVEKKMWNVIVVGDGESKSIVSGRLNVIDGTPTFKTATFAVFGKGFMARDMGF 350
Query: 366 ENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDS 425
N+AGP KHQAVAL V +D + FY+C M+ YQDTLY H RQFYR+C I GT+DF+FG+S
Sbjct: 351 INTAGPSKHQAVALMVSADLTAFYRCTMNAYQDTLYVHAQRQFYRECTIIGTVDFIFGNS 410
Query: 426 IAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYL 485
+VLQ+C + R+P++ QQ +TAQGR + N TG+ I +I V +L
Sbjct: 411 ASVLQSCRILPRRPMKGQQNTITAQGRTDPNMNTGISIHRCNISPLGDLTDV----MTFL 466
Query: 486 ARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVK 545
RPWK+FS T+ +D+Y+ I +G++PW G + DT +YGEY N GPG+ K RVK
Sbjct: 467 GRPWKNFSTTVIMDSYLHGFIDRKGWLPW---TGDSAPDTIFYGEYKNTGPGASTKNRVK 523
Query: 546 WQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
W+G++ ++++ A F F G W+ T+VP+ G
Sbjct: 524 WKGLRFLSTKEANRFTVKPFIDGGRWLPATKVPFRSG 560
>At2g26450 putative pectinesterase
Length = 496
Score = 370 bits (951), Expect = e-103
Identities = 199/500 (39%), Positives = 297/500 (58%), Gaps = 25/500 (5%)
Query: 83 ITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQRSL 142
+ P +K A + L+K L+ +D+ K A++ CK +++ + +E SL
Sbjct: 18 LDNPTTFLKSAIEAVNEDLDLVLEKVLSLKTENQDD--KDAIEQCKLLVEDAKEETVASL 75
Query: 143 ERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKEVLQTSMHMSSN 202
+ + ++NS ++V+ L+ WLS ++YQETCLD FE + ++K + +S ++SN
Sbjct: 76 NKINVTEVNSFEKVVPDLESWLSAVMSYQETCLDGFEEGNLKS--EVKTSVNSSQVLTSN 133
Query: 203 GLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKM 262
L++I KTF E P + + + +D ++P WV + R++L +
Sbjct: 134 SLALI----KTFTENLSPVMKVVERHLLD------------DIPSWVSNDD--RRMLRAV 175
Query: 263 TGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVV 322
+ L+ + VAKDGSG+FTTI +AL+ +P+K ++IY+K+G+Y EYV V K ++
Sbjct: 176 DVKALKPNATVAKDGSGDFTTINDALRAMPEKYEGRYIIYVKQGIYDEYVTVDKKKANLT 235
Query: 323 FIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQ 382
+GDG +KT +TGNK+ + TF TA+ G+ F+ +GF N+AGPE HQAVA+RVQ
Sbjct: 236 MVGDGSQKTIVTGNKSHAKKIRTFLTATFVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQ 295
Query: 383 SDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLEN 442
SDRSIF CR +GYQDTLYA+T RQ+YR C+I GTIDF+FGD+ A+ QNC +RK L
Sbjct: 296 SDRSIFLNCRFEGYQDTLYAYTHRQYYRSCVIVGTIDFIFGDAAAIFQNCNIFIRKGLPG 355
Query: 443 QQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYI 502
Q+ VTAQGR +K Q TG ++ I A+ PV+ + K+YL RPWK++SRTI +++ I
Sbjct: 356 QKNTVTAQGRVDKFQTTGFVVHNCKIAANEDLKPVKEEYKSYLGRPWKNYSRTIIMESKI 415
Query: 503 GDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAASFVP 562
++I P G++ WQ DT YY EYNN+G D RVKW G K I E A ++
Sbjct: 416 ENVIDPVGWLRWQETD--FAIDTLYYAEYNNKGSSGDTTSRVKWPGFKVINKEEALNYTV 473
Query: 563 IRFFHGDDWIRVTRVPYSPG 582
F G DWI + P G
Sbjct: 474 GPFLQG-DWISASGSPVKLG 492
>At1g53840 unknown protein
Length = 586
Score = 369 bits (946), Expect = e-102
Identities = 223/586 (38%), Positives = 324/586 (55%), Gaps = 44/586 (7%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTV-NVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPT 66
K KRL ++ +S +L+A+++A V V NK E E S S+KA+ C+ T
Sbjct: 24 KTRKRLLLLSISVVVLIAVIIAAVVATVVHKNKNESTPSPPPELTPSTSLKAI---CSVT 80
Query: 67 DYKKECEDSLIA-HAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
+ + C S+ + N T+P+ L K++ + I ++ L + +DER K AL
Sbjct: 81 RFPESCISSISKLPSSNTTDPETLFKLSLKVIIDELDSISDLPEKLSKETEDERIKSALR 140
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTS-----LKVWLSGAITYQETCLDAFE- 179
C +++ ++D ++ S D + L+S LK WLS +T ETC D+ +
Sbjct: 141 VCGDLIEDALDRLNDTV---SAIDDEEKKKTLSSSKIEDLKTWLSATVTDHETCFDSLDE 197
Query: 180 ---NTTTDAG----KKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDG 232
N T A + +K + S +SN L+I++++ ++ P RR S
Sbjct: 198 LKQNKTEYANSTITQNLKSAMSRSTEFTSNSLAIVSKILSALSDLGIPIHRRRRLMSHHH 257
Query: 233 EEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVP 292
++ V DFE +W R+ L + G L+ V VA DG+G+ T+ EA+ VP
Sbjct: 258 QQSV-----DFE--KWA------RRRLLQTAG--LKPDVTVAGDGTGDVLTVNEAVAKVP 302
Query: 293 KKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVA 352
KK+LK FVIY+K G Y E V + K+ +V+ GDG KT I+G+KNF+DG T++TA+ A
Sbjct: 303 KKSLKMFVIYVKSGTYVENVVMDKSKWNVMIYGDGKGKTIISGSKNFVDGTPTYETATFA 362
Query: 353 ITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDC 412
I G F+ IG N+AG KHQAVA R SD S++Y+C DG+QDTLY H+ RQFYRDC
Sbjct: 363 IQGKGFIMKDIGIINTAGAAKHQAVAFRSGSDFSVYYQCSFDGFQDTLYPHSNRQFYRDC 422
Query: 413 IISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADP 472
++GTIDF+FG + V Q C + R+PL NQ +TAQG+K+ NQ +G+ IQ +I A+
Sbjct: 423 DVTGTIDFIFGSAAVVFQGCKIMPRQPLSNQFNTITAQGKKDPNQSSGMSIQRCTISANG 482
Query: 473 KYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYN 532
YL RPWK+FS T+ ++T IG ++ P G+M W +G+ + YGEY
Sbjct: 483 NVIA-----PTYLGRPWKEFSTTVIMETVIGAVVRPSGWMSW--VSGVDPPASIVYGEYK 535
Query: 533 NRGPGSDVKQRVKWQGVKTITSEG-AASFVPIRFFHGDDWIRVTRV 577
N GPGSDV QRVKW G K + S+ AA F HG DWI T V
Sbjct: 536 NTGPGSDVTQRVKWAGYKPVMSDAEAAKFTVATLLHGADWIPATGV 581
>At4g33230 pectinesterase - like protein
Length = 609
Score = 365 bits (938), Expect = e-101
Identities = 221/606 (36%), Positives = 329/606 (53%), Gaps = 51/606 (8%)
Query: 8 KRNKRLAIIGVSTFLLVAMVV--AVTVNVGFNNK-KEPGEETSKESHVS----------- 53
K KR+ I+GV + L+VA + V + NK +E G+ T+ +S S
Sbjct: 20 KFRKRI-ILGVVSVLVVAAAIIGGAFAYVTYENKTQEQGKTTNNKSKDSPTKSESPSPKP 78
Query: 54 -----QSVKA------VKTLCAPTDYKKECEDSLIAHAGNIT---EPKELIKIAFNITIA 99
Q+VKA ++TLC T YK C+++L T +P+ L+K A
Sbjct: 79 PSSAAQTVKAGQVDKIIQTLCNSTLYKPTCQNTLKNETKKDTPQTDPRSLLKSAIVAVND 138
Query: 100 KISEGLKKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTS 159
+ + K+ L+ KD+ K A+ CK ++ + +E S++R ++ ++N+ +++
Sbjct: 139 DLDQVFKRVLSLKTENKDD--KDAIAQCKLLVDEAKEELGTSMKRINDSEVNNFAKIVPD 196
Query: 160 LKVWLSGAITYQETCLDAFENTTTDAGKKMKEV---LQTSMHMSSNGLSIINQLSKTFEE 216
L WLS ++YQETC+D FE GK E+ +S ++SN L++I L
Sbjct: 197 LDSWLSAVMSYQETCVDGFEE-----GKLKTEIRKNFNSSQVLTSNSLAMIKSLDGYLSS 251
Query: 217 MKQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKD 276
+ + R LL+ +E + W+ ++ R++L + + L+ + VAKD
Sbjct: 252 VPKVKTRLLLEARSSAKETD-------HITSWLSNKE--RRMLKAVDVKALKPNATVAKD 302
Query: 277 GSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGN 336
GSGNFTTI ALK +P K + IYIK G+Y E V + K +V +GDG +KT +TGN
Sbjct: 303 GSGNFTTINAALKAMPAKYQGRYTIYIKHGIYDESVIIDKKKPNVTMVGDGSQKTIVTGN 362
Query: 337 KNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGY 396
K+ + TF TA+ G+ F+ +GF N+AGPE HQAVA+RVQSDRS+F CR +GY
Sbjct: 363 KSHAKKIRTFLTATFVAQGEGFMAQSMGFRNTAGPEGHQAVAIRVQSDRSVFLNCRFEGY 422
Query: 397 QDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKN 456
QDTLYA+T RQ+YR C+I GT+DF+FGD+ A+ QNC +RK L Q+ VTAQGR +K
Sbjct: 423 QDTLYAYTHRQYYRSCVIIGTVDFIFGDAAAIFQNCDIFIRKGLPGQKNTVTAQGRVDKF 482
Query: 457 QPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQT 516
Q TG +I ++ + PV+ + K+YL RPWK SRT+ +++ I D+I P G++ WQ
Sbjct: 483 QTTGFVIHNCTVAPNEDLKPVKAQFKSYLGRPWKPHSRTVVMESTIEDVIDPVGWLRWQE 542
Query: 517 PAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTR 576
DT Y EY N GP RVKW G + + E A F F G +WI+
Sbjct: 543 TD--FAIDTLSYAEYKNDGPSGATAARVKWPGFRVLNKEEAMKFTVGPFLQG-EWIQAIG 599
Query: 577 VPYSPG 582
P G
Sbjct: 600 SPVKLG 605
>At1g53830 unknown protein
Length = 587
Score = 363 bits (932), Expect = e-100
Identities = 231/594 (38%), Positives = 328/594 (54%), Gaps = 41/594 (6%)
Query: 6 DAKRNKRLAIIGVS-TFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCA 64
D K NK+L + + LL+A +V + NK + S SH +K++C+
Sbjct: 13 DFKNNKKLILSSAAIALLLLASIVGIAATTTNQNKNQKITTLSSTSHA-----ILKSVCS 67
Query: 65 PTDYKKECEDSLIAHAGN-ITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKD--ERTK 121
T Y + C ++ A G +T KE+I+ + N+T + L K R
Sbjct: 68 STLYPELCFSAVAATGGKELTSQKEVIEASLNLTTKAVKHNYFAVKKLIAKRKGLTPREV 127
Query: 122 QALDTCKQVMQLSIDEFQRSLERFSNFDLN-SLDRVLTSLKVWLSGAITYQETCLDAFEN 180
AL C + + ++DE ++E + SL + LK +S AIT Q TCLD F
Sbjct: 128 TALHDCLETIDETLDELHVAVEDLHQYPKQKSLRKHADDLKTLISSAITNQGTCLDGF-- 185
Query: 181 TTTDAGKKMKEVLQTSM----HMSSNGLSIINQLSKT----FEEMKQPA-----GRRLLK 227
+ DA +K+++ L HM SN L++I +++T FE + + R LK
Sbjct: 186 SYDDADRKVRKALLKGQVHVEHMCSNALAMIKNMTETDIANFELRDKSSTFTNNNNRKLK 245
Query: 228 ESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEA 287
E V G+ D G P+W+ G R+LL G ++A VA DGSG+FTT+ A
Sbjct: 246 E-VTGDLDSDGW------PKWLS--VGDRRLLQ---GSTIKADATVADDGSGDFTTVAAA 293
Query: 288 LKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFK 347
+ P+K+ K FVI+IK GVY+E VEVTK T+++F+GDG KT ITG++N +DG TF
Sbjct: 294 VAAAPEKSNKRFVIHIKAGVYRENVEVTKKKTNIMFLGDGRGKTIITGSRNVVDGSTTFH 353
Query: 348 TASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQ 407
+A+VA G+ F+ I F+N+AGP KHQAVALRV SD S FY+C M YQDTLY H+ RQ
Sbjct: 354 SATVAAVGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYQCDMFAYQDTLYVHSNRQ 413
Query: 408 FYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGS 467
F+ C I+GT+DF+FG++ AVLQ+C R+P Q+ +VTAQGR + NQ TG++IQ
Sbjct: 414 FFVKCHITGTVDFIFGNAAAVLQDCDINARRPNSGQKNMVTAQGRSDPNQNTGIVIQNCR 473
Query: 468 IVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCY 527
I V+ YL RPWK++SRT+ + + I D+I PEG+ W +G DT
Sbjct: 474 IGGTSDLLAVKGTFPTYLGRPWKEYSRTVIMQSDISDVIRPEGWHEW---SGSFALDTLT 530
Query: 528 YGEYNNRGPGSDVKQRVKWQGVKTITSEGAAS-FVPIRFFHGDDWIRVTRVPYS 580
Y EY NRG G+ RVKW+G K ITS+ A F +F G W+ T P+S
Sbjct: 531 YREYLNRGGGAGTANRVKWKGYKVITSDTEAQPFTAGQFIGGGGWLASTGFPFS 584
>At3g14310 pectin methylesterase like protein
Length = 592
Score = 352 bits (903), Expect = 3e-97
Identities = 221/595 (37%), Positives = 324/595 (54%), Gaps = 38/595 (6%)
Query: 8 KRNKRLAIIGVSTFLL-VAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPT 66
K+NK+L ++ + LL VA V ++ N+K S SH +++ C+ T
Sbjct: 15 KKNKKLVLLSAAVALLFVAAVAGISAGASKANEKRT---LSPSSHA-----VLRSSCSST 66
Query: 67 DYKKECEDSLIAHAG-NITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKD--ERTKQA 123
Y + C +++ G +T K++I+ + N+TI + L + K R K A
Sbjct: 67 RYPELCISAVVTAGGVELTSQKDVIEASVNLTITAVEHNYFTVKKLIKKRKGLTPREKTA 126
Query: 124 LDTCKQVMQLSIDEFQRSLERFSNFDLN-SLDRVLTSLKVWLSGAITYQETCLDAFENTT 182
L C + + ++DE ++E + +L LK +S AIT QETCLD F +
Sbjct: 127 LHDCLETIDETLDELHETVEDLHLYPTKKTLREHAGDLKTLISSAITNQETCLDGFSHD- 185
Query: 183 TDAGKKMKEVLQTSM----HMSSNGLSIINQLSKT----FEEM-KQPAGRRLLKESVDGE 233
DA K++++ L HM SN L++I ++ T FE+ K + R LKE
Sbjct: 186 -DADKQVRKALLKGQIHVEHMCSNALAMIKNMTDTDIANFEQKAKITSNNRKLKEENQET 244
Query: 234 EDVLGHGGDFEL-----PEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEAL 288
+ G EL P W+ AG R+LL G ++A VA DGSG F T+ A+
Sbjct: 245 TVAVDIAGAGELDSEGWPTWLS--AGDRRLLQ---GSGVKADATVAADGSGTFKTVAAAV 299
Query: 289 KHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKT 348
P+ + K +VI+IK GVY+E VEV K +++F+GDG +T ITG++N +DG TF +
Sbjct: 300 AAAPENSNKRYVIHIKAGVYRENVEVAKKKKNIMFMGDGRTRTIITGSRNVVDGSTTFHS 359
Query: 349 ASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQF 408
A+VA G+ F+ I F+N+AGP KHQAVALRV SD S FY C M YQDTLY H+ RQF
Sbjct: 360 ATVAAVGERFLARDITFQNTAGPSKHQAVALRVGSDFSAFYNCDMLAYQDTLYVHSNRQF 419
Query: 409 YRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSI 468
+ C+I+GT+DF+FG++ VLQ+C R+P Q+ +VTAQGR + NQ TG++IQ I
Sbjct: 420 FVKCLIAGTVDFIFGNAAVVLQDCDIHARRPNSGQKNMVTAQGRTDPNQNTGIVIQKCRI 479
Query: 469 VADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYY 528
A V+ YL RPWK++S+T+ + + I D+I PEG+ W G +T Y
Sbjct: 480 GATSDLQSVKGSFPTYLGRPWKEYSQTVIMQSAISDVIRPEGWSEW---TGTFALNTLTY 536
Query: 529 GEYNNRGPGSDVKQRVKWQGVKTITSEG-AASFVPIRFFHGDDWIRVTRVPYSPG 582
EY+N G G+ RVKW+G K IT+ A + +F G W+ T P+S G
Sbjct: 537 REYSNTGAGAGTANRVKWRGFKVITAAAEAQKYTAGQFIGGGGWLSSTGFPFSLG 591
>At5g53370 pectinesterase
Length = 587
Score = 350 bits (898), Expect = 1e-96
Identities = 209/571 (36%), Positives = 312/571 (54%), Gaps = 26/571 (4%)
Query: 21 FLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTDYKKECEDSLIAHA 80
F L +VV V F + ++ + +A+ C+ + Y C D+L+
Sbjct: 37 FTLAVLVVGVVCFGIFAGIRAVDSGKTEPKLTRKPTQAISRTCSKSLYPNLCIDTLLDFP 96
Query: 81 GNIT-EPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQ 139
G++T + ELI I+FN T+ K S+ L + + + R + A D+C +++ S+D
Sbjct: 97 GSLTADENELIHISFNATLQKFSKALYTSSTITYTQMPPRVRSAYDSCLELLDDSVDALT 156
Query: 140 RSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDAGKKMKEVLQTSMHM 199
R+L S+ + S D + + WLS A+T +TC D F+ G+ +V+ +
Sbjct: 157 RAL---SSVVVVSGDESHSDVMTWLSSAMTNHDTCTDGFDEIEGQGGEVKDQVIGAVKDL 213
Query: 200 S---SNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVR 256
S SN L+I K + R+LL G E+ ELP W+ + R
Sbjct: 214 SEMVSNCLAIFAGKVKDLSGVPVVNNRKLL-----GTEETE------ELPNWL--KREDR 260
Query: 257 KLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEY-VEVT 315
+LL T +QA + V+KDGSG F TI EA+K P+ + + FVIY+K G Y+E ++V
Sbjct: 261 ELLGTPTSA-IQADITVSKDGSGTFKTIAEAIKKAPEHSSRRFVIYVKAGRYEEENLKVG 319
Query: 316 KTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQ 375
+ T+++FIGDG KT ITG K+ D + TF TA+ A TG F+ + FEN AGP KHQ
Sbjct: 320 RKKTNLMFIGDGKGKTVITGGKSIADDLTTFHTATFAATGAGFIVRDMTFENYAGPAKHQ 379
Query: 376 AVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFV 435
AVALRV D ++ Y+C + GYQD LY H+ RQF+R+C I GT+DF+FG++ +LQ+C
Sbjct: 380 AVALRVGGDHAVVYRCNIIGYQDALYVHSNRQFFRECEIYGTVDFIFGNAAVILQSCNIY 439
Query: 436 VRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRT 495
RKP+ Q+ +TAQ RK+ NQ TG+ I ++A P + YL RPWK +SR
Sbjct: 440 ARKPMAQQKITITAQNRKDPNQNTGISIHACKLLATPDLEASKGSYPTYLGRPWKLYSRV 499
Query: 496 IFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSE 555
+++ + +GD I P G++ W P + D+ YYGEY N+G GS + QRVKW G ITS
Sbjct: 500 VYMMSDMGDHIDPRGWLEWNGPFAL---DSLYYGEYMNKGLGSGIGQRVKWPGYHVITST 556
Query: 556 GAAS-FVPIRFFHGDDWIRVTRVPYSPGATK 585
AS F +F G W+ T V + G ++
Sbjct: 557 VEASKFTVAQFISGSSWLPSTGVSFFSGLSQ 587
>At3g49220 pectinesterase - like protein
Length = 598
Score = 345 bits (884), Expect = 5e-95
Identities = 201/581 (34%), Positives = 316/581 (53%), Gaps = 30/581 (5%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTD 67
+R+K+ ++ S L +++++A + G ++ + + S + +A+ C T
Sbjct: 39 RRSKKKLVVS-SIVLAISLILAAAIFAGVRSRLKLNQ--SVPGLARKPSQAISKACELTR 95
Query: 68 YKKECEDSLIAHAGNI--TEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
+ + C DSL+ G++ + K+LI + N+T+ S L + L + R + A D
Sbjct: 96 FPELCVDSLMDFPGSLAASSSKDLIHVTVNMTLHHFSHALYSSASLSFVDMPPRARSAYD 155
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLDAFENTTTDA 185
+C +++ S+D R+L + D + WLS A+T +TC + F+ D
Sbjct: 156 SCVELLDDSVDALSRALSSVVSSSAKPQD-----VTTWLSAALTNHDTCTEGFDGVD-DG 209
Query: 186 GKK--MKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGDF 243
G K M LQ + SN L+I + + P R L + EE
Sbjct: 210 GVKDHMTAALQNLSELVSNCLAIFSASHDGDDFAGVPIQNRRLLGVEEREE--------- 260
Query: 244 ELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIYI 303
+ P W+ R R++L +M ++QA ++V+KDG+G TI+EA+K P+ + + +IY+
Sbjct: 261 KFPRWM--RPKEREIL-EMPVSQIQADIIVSKDGNGTCKTISEAIKKAPQNSTRRIIIYV 317
Query: 304 KEGVYKEY-VEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
K G Y+E ++V + +++F+GDG KT I+G K+ D + TF TAS A TG F+
Sbjct: 318 KAGRYEENNLKVGRKKINLMFVGDGKGKTVISGGKSIFDNITTFHTASFAATGAGFIARD 377
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
I FEN AGP KHQAVALR+ +D ++ Y+C + GYQDTLY H+ RQF+R+C I GT+DF+F
Sbjct: 378 ITFENWAGPAKHQAVALRIGADHAVIYRCNIIGYQDTLYVHSNRQFFRECDIYGTVDFIF 437
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
G++ VLQNC+ RKP++ Q+ +TAQ RK+ NQ TG+ I ++A +
Sbjct: 438 GNAAVVLQNCSIYARKPMDFQKNTITAQNRKDPNQNTGISIHASRVLAASDLQATNGSTQ 497
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
YL RPWK FSRT+++ +YIG + G++ W T + DT YYGEY N GPGS + Q
Sbjct: 498 TYLGRPWKLFSRTVYMMSYIGGHVHTRGWLEWNTTFAL---DTLYYGEYLNSGPGSGLGQ 554
Query: 543 RVKWQGVKTITSEGAAS-FVPIRFFHGDDWIRVTRVPYSPG 582
RV W G + I S A+ F F +G W+ T V + G
Sbjct: 555 RVSWPGYRVINSTAEANRFTVAEFIYGSSWLPSTGVSFLAG 595
>At3g05620 putative pectinesterase
Length = 543
Score = 341 bits (874), Expect = 8e-94
Identities = 202/580 (34%), Positives = 309/580 (52%), Gaps = 53/580 (9%)
Query: 16 IGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKAVKTLCAPTDYKKECEDS 75
+G++T LL+ M+++V + +P +E + S V+++ C D + C +
Sbjct: 1 MGITTALLLVMLMSVHTSSYETTILKPYKEDNFRSLVAKA-------CQFIDAHELCVSN 53
Query: 76 LIAH---AGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALDTCKQVMQ 132
+ H +G+ P +++ A K +++ + R + A++ CK+++
Sbjct: 54 IWTHVKESGHGLNPHSVLRAAVKEAHDKAKLAMERIPTVMMLSIRSREQVAIEDCKELVG 113
Query: 133 LSIDEFQRSLERFS--------NFDLNSLDRVLT--SLKVWLSGAITYQETCLDAFENTT 182
S+ E S+ + + D S D +LK WLS A++ Q+TCL+ FE T
Sbjct: 114 FSVTELAWSMLEMNKLHGGGGIDLDDGSHDAAAAGGNLKTWLSAAMSNQDTCLEGFEGTE 173
Query: 183 TDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDVLGHGGD 242
+ +K L+ + SN L + QL+ L E V+
Sbjct: 174 RKYEELIKGSLRQVTQLVSNVLDMYTQLNA-------------LPFKASRNESVIAS--- 217
Query: 243 FELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNLKPFVIY 302
PEW+ + L+ + + + VVA DG G + TI EA+ P + K +VIY
Sbjct: 218 ---PEWLTETD--ESLMMRHDPSVMHPNTVVAIDGKGKYRTINEAINEAPNHSTKRYVIY 272
Query: 303 IKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGDFFVGIG 362
+K+GVYKE +++ K T+++ +GDG +T ITG++NF+ G+ TF+TA+VA++G F+
Sbjct: 273 VKKGVYKENIDLKKKKTNIMLVGDGIGQTIITGDRNFMQGLTTFRTATVAVSGRGFIAKD 332
Query: 363 IGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISGTIDFVF 422
I F N+AGP+ QAVALRV SD+S FY+C ++GYQDTLYAH++RQFYRDC I GTIDF+F
Sbjct: 333 ITFRNTAGPQNRQAVALRVDSDQSAFYRCSVEGYQDTLYAHSLRQFYRDCEIYGTIDFIF 392
Query: 423 GDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYPVRLKNK 482
G+ AVLQNC R PL Q+ +TAQGRK NQ TG +IQ ++A
Sbjct: 393 GNGAAVLQNCKIYTRVPLPLQKVTITAQGRKSPNQNTGFVIQNSYVLA---------TQP 443
Query: 483 AYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGPGSDVKQ 542
YL RPWK +SRT++++TY+ ++ P G++ W G DT +YGEYNN GPG
Sbjct: 444 TYLGRPWKLYSRTVYMNTYMSQLVQPRGWLEW---FGNFALDTLWYGEYNNIGPGWRSSG 500
Query: 543 RVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
RVKW G + A SF F G W+ T V ++ G
Sbjct: 501 RVKWPGYHIMDKRTALSFTVGSFIDGRRWLPATGVTFTAG 540
>At3g43270 pectinesterase like protein
Length = 527
Score = 338 bits (867), Expect = 5e-93
Identities = 187/467 (40%), Positives = 273/467 (58%), Gaps = 21/467 (4%)
Query: 119 RTKQALDTCKQVMQLSIDEFQR--SLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLD 176
R A+ C ++ + +E S + N NS V + L+ W+S A++ Q+TCLD
Sbjct: 76 RVSNAIVDCVDLLDSAAEELSWIISASQSPNGKDNSTGDVGSDLRTWISAALSNQDTCLD 135
Query: 177 AFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDV 236
FE T +K+++ + S G ++ N L+ +P + + +++
Sbjct: 136 GFEGTNGI----IKKIVAGGL--SKVGTTVRNLLTMVHSPPSKPKPKPIKAQTM-----T 184
Query: 237 LGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKKNL 296
H G + P WV + G RKLL T A VVA DG+GNFTTI++A+ P +
Sbjct: 185 KAHSGFSKFPSWV--KPGDRKLLQ--TDNITVADAVVAADGTGNFTTISDAVLAAPDYST 240
Query: 297 KPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITGD 356
K +VI++K GVY E VE+ K +++ +GDG T ITGN++FIDG TF++A+ A++G
Sbjct: 241 KRYVIHVKRGVYVENVEIKKKKWNIMMVGDGIDATVITGNRSFIDGWTTFRSATFAVSGR 300
Query: 357 FFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIISG 416
F+ I F+N+AGPEKHQAVA+R +D +FY+C M GYQDTLYAH+MRQF+R+CII+G
Sbjct: 301 GFIARDITFQNTAGPEKHQAVAIRSDTDLGVFYRCAMRGYQDTLYAHSMRQFFRECIITG 360
Query: 417 TIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYYP 476
T+DF+FGD+ AV Q+C ++ L NQ+ +TAQGRK+ N+PTG IQ +I AD
Sbjct: 361 TVDFIFGDATAVFQSCQIKAKQGLPNQKNSITAQGRKDPNEPTGFTIQFSNIAADTDLLL 420
Query: 477 VRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRGP 536
YL RPWK +SRT+F+ Y+ D I P G++ W G DT YYGEY N GP
Sbjct: 421 NLNTTATYLGRPWKLYSRTVFMQNYMSDAINPVGWLEWN---GNFALDTLYYGEYMNSGP 477
Query: 537 GSDVKQRVKWQGVKTI-TSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
G+ + +RVKW G + TS A +F + G+ W+ T + + G
Sbjct: 478 GASLDRRVKWPGYHVLNTSAEANNFTVSQLIQGNLWLPSTGITFIAG 524
>At3g14300 putative pectin methylesterase
Length = 968
Score = 337 bits (865), Expect = 8e-93
Identities = 202/536 (37%), Positives = 289/536 (53%), Gaps = 37/536 (6%)
Query: 48 KESHVSQSVKAVKTLCAPTDYKKECEDSL--IAHAGNITEPKELIKIAFNITIAKISEGL 105
K H + S ++T+C T+Y C S+ + + T+PK L +++ +T +++ +
Sbjct: 449 KSPHPTPS-SVLRTVCNVTNYPASCISSISKLPLSKTTTDPKVLFRLSLQVTFDELNSIV 507
Query: 106 KKTHLLQEAEKDERTKQALDTCKQVMQLSIDEFQRSL----ERFSNFDLNSLDRVLTSLK 161
L E DE K AL C V L++D ++ E S N + L
Sbjct: 508 GLPKKLAEETNDEGLKSALSVCADVFDLAVDSVNDTISSLDEVISGGKKNLNSSTIGDLI 567
Query: 162 VWLSGAITYQETCLDAFE--NTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQ 219
WLS A+T TC D + N + +K+K + S +SN L+I+ Q+ K + +
Sbjct: 568 TWLSSAVTDIGTCGDTLDEDNYNSPIPQKLKSAMVNSTEFTSNSLAIVAQVLKKPSKSRI 627
Query: 220 PA-GRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDGS 278
P GRRLL + P WV R GVR+LL + L HV VA DGS
Sbjct: 628 PVQGRRLLNSN--------------SFPNWV--RPGVRRLLQ---AKNLTPHVTVAADGS 668
Query: 279 GNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKN 338
G+ T+ EA+ VPKK FVIY+K G Y E V + K +V GDG KT I+G+ N
Sbjct: 669 GDVRTVNEAVWRVPKKGKTMFVIYVKAGTYVENVLMKKDKWNVFIYGDGRDKTIISGSTN 728
Query: 339 FIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQD 398
+DGV TF T++ A G F+ +G N+AGPEKHQAVA R SDRS++Y+C DGYQD
Sbjct: 729 MVDGVRTFNTSTFATEGKGFMMKDMGIINTAGPEKHQAVAFRSDSDRSVYYRCSFDGYQD 788
Query: 399 TLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQP 458
TLY H+ RQ+YR+C ++GT+DF+FG V Q C+ R+PL NQ +TA+G +E NQ
Sbjct: 789 TLYTHSNRQYYRNCDVTGTVDFIFGAGTVVFQGCSIRPRQPLPNQFNTITAEGTQEANQN 848
Query: 459 TGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPA 518
TG+ I +I + + YL RPWK FS+T+ + + IG + P G++ W +
Sbjct: 849 TGISIHQCTISPNG-----NVTATTYLGRPWKLFSKTVIMQSVIGSFVNPAGWIAWNSTY 903
Query: 519 GITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTITSEGAASFVPIRFF-HGDD-WI 572
T +Y EY N GPGSD+ +RVKW G K I+S+ A+ +++F GDD WI
Sbjct: 904 D-PPPRTIFYREYKNSGPGSDLSKRVKWAGYKPISSDDEAARFTVKYFLRGDDNWI 958
Score = 101 bits (252), Expect = 1e-21
Identities = 67/230 (29%), Positives = 115/230 (49%), Gaps = 8/230 (3%)
Query: 8 KRNKRLAIIGVSTFLLVAMVVAVTVNVGFNNKKEPGEETSKESHVSQSVKA-VKTLCAPT 66
K KRL IG+S +LVA++++ TV + +++K + S + A +KT+C+ T
Sbjct: 25 KTRKRLYQIGISVAVLVAIIISSTVTIAIHSRKGNSPHPTPSSVPELTPAASLKTVCSVT 84
Query: 67 DYKKECEDSLIA-HAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKDERTKQALD 125
+Y C S+ N T+P+ + +++ + I +++ ++ L E DE K AL
Sbjct: 85 NYPVSCFSSISKLPLSNTTDPEVIFRLSLQVVIDELNSIVELPKKLAEETDDEGLKSALS 144
Query: 126 TCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLT--SLKVWLSGAITYQETCLDAFE---N 180
C+ ++ L+ID ++ D + T L WLS A+TY TCLDA + +
Sbjct: 145 VCEHLLDLAIDRVNETVSAMEVVDGKKILNAATIDDLLTWLSAAVTYHGTCLDALDEISH 204
Query: 181 TTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPA-GRRLLKES 229
T + K+K + S +SN L+I+ ++ T + P GRRLL S
Sbjct: 205 TNSAIPLKLKSGMVNSTEFTSNSLAIVAKILSTISDFGIPIHGRRLLNSS 254
Score = 61.6 bits (148), Expect = 1e-09
Identities = 43/162 (26%), Positives = 75/162 (45%), Gaps = 12/162 (7%)
Query: 58 AVKTLCAPTDYKKECEDSLIA-HAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEK 116
+++ +C+ T Y C S+ + N T+P+ L +++ + I +++ L E
Sbjct: 270 SLRNVCSVTRYPASCVSSISKLPSSNTTDPEALFRLSLQVVINELNSIAGLPKKLAEETD 329
Query: 117 DERTKQALDTCKQVMQLSID---EFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQET 173
DER K +L C V +ID + ++E + + ++ WLS A+T +T
Sbjct: 330 DERLKSSLSVCGDVFNDAIDIVNDTISTMEEVGDGKKILKSSTIDEIQTWLSAAVTDHDT 389
Query: 174 CLDAF----ENTTTDAGK----KMKEVLQTSMHMSSNGLSII 207
CLDA +N T A K+K + S +SN L+II
Sbjct: 390 CLDALDELSQNKTEYANSPISLKLKSAMVNSRKFTSNSLAII 431
>At1g11580 unknown protein
Length = 557
Score = 325 bits (832), Expect = 6e-89
Identities = 181/465 (38%), Positives = 265/465 (56%), Gaps = 33/465 (7%)
Query: 119 RTKQALDTCKQVMQLSIDEFQRSLE--RFSNFDLNSLDRVLTSLKVWLSGAITYQETCLD 176
R K C+++M +S D S+E R N++L S V T WLS +T TCL+
Sbjct: 118 RDKAGFADCEEMMDVSKDRMMSSMEELRGGNYNLESYSNVHT----WLSSVLTNYMTCLE 173
Query: 177 AFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDV 236
+ + + ++ + +K L+ + + L+I + +++K R
Sbjct: 174 SISDVSVNSKQIVKPQLEDLVSRARVALAIFVSVLPARDDLKMIISNRF----------- 222
Query: 237 LGHGGDFELPEWVDDRAGVRKLLNKMTGR-KLQAHVVVAKDGSGNFTTITEALKHVPKKN 295
P W+ A RKLL K+ A+VVVAKDG+G F T+ EA+ P+ +
Sbjct: 223 ---------PSWLT--ALDRKLLESSPKTLKVTANVVVAKDGTGKFKTVNEAVAAAPENS 271
Query: 296 LKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAITG 355
+VIY+K+GVYKE +++ K +++ +GDG T ITG+ N IDG TF++A+VA G
Sbjct: 272 NTRYVIYVKKGVYKETIDIGKKKKNLMLVGDGKDATIITGSLNVIDGSTTFRSATVAANG 331
Query: 356 DFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCIIS 415
D F+ I F+N+AGP KHQAVALRV +D+++ +CR+D YQDTLY HT+RQFYRD I+
Sbjct: 332 DGFMAQDIWFQNTAGPAKHQAVALRVSADQTVINRCRIDAYQDTLYTHTLRQFYRDSYIT 391
Query: 416 GTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKYY 475
GT+DF+FG+S V QNC V R P Q+ ++TAQGR+++NQ T + IQ I A
Sbjct: 392 GTVDFIFGNSAVVFQNCDIVARNPGAGQKNMLTAQGREDQNQNTAISIQKCKITASSDLA 451
Query: 476 PVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNRG 535
PV+ K +L RPWK +SRT+ + ++I + I P G+ PW G T YYGEY N G
Sbjct: 452 PVKGSVKTFLGRPWKLYSRTVIMQSFIDNHIDPAGWFPWD---GEFALSTLYYGEYANTG 508
Query: 536 PGSDVKQRVKWQGVKTI-TSEGAASFVPIRFFHGDDWIRVTRVPY 579
PG+D +RV W+G K I S+ A F + G W++ T V +
Sbjct: 509 PGADTSKRVNWKGFKVIKDSKEAEQFTVAKLIQGGLWLKPTGVTF 553
>At4g33220 pectinesterase like protein
Length = 404
Score = 314 bits (805), Expect = 8e-86
Identities = 169/366 (46%), Positives = 228/366 (62%), Gaps = 18/366 (4%)
Query: 218 KQPAGRRLLKESVDGEEDVLGHGGDFELPEWVDDRAGVRKLLNKMTGRKLQAHVVVAKDG 277
K P GR+L D +ED + P+WV R RKLL GR V VA DG
Sbjct: 53 KAPPGRKLR----DTDEDE-----SLQFPDWV--RPDDRKLLES-NGRTYD--VSVALDG 98
Query: 278 SGNFTTITEALKHVPKKNLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNK 337
+GNFT I +A+K P + FVIYIK+G+Y E VE+ K ++V +GDG T I+GN+
Sbjct: 99 TGNFTKIMDAIKKAPDYSSTRFVIYIKKGLYLENVEIKKKKWNIVMLGDGIDVTVISGNR 158
Query: 338 NFIDGVGTFKTASVAITGDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQ 397
+FIDG TF++A+ A++G F+ I F+N+AGPEKHQAVALR SD S+F++C M GYQ
Sbjct: 159 SFIDGWTTFRSATFAVSGRGFLARDITFQNTAGPEKHQAVALRSDSDLSVFFRCAMRGYQ 218
Query: 398 DTLYAHTMRQFYRDCIISGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQ 457
DTLY HTMRQFYR+C I+GT+DF+FGD V QNC + ++ L NQ+ +TAQGRK+ NQ
Sbjct: 219 DTLYTHTMRQFYRECTITGTVDFIFGDGTVVFQNCQILAKRGLPNQKNTITAQGRKDVNQ 278
Query: 458 PTGLIIQGGSIVADPKYYPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTP 517
P+G IQ +I AD P + YL RPWK +SRT+F+ + D++ PEG++ W
Sbjct: 279 PSGFSIQFSNISADADLVPYLNTTRTYLGRPWKLYSRTVFIRNNMSDVVRPEGWLEWNAD 338
Query: 518 AGITGTDTCYYGEYNNRGPGSDVKQRVKWQGVKTI-TSEGAASFVPIRFFHGDDWIRVTR 576
+ DT +YGE+ N GPGS + RVKW G S+ A +F +F G+ W+ T
Sbjct: 339 FAL---DTLFYGEFMNYGPGSGLSSRVKWPGYHVFNNSDQANNFTVSQFIKGNLWLPSTG 395
Query: 577 VPYSPG 582
V +S G
Sbjct: 396 VTFSDG 401
>At2g26440 putative pectinesterase
Length = 547
Score = 313 bits (801), Expect = 2e-85
Identities = 190/528 (35%), Positives = 274/528 (50%), Gaps = 26/528 (4%)
Query: 61 TLCAPTDYKKECEDSLIAHAGNITEPKELIKIAFNITIAKISEGLKKTHLLQEAEKD--- 117
+ C T Y C SL P L + + A +SE K T LL A
Sbjct: 39 SFCKNTPYPDACFTSLKLSISINISPNILSFLLQTLQTA-LSEAGKLTDLLSGAGVSNNL 97
Query: 118 -ERTKQALDTCKQVMQLSIDEFQRSLERFSNFDLNSLDRVLTSLKVWLSGAITYQETCLD 176
E + +L CK + ++ +RS+ + + +S R L + +LS A+T + TCL+
Sbjct: 98 VEGQRGSLQDCKDLHHITSSFLKRSISKIQDGVNDS--RKLADARAYLSAALTNKITCLE 155
Query: 177 AFENTTTDAGKKMKEVLQTSMHMSSNGLSIINQLSKTFEEMKQPAGRRLLKESVDGEEDV 236
E+ + K+ T+ SN LS + P RR G
Sbjct: 156 GLESASGPLKPKLVTSFTTTYKHISNSLSAL------------PKQRRTTNPKTGGNTKN 203
Query: 237 LGHGGDFELPEWV--DDRAGVRKLLNKMTGRKLQAHVVVAKDGSGNFTTITEALKHVPKK 294
G F P+WV D + + +VVA DG+GNF+TI EA+ P
Sbjct: 204 RRLLGLF--PDWVYKKDHRFLEDSSDGYDEYDPSESLVVAADGTGNFSTINEAISFAPNM 261
Query: 295 NLKPFVIYIKEGVYKEYVEVTKTMTHVVFIGDGGRKTRITGNKNFIDGVGTFKTASVAIT 354
+ +IY+KEGVY E +++ T++V IGDG T ITGN++ DG TF++A++A++
Sbjct: 262 SNDRVLIYVKEGVYDENIDIPIYKTNIVLIGDGSDVTFITGNRSVGDGWTTFRSATLAVS 321
Query: 355 GDFFVGIGIGFENSAGPEKHQAVALRVQSDRSIFYKCRMDGYQDTLYAHTMRQFYRDCII 414
G+ F+ I N+AGPEKHQAVALRV +D Y+C +DGYQDTLY H+ RQFYR+C I
Sbjct: 322 GEGFLARDIMITNTAGPEKHQAVALRVNADFVALYRCVIDGYQDTLYTHSFRQFYRECDI 381
Query: 415 SGTIDFVFGDSIAVLQNCTFVVRKPLENQQCIVTAQGRKEKNQPTGLIIQGGSIVADPKY 474
GTID++FG++ V Q C V + P+ Q ++TAQ R +++ TG+ +Q SI+A
Sbjct: 382 YGTIDYIFGNAAVVFQGCNIVSKLPMPGQFTVITAQSRDTQDEDTGISMQNCSILASEDL 441
Query: 475 YPVRLKNKAYLARPWKDFSRTIFLDTYIGDMITPEGYMPWQTPAGITGTDTCYYGEYNNR 534
+ K K+YL RPW++FSRT+ +++YI + I G+ W G DT YYGEYNN
Sbjct: 442 FNSSNKVKSYLGRPWREFSRTVVMESYIDEFIDGSGWSKWN---GGEALDTLYYGEYNNN 498
Query: 535 GPGSDVKQRVKWQGVKTITSEGAASFVPIRFFHGDDWIRVTRVPYSPG 582
GPGS+ +RV W G + E A +F F GD W+ T PY G
Sbjct: 499 GPGSETVKRVNWPGFHIMGYEDAFNFTATEFITGDGWLGSTSFPYDNG 546
Database: ara_mips
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,978,382
Number of sequences in database: 6832
Database: /data/blast2/ara_mips_chr2
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,737,135
Number of sequences in database: 4184
Database: /data/blast2/ara_mips_chr3
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,236,886
Number of sequences in database: 5377
Database: /data/blast2/ara_mips_chr4
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 1,748,816
Number of sequences in database: 4030
Database: /data/blast2/ara_mips_chr5
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 2,569,679
Number of sequences in database: 6098
Database: /data/blast2/ara_mips_chl
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 25,951
Number of sequences in database: 85
Database: /data/blast2/ara_mips_mit
Posted date: Jul 15, 2004 10:29 AM
Number of letters in database: 21,747
Number of sequences in database: 113
Lambda K H
0.318 0.135 0.396
Gapped
Lambda K H
0.267 0.0410 0.140
Matrix: BLOSUM62
Gap Penalties: Existence: 11, Extension: 1
Number of Hits to DB: 13,377,485
Number of Sequences: 26719
Number of extensions: 583134
Number of successful extensions: 1860
Number of sequences better than 10.0: 108
Number of HSP's better than 10.0 without gapping: 83
Number of HSP's successfully gapped in prelim test: 25
Number of HSP's that attempted gapping in prelim test: 1511
Number of HSP's gapped (non-prelim): 124
length of query: 593
length of database: 11,318,596
effective HSP length: 105
effective length of query: 488
effective length of database: 8,513,101
effective search space: 4154393288
effective search space used: 4154393288
T: 11
A: 40
X1: 16 ( 7.3 bits)
X2: 38 (14.6 bits)
X3: 64 (24.7 bits)
S1: 41 (21.7 bits)
S2: 63 (28.9 bits)
Medicago: description of AC144541.15


